Abstract
The study of macroautophagy in mammalian cells has described induction, vesicle nucleation, and membrane elongation complexes as key signaling intermediates driving autophagosome biogenesis. How these components are recruited to nascent autophagosomes is poorly understood, and although much is known about signaling mechanisms that restrain autophagy, the nature of positive inductive signals that can promote autophagy remain cryptic. We find that the Ras-like small G-protein, RalB, is localized to nascent autophagosomes and is activated upon nutrient deprivation. RalB and its effector Exo84 are required for nutrient starvation-induced autophagocytosis, and RalB activation is sufficient to promote autophagosome formation. Through direct binding to Exo84, RalB induces the assembly of catalytically active ULK1 and Beclin1-VPS34 complexes on the exocyst, which are required for isolation membrane formation and maturation. Thus, RalB signaling is a primary adaptive response to nutrient limitation that directly engages autophagocytosis through mobilization of the core vesicle nucleation machinery.
Introduction
The critical role of macroautophagy (herein referred to as autophagy) in tissue homeostasis, cellular adaptation to nutrient restriction and in clearance of pathogens and dysfunctional organelles suggests de novo generation of the double-membrane autophagosome requires responsiveness to inductive signals that specify location, contents, and duration (Kissova et al., 2004; Noda et al., 1995; Yang et al., 2006). A number of key signaling events have been identified that specify autophagosome biogenesis. Among the earliest is the de-phosphorylation of inhibitory mTOR-dependent sites on the ULK1-Atg13-FIP200 induction complex (Hosokawa et al., 2009a; Hosokawa et al., 2009b). This presumably releases ULK1 activity to facilitate auto-phosphorylation of the ULK1-ATG13-FIP200 complex and assembly with the vertabrate-specific autophagy protein ATG101 (Hosokawa et al., 2009a; Hosokawa et al., 2009b; Jung et al., 2009; Mercer et al., 2009). Through currently undescribed mechanisms, this leads to the activation of an autophagy specific class III PI(3)K complex, the Beclin1-ATG14L-VPS34-VPS15 complex. This activity coats a cup-shaped isolation membrane with phosphatidylinositol-3-phosphate, PI(3)P, which serves as a recruitment signal for the ATG16-ATG5/ATG12 component of the isolation membrane elongation machinery (Suzuki et al., 2001). Two ubiquitin-like molecules, ATG12 and LC3, undergo conjugation to ATG5 and phosphatidylethanolamine respectively to promote autophagosome formation. ATG12 is activated by ATG7 (E1), transferred to ATG10 (E2), followed by covalent linkage to an internal lysine on Atg5 (Mizushima et al., 1998a; Mizushima et al., 1998b). In the second conjugation system, LC3 is first cleaved by the cysteine protease, ATG4, which exposes a C-terminal glycine residue. ATG7 (E1) activates LC3 and transfers it to ATG3 (E2) (Amar et al., 2006; Tanida et al., 2004; Tanida et al., 2002). LC3 is then conjugated to phosphatidylethanolamine with assistance of ATG5/12 conjugates (Fujita et al., 2008; Hanada et al., 2007; Kabeya et al., 2000). The lipidated LC3, LC3-II, coats the inner and outer surfaces of the autophagosome, and along with ATG5, serves as a discrete marker of autophagosomes and autophagosome precursors, respectively (George et al., 2000; Kabeya et al., 2000; Kabeya et al., 2004; Mizushima et al., 2001). These key signaling events are coordinated with dynamic membrane events to culminate in the formation of a double-membrane autophagosome. The autophagosome ultimately fuses with a lysosome that facilitates the turnover of engulfed material by lysosomal/vacuolar acid hydrolases. How signaling intermediates are coordinated with the dynamic membrane events during the autophagosome biogenesis is currently unknown.
RalA and RalB are close relatives to the founding members of the Ras GTPase superfamily. They are engaged in response to mitogenic, trophic, and hormonal signals by a diverse group of guanyl nucleotide exchange factors that fall into two major groups: those that are directly Ras-responsive via a carboxyterminal Ras binding domain and those that are apparently mobilized by phosphoinositide second messengers via a carboxyterminal pleckstrin homology domain (Bodemann and White, 2008; Feig, 2003). While a number of RalGTP effector proteins have been identified that couple RalA/B activation to dynamic cell biological processes, an overarching occupation of the Ral GTPases is the direct regulation of the Sec6/8, or exocyst, complex (Bodemann and White, 2008; Feig, 2003). Two members of the heteroctomeric exocyst complex, Sec5 (EXOC2) and Exo84 (EXOC8), are bona fide effector molecules that mediate RalA/B regulation of dynamic secretory vesicle targeting and tethering processes (Bodemann and White, 2008; Moskalenko et al., 2002; Moskalenko et al., 2003; Sugihara et al., 2002). RalA-dependent mobilization of exocyst holocomplex assembly is critical for maintenance of apical/basolateral membrane identity in polarized epithelial cells (Moskalenko et al., 2002; Moskalenko et al., 2003) and for insulin-stimulated Glut4 delivery to the plasma membrane in adipocytes (Chen et al., 2007). Distinct from regulation of membrane trafficking, RalB has been demonstrated to mediate signal transduction cascades supporting the host defense response. Upon Toll-like receptor activation, RalB/Sec5 complex assembly directly participates in activation of the innate immune signaling kinase TBK1 to facilitate an interferon response (Chien et al., 2006). This combination of roles, vesicle trafficking/tethering and signal cascade assembly/activation, suggests that Ral/exocyst effector complexes may coordinate dynamic membrane trafficking events with stimulus-dependent signaling events.
Here we show that the small G-protein, RalB, and an Exo84-dependent subcomplex of the exocyst are critical for nutrient starvation and pathogen-induced autophagosome formation. Native RalB proteins localize to sites of nascent autophagosome formation and accumulate in the “active” GTP-bound state under nutrient limited conditions. RalB, but not its close homolog RalA, is required for autophagosome biogenesis and is sufficient to activate autophagy in human epithelial cells. The mechanism of action is through direct triggering of vesicle nucleation by assembly of an active ULK1-Beclin1-VPS34 initiation complex on the RalB effector protein Exo84. Thus, the RalB-Exo84 effector complex defines a key proximal regulatory component of the cellular response to nutrient deprivation.
Results
Association of the exocyst with autophagosome assembly machinery
Accumulating observations indicate direct participation of the heterooctomeric exocyst (aka Sec6/8) complex in adaptive responses to pathogen challenge (Bhuvanakantham et al., 2010; Chien et al., 2006; Ishikawa and Barber, 2008; Ishikawa et al., 2009). Most strikingly, core innate immune signaling through TBK1 and STING is supported by the Sec5 subunit of the exocyst (Chien et al., 2006; Ishikawa and Barber, 2008; Ishikawa et al., 2009). To help generate molecular leads that may account for the participation of exocyst components in host defense signaling, we used high throughput yeast two-hybrid screening to isolate a cohort of proteins that can associate with exocyst subunits (Formstecher et al., 2005). Among this cohort, both negative (RUBICON) and positive (FIP200, ATG14L) modulators of autophagy were isolated in the first-degree interaction neighborhood of Sec3 (see methods). Given the functional convergence of Ral/exocyst signaling and autophagy in pathogen recognition and clearance, we examined the association of exocyst components and autophagy proteins in human epithelial cell cultures. The interaction of Sec3 with RUBICON and ATG14L was validated by expression co-IP (Figure 1A,B). In addition Exo84 and Sec5 could interact with RUBICON and ATG14L, as would be expected if autophagosome machinery/exocyst interactions occur in the context of multisubunit exocyst complexes (Figure 1C,D,E,F). Immunoprecipitation of the core exocyst subunit, Sec8, recovers all characterized components of the exocyst complex (Grindstaff et al., 1998). Therefore, to examine if the exocyst may be associated with the LC3-modification machinery that drives elongation of isolation membranes, we probed Sec8 complexes for the presence of ATG5/ATG12 conjugates. As shown, Sec8-ATG5/ATG12 complexes were recovered from both overexpression co-IPs (Figure 1G) and by co-immunoprecipitation of endogenous proteins (Figure 1H), indicating a physical integration of the exocyst and autophagosome assembly machinery.
Figure 1. Physical and Functional interaction of the Ral-Exocyst complex with autophagy machinery.
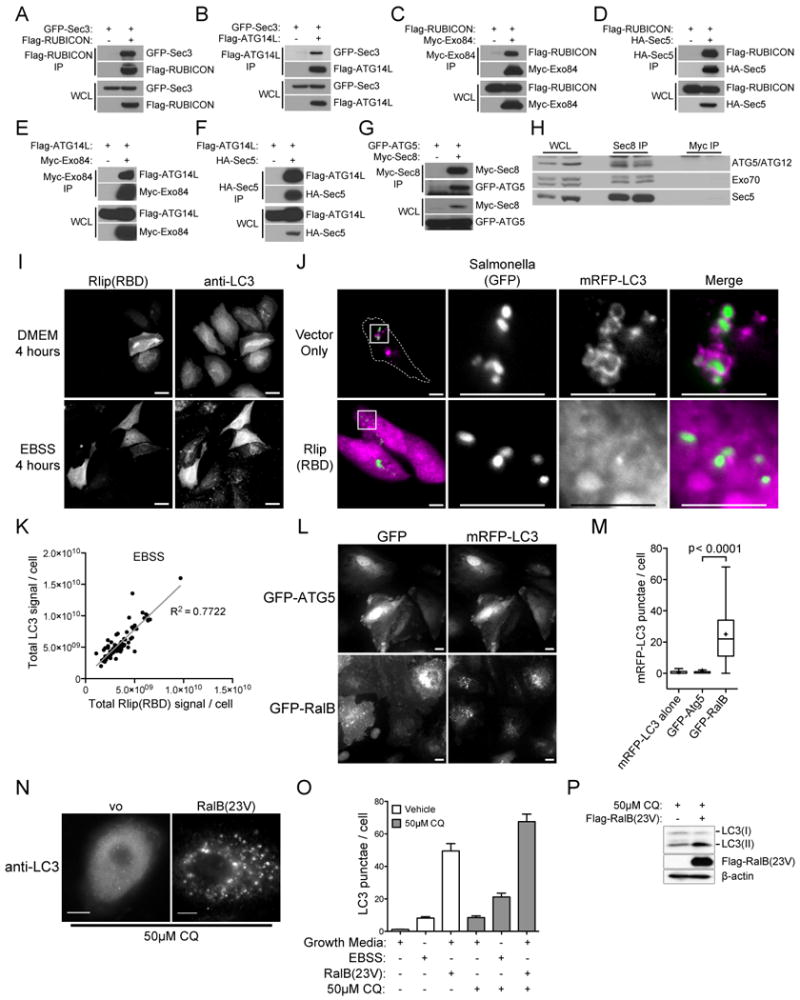
A-G: Exocyst subunits interact with autophagy proteins. The indicated proteins were over-expressed in HEK-293 cells, then immunoprecipitated with an antibody directed to the specified tag. Immunoprecipitates were analyzed for coprecipitation with (A,B) GFP-Sec3; (C,D) Flag-RUBICON; (E,F) Flag-ATG14L; and (G) GFP-ATG5 as indicated. Whole cell lysate (WCL), Immunoprecipitation (IP).
H: Endogenous Sec8 complexes contain ATG5-12 conjugates. The endogenous exocyst complex was immunoprecipitated from HEK-293 cells with anti-Sec8 antibody and analyzed for coprecipitation of ATG5/ATG12 conjugates (Sec8 IP) using anti-ATG5 antibody. Anti-Myc immunoprecipitates served as a negative control (Myc IP). Two independent experiments are shown. Representation of the examined proteins in the input whole cell non denaturing lysates is shown (WCL).
I: Inhibition of Ral signaling blocks the LC3 response to amino-acid starvation. 48 hours post-transfection with Myc-Rlip(RBD) Hela cells were incubated in DMEM or EBSS for an additional 4 hours as indicated. Myc-Rlip(RBD) and LC3 were detected by immunofluorescence using anti-myc and anti-LC3 antibodies, respectively. Vector control cells were similar to untransfected cells. Scale bar 20μm.
J: Inhibition of Ral signaling blocks the LC3 response to pathogen infection. Hela cells were transfected with monomeric RFP-LC3 together with Myc-Rlip(RBD) or an empty vector control as indicated. 48 hours post-transfection, cells were infected with Salmonella typhimurium-GFP for 1 hour followed by 3 hours of post-infection selection for intracellular Salmonella. Internalized Salmonella and LC3 were visualized using their respective fluorescent fusions. High magnification of the subcellular regions indicated by the boxes are shown in the panels on the right. Dashed lines indicated cell borders as visualized in a saturated exposure. Scale bar 10μm.
K: Total fluorescence intensity corresponding to Myc-Rlip(RBD) (anti-myc) and endogenous LC3 (anti-LC3) at single-cell resolution for EBSS-treated cells as shown in (I) (n=82, R2=0.7722).
L: RalB is sufficient to induce accumulation of LC3 punctae. Hela cells expressing monomeric RFP-LC3 together with GFP-ATG5 or GFP-RalB are shown as indicated. Scale bar 10μm.
M: mRFP-LC3 punctae in cells treated as in (L) were quantitated. The distribution of mRFP-LC3 punctae/cell is displayed as box-and-whisker plots. The three bands of the box illustrate the 25th (lower), 50th (middle), and 75th (upper) quartiles. The whiskers go 1.5 times the interquartile distance or to the highest or lowest point, whichever is shorter. The + designates the mean. P-values were calculated using the student's t-test.
N: HBEC3-KT cells expressing RalB(23V) or transfected with vector control were incubated in growth medium containing 50μM Chloroquine (CQ), to prevent LC3 turnover by autophagolysosomes, for 4 hours followed by detection of endogenous LC3 with anti-LC3 antibody.
O: RalB is sufficient to induce autophagic flux. HBEC3-KT cells treated as in (N) were incubated in growth media or amino-acid free EBSS (Earle's balanced salt solution) for 4 hours with or without 50μM Chloroquine (CQ), to prevent LC3 turnover in autophagolysosomes, as indicated. Immunofluorescence was performed with anti-LC3 antibody and LC3 punctae were quantitated. Data are represented as mean +/- SEM.
P: Whole cell lysates from HBEC3-KT cells transfected with Flag-RalB(G23V) or vector control were analyzed for the relative accumulation of LC3(I) and LC3(II) when incubated in growth medium containing 50μM Chloroquine (CQ) for 4 hours. β-actin is shown as a loading control.
See also Table S1.
RalB signaling is required and sufficient for induction of autophagosome formation
Mobilization of exocyst assembly in response to regulatory inputs is a major occupation of the Ras-like GTPases RalA and RalB (Balakireva et al., 2006; Cascone et al., 2008; Chen et al., 2007; Chien et al., 2006; Frische et al., 2007; Hase et al., 2009; Jin et al., 2005; Lalli and Hall, 2005; Moskalenko et al., 2002; Moskalenko et al., 2003; Rosse et al., 2006; Spiczka and Yeaman, 2008; Sugihara et al., 2002). To examine the potential participation of Ral GTPase signaling in the regulation of autophagy, we first tested the consequence of blocking Ral-GTP/effector interactions on amino acid starvation-induced autophagosome accumulation and on isolation membrane encapsulation of bacterial pathogens. Expression of the minimal Ral-binding domain of the Ral effector RalBP1/RLIP76 (RLIP(RBD)) is dominant inhibitory to the action of endogenous RalA and RalB proteins through direct competition with Ral effector molecules (Chien et al., 2006; Moskalenko et al., 2002). As previously demonstrated (Fass et al., 2006; Pattingre et al., 2005), serum and amino-acid starvation of HeLa cells with Earle's Basic Salt Solution (EBSS) induced re-localization of endogenous LC3 protein from a diffuse cytosolic distribution to a condensed punctate pattern, and significantly decreased the total LC3 signal; consistent with starvation-induced autophagosome formation and maturation. RLIP(RBD) expression blocked both LC3 punctae formation and LC3 turnover (Figure 1I). As a surrogate measure for autophagic flux, we quantitated the total endogenous LC3 signal of individual cells following amino acid starvation, and found that RLIP(RBD) expression inhibited LC3 protein turnover in a dose-dependent fashion (Figure 1K). To investigate the contribution of Ral signaling to pathogen-responsive LC3 modification of membranes, mRFP-LC3 expressing Hela cells were infected with GFP-labeled Salmonellae typhimurium. As expected if Ral signaling supports this response, RLIP(RBD) expression blocked recruitment of LC3 to internalized Salmonellae (Figure 1J). In addition, we found that ectopic expression of RalB was sufficient to induce the accumulation of LC3 punctae in cervical cancer cells (Figure 1L,M) and in immortalized bronchial epithelial cells (Figure 1N,O) in the absence of amino acid starvation or pathogen exposure. Remarkably, RalB(G23V) expression in nutrient rich conditions was sufficient to induce an accumulation of LC3 punctae that was 4-5 fold higher than that induced by amino acid deprivation (Figure 1O). This accumulation is likely associated with increased autophagic flux as RalB(G23V)-induced LC3 punctae were further increased by chloroquine-mediated inhibition of autophagosome turnover (Figure 1O; P=0.011, student's T-test), and this correlated with accumulation of phosphatidylethanolamine-conjugated LC3 (Figure 1P). Thus Ral signaling appears to be necessary and sufficient to engage autophagy. Evaluation of interactions between Ral signaling and autophagy in animals was carried out in Drosophila dRal hypomorphs (Ral35d), which have a weak loss-of-bristle phenotype associated with post-mitotic cell-specific apoptosis (Balakireva et al., 2006). Depletion of ATG14L, ATG1 (ULK1), ATG8a (LC3), ATG6 (Beclin) or VPS34, by in vivo expression of corresponding dsRNA, significantly enhanced the Ral35d phenotype (Table S1).
To directly investigate the individual contributions of human Ral GTPases and exocyst proteins to regulated autophagosome biogenesis, we next tested the consequence of siRNA-mediated RalA, RalB, and exocyst subunit depletion on nutrient-starvation induced autophagy. Depletion of RalA, in a stable GFP-LC3 expressing cell line, had no consequence on GFP-LC3 signal accumulation or punctae formation. In contrast, RalB depletion significantly impaired starvation-induced LC3 punctae formation and LC3 turnover (Figure 2A). The extent of autophagosome inhibition observed upon RalB depletion, as monitored by quantitation of GFP-LC3 punctae and total GFP-LC3 signal intensity, was equivalent to that seen upon depletion of the known components of autophagosome formation, ATG5 or Beclin1 (Figure 2B,C). An equivalent analysis of the exocyst subunits Sec8, Sec5 and Exo84 indicated selective contributions of exocyst components to presumed autophagosome formation. Depletion of Sec8, a central exocyst subunit, had equivalent consequences as depletion of RalB, ATG5 or Beclin1. In contrast, among the two Ral effectors in the exocyst, Sec5 and Exo84, only Exo84 depletion impaired starvation-induced LC3 punctae formation and increased LC3 accumulation (Figure 2B,C). Evaluation of each of the 8 exocyst subunits suggested that in addition to Sec8 and Exo84, Sec3 and Exo70 are limiting for support of autophagocytosis (Figure 2D, and Figure S1A). The selective requirement for Exo84 versus Sec5 indicates that RalB regulation of autophagy is likely independent of the previously characterized RalB/Sec5/TBK1 signaling pathway (Chien et al., 2006). Additional observations supporting RalB-selective support of autophagosome formation were made through examination of starvation-induced changes in endogenous LC3 localization and LC3 post translational modification (Figure 2E,F,G). These combined observations indicate that RalB and discrete components of the exocyst are required for autophagosome formation in multiple biological contexts. Activation of endogenous Ral GTPases may also be sufficient to induce autophagy, as depletion of endogenous RalGAP in the absence of nutrient limitation was sufficient to activate RalB and induce autophagic flux (Figure S1B,C,D,E).
Figure 2. RalB and an Exo84-containing subcomplex of the exocyst are necessary for amino acid starvation induced autophagy.
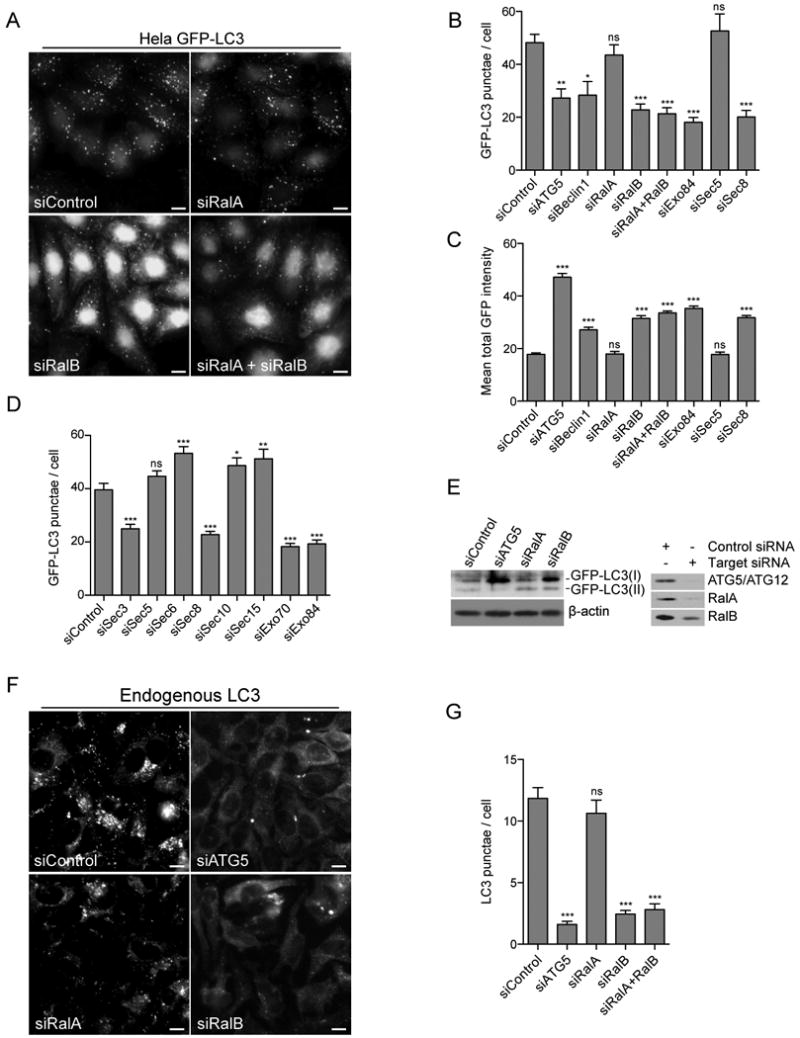
A: RalB depletion inhibits accumulation of GFP-LC3 punctae. Hela cells stably expressing GFP-LC3 were depleted of the indicated proteins by siRNA transfection. Cells were imaged by GFP fluorescence 96 hours after transfection. Scale bar 10μm.
B: Sec5 and Exo84 selectively participate in accumulation of GFP-LC3 punctae. GFP-LC3 punctae in cells treated as in (A) were quantitated. The mean distribution of GFP-LC3 punctae/cell is displayed as a bar graph, data are represented as mean +/- SEM. P-values were calculated by one-way ANOVA followed by Dunnett's Multiple Comparison Test.
C: Inhibition of GFP-LC3 punctae correlates with accumulation of LC3 protein. The mean total intensity of GFP-LC3 in cells treated as in (A) was quantitated. The distribution of the mean total GFP intensity is displayed as a bar graph, data are represented as mean +/- SEM. P-values were calculated by one-way ANOVA followed by Dunnett's Multiple Comparison Test.
D: A subset of exocyst subunits are limiting for accumulation of GFP-LC3 punctae. The indicated siRNAs were evaluated as in (B).
E: RalB depletion inhibits accumulation of LC3-lipid conjugates. Whole cell lysates from HBEC3-KT cells stably expressing GFP-LC3 transfected with the indicated siRNAs were assayed for the relative accumulation of GFP-LC3(I) and GFP-LC3(II). β-actin is shown as a loading control. siRNA-mediated target depletion is shown 96 hours post transfection (right panels).
F: RalB participates in accumulation of endogenous LC3 punctae. Hela cells were depleted of the indicated proteins by siRNA transfection. 96 hours after transfection, cells were incubated in amino acid free EBSS for 4 hours. Endogenous LC3 was imaged by anti-LC3 immunofluorescence. Scale bar 10μm.
G: Endogenous LC3 punctae in cells treated as in (E) were quantitated. The mean distribution of LC3 punctae/cell is displayed as a bar graph, data are represented as mean +/- SEM. P-values were calculated by one-way ANOVA followed by Dunnett's Multiple Comparison Test.
See also Figure S1.
RalB is recruited to sites of nascent autophagosome formation
To investigate the physical proximity of RalB to autophagosome formation, we examined the subcellular localization of endogenous RalB and components of the autophagosome initiation and elongation machinery. In telomerase and CDK4-immortalized normal human airway epithelial cells (HBEC30-KT), we noticed conspicuous co-localization of endogenous Beclin1 and endogenous RalB in perinuclear structures. Upon amino acid starvation (EBSS for 30 minutes), we observed re-distribution of both RalB and Beclin1 to vesicular structures throughout the cell body (Figure 3A). The majority of these RalB positive structures co-labeled with a GFP-2X-FYVE reporter that localizes to sites of PI-(3)-P enrichment (Gillooly et al., 2000), the product of the Beclin1-associated class III PI3K VPS34 (Figure 3B). In addition, we found marked colocalization of endogenous RalB with GFP-ATG5 after a 90-minute incubation in starvation media (Figure 3C). By 4 hours, GFP-LC3 punctae had accumulated, many of which were RalB positive (Figure 3D).
Figure 3. Native RalB colocalizes with autophagy machinery.
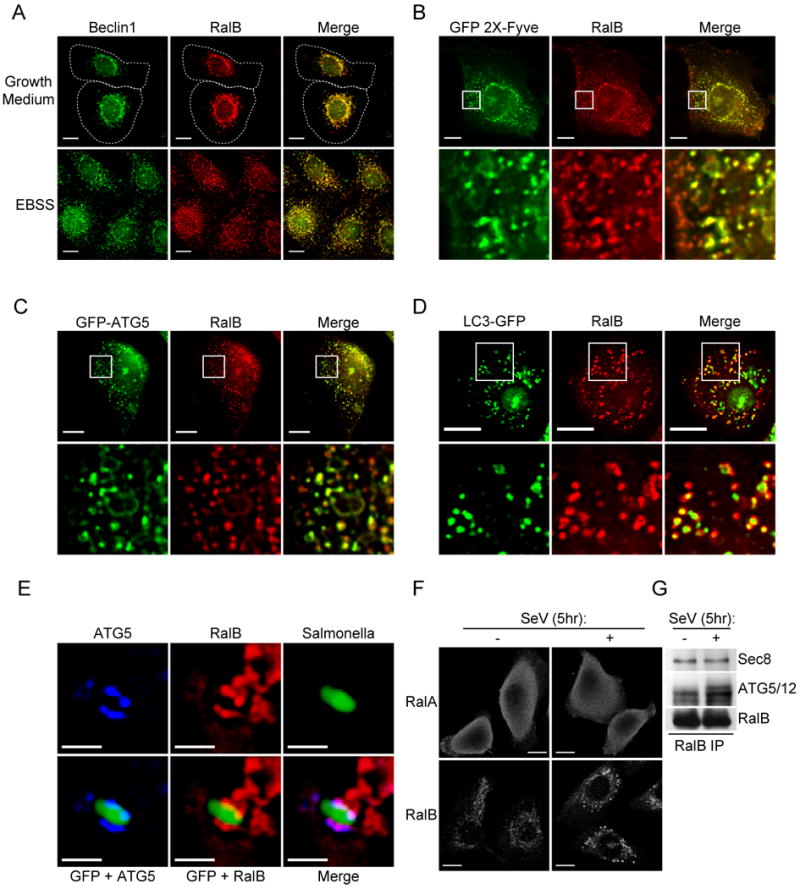
A: Beclin1 and RalB colocalize in cells pre and post induction of autophagy. Endogenous immunofluorescence of Beclin1 (anti-Beclin1) and RalB (anti-RalB) in HBEC30-KT cells incubated for 90 minutes in fresh growth medium or EBSS as indicated. Dashed line indicates cell outline. Scale bar 10μm.
B-D: RalB colocalizes with early and late markers of autophagosome biogenesis. HBEC30-KT cells were transfected with (B) GFP-2X-Fyve; (C) GFP-ATG5; and (D) GFP-LC3. Cells were incubated in EBSS for (B) 30 minutes; (C) 90 minutes; or (D) 3 hours. GFP fluorescence and endogenous RalB (anti-RalB) immunofluorescence is shown. High magnification of 10μm × 10μm regions indicated by the boxes are shown in the bottom panels. Scale bar 10μm.
E: ATG5 and RalB are recruited to sites of incipient isolation membrane formation. Endogenous immunofluorescence of ATG5 (anti-ATG5) and RalB (anti-RalB) in HBEC30-KT cells infected with Salmonella typhimurium-GFP. Cells were exposed to Salmonella typhimurium-GFP for 1 hour followed by 3 hours of post-infection antibiotic selection against extracellular Salmonella. Scale bar 2μm.
F: SenV infection selectively alters the subcellular distribution of RalB versus RalA. Endogenous immunofluorescence of RalA (anti-RalA) and RalB (anti-RalB) in HBEC3-KT cells mock infected or infected with Sendai virus for 5 hours. Scale bar 10μm.
G: SenV infection induces accumulation of endogenous RalB/ATG5-12 complexes. Endogenous RalB complexes were immunoprecipitated from mock infected or Sendai virus infected HBEC3-KT cells with anti-RalB antibodies and analyzed for coprecipitation of ATG5/ATG12 conjugates.
To investigate the recruitment of RalB to a discrete membrane site undergoing LC3-modification, we utilized GFP-expressing Salmonellae typhimurium as a detectable, proximal signal for LC3-modification of the vacuole. Three hours after post-infection antibiotic selection to remove extracellular Salmonellae, we found that endogenous ATG5 was present along the surface of internalized GFP-Salmonellae, which colocalized with RalB (Figure 3E). Finally, an autophagic response of HBEC cells to Sendai virus exposure induced a re-distribution of RalB but not RalA to cytosolic vesicular structures and promoted accumulation of endogenous RalB-ATG5/ATG12 protein complexes (Figure 3F,G).
Nutrient-starvation and RalB drive assembly of Exo84-Beclin1 complexes
Given that Beclin1, a central regulatory node engaged to initiate autophagic responses to diverse stimuli, colocalized with RalB, we examined the relationship between Beclin1 and exocyst subunits. We found that nutrient starvation induced a dramatic assembly of Exo84/Beclin1 complexes in HEK293 cells (Figure 4A). In stark contrast, abundant Sec5/Beclin1 complexes present under nutrient-rich growth conditions were disassembled within 90 minutes of nutrient deprivation (Figure 4B), which could be reversed by addition of nonessential amino acids (Figure 4C). Sec8/Beclin1 complexes, on the other hand, were present under both nutrient-rich and nutrient-poor growth conditions (Figure 4D). Analysis of Beclin1 deletion constructs indicated that both Exo84 and Sec5 required the amino-terminal BCL2-interacting domain for interaction with Beclin1 (88-150), while the evolutionarily conserved domain (244-337) was dispensable (Figure S2A). However, Exo84 and Sec5 likely have distinct binding determinants within the BCL2-interacting domain, as Beclin1(F123A), which fails to bind BCL2, interferes with Sec5 but not Exo84 association (Figure 4E,F). Importantly, we found that immunoprecipitation of endogenous Beclin1 from nutrient-deprived versus nutrient replete cells resulted in selective coprecipitation of endogenous Exo84 under starvation conditions (Figure 4G). These observations suggest that Beclin1 is recruited to distinct exocyst subcomplexes in response to nutrient availability. Previous observations from our group indicated that discrete macromolecular Exo84 and Sec5 complexes can be detected by density gradient centrifugation in pheochromocytoma cells (Moskalenko et al., 2003). Accordingly we found that endogenous Exo84 and Sec5 display very distinct localization patterns in epithelial cells (Figure 4H). Likewise, ectopic expression of Exo84 or Sec5 was sufficient to differentially recruit Beclin1 to these distinct subcellular compartments (Figure 4 I,J). The Exo84 compartment and the Sec5 compartment were reminiscent of the staining patterns observed with endogenous Beclin1 in the nutrient starved versus fed states respectively (Figure 3A). Size exclusion chromatography of cleared lysates, from proliferating cells, indicated the bulk of endogenous Exo84 and Sec5 eluted in separate fractions of ∼500 kDa and > 700 kDA respectively, both of which partially cofractionated with Beclin1. Intriguingly, the ATG1 ortholog, ULK1, displayed a bimodal distribution presumably representative of distinct high and low molecular weight complexes (Figure S2B).
Figure 4. Nutrient deprivation drives assembly of Exo84/Beclin1 complexes.
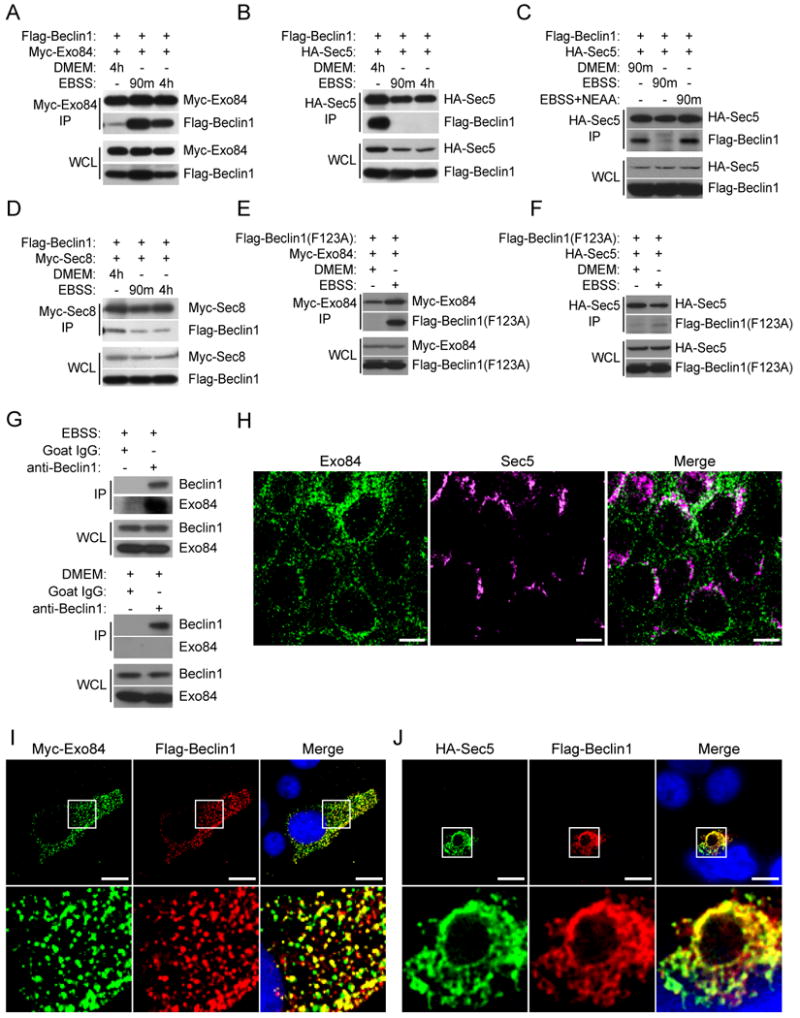
A-D: Nutrient limitation induces Beclin1/Exo84 interactions and inhibits Beclin1/Sec5 interactions. 48 hours post-transfection with tagged Beclin1 and exocyst expression constructs, HEK-293 cells were incubated in DMEM, EBSS, or EBSS with 1× Non-Essential amino acids for 90 minutes or 4 hours as shown. The indicated proteins were then immunoprecipitated with antibodies directed to the specified tag. Immunoprecipitates were analyzed for coprecipitation with Flag-Beclin1. Whole cell lysate (WCL), Immunoprecipitates (IP).
E,F: Beclin1(F123A) mutant interacts with Exo84 but not Sec5. Co-expression, co-IPs with the indicated proteins were performed as in (A-D).
G: Endogenous Beclin1/Exo84 complexes accumulate in response to nutrient deprivation. Endogenous Beclin1 was immunoprecipitated from HEK-293 cells incubated in EBSS (top panels) or DMEM (bottom panels) for 90 minutes and analyzed for coprecipitation of Exo84 (IP). Host species-matched non-specific IgG immunoprecipitates served as negative controls. Representation of the examined proteins in the input whole cell non-denaturing lysates is shown (WCL).
H: Exo84 and Sec5 are enriched in distinct subcellular compartments. Endogenous immunofluorescence of Sec5 (anti-Sec5) and Exo84 (anti-Exo84) in MDCK cells. Scale bar 10μm.
I,J: Exo84 and Sec5 can recruit Beclin1 to distinct subcellular compartments. HEK-293 cells were transfected with (F) Flag-Beclin1 and Myc-Exo84; (G) Flag-Beclin1 and HA-Sec5. Immunofluorescence of the indicated fusion tags was performed. High magnification of 10μm × 10μm regions indicated by the boxes are shown in the bottom panels. Scale bar 10μm.
See also Figure S2.
Consistent with a sentinel role in the cellular response to nutrient deprivation, RalB was activated by nutrient deprivation as indicated by accumulation of the GTP-bound conformation. RalA, on the other hand, was unaffected (Figure 5A). Accordingly, expression of a constitutively active RalB variant (RalB(23V)) was sufficient to induce Beclin1/Exo84 complex formation in the absence of nutrient deprivation (Figure 5B). To examine whether a direct RalB-Exo84 effector interaction was necessary for RalB to drive Exo84-Beclin1 association, we employed partial loss of function RalB variants selectively uncoupled from Exo84 versus Sec5 (Cascone et al., 2008; Jin et al., 2005). RalB(G23V,A48W) has a 43-fold higher affinity for Sec5 versus Exo84, and RalB(G23V,E38R) has a 104-fold higher affinity for Exo84 versus Sec5 (Jin et al., 2005). As shown, RalB(G23V,E38R) was considerably more effective at promoting Exo84/Beclin1 complex formation as compared to its Exo84-binding defective counterpart, RalB(G23V,A48W) (Figure 5C). In contrast to Exo84, Beclin1/Sec5 complexes can be isolated under nutrient rich conditions. Interestingly, inhibition of Ral signaling by RLIP(RBD) expression eliminates the accumulation of Beclin1/Sec5 complexes suggesting that RalA/B signaling under nutrient rich conditions is required for this interaction (Figure 5D). A point mutation of the Ral-binding domain of Sec5(T11A), which abolishes binding to Ral-GTP, also abolished the Sec5-Beclin1 interaction (Figure 5E). No interaction of Beclin1 with RLIP76, an exocyst-independent Ral effector, was observed (Figure 5F); indicating that Ral family modulation of Sec5/Beclin1 and Exo84/Beclin1 complexes is specific. The sufficiency of RalB interactions to drive both Sec5-Beclin1 and Exo84-Beclin1 complexes, coupled with our observations of the selective responsiveness of these complexes to nutrient status, suggested the possibility that nutrient availability results in distinct RalB-effector coupling. Indeed, endogenous RalB preferentially associated with Exo84 in nutrient poor conditions and Sec5 under nutrient rich conditions (Figure 5 G,H). Importantly, endogenous ULK1, a key kinase that promotes initiation of autophagy (Chan et al., 2007; Hosokawa et al., 2009a; Jung et al., 2009), was selectively enriched in RalB immunoprecipitates upon nutrient depletion (Figure 5G). These observations indicate that direct RalB/exocyst effector interactions differentially deliver Beclin1 to Sec5 or Exo84-containing subcomplexes in response to nutrient availability and that RalB/Exo84 complexes may specify activation of autophagosome formation. Consistent with this, we found that inhibition of Ral signaling by Rlip(RBD), which inhibits autophagy, promoted the association of the autophagy inhibitor protein RUBICON with Exo84 (Figure 5I).
Figure 5. RalB drives assembly of Exo84/Beclin1 complexes through direct RalB-Exo84 effector binding.
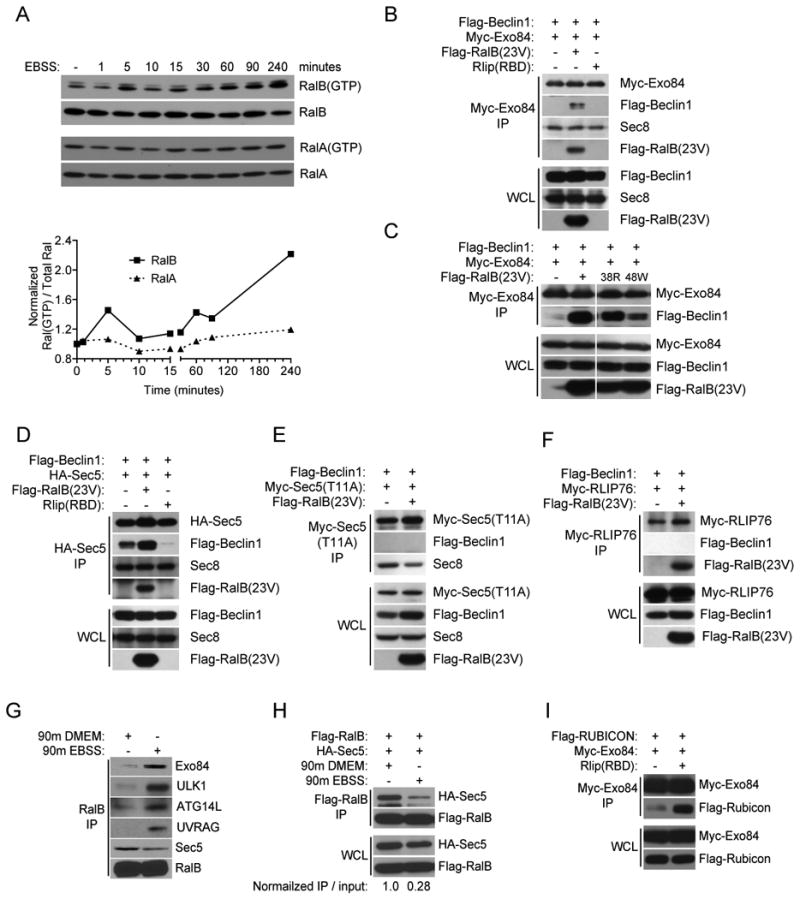
A: Amino-acid depletion activates RalB. Endogenous GTP-bound RalA and RalB were collected by GST-Sec5-RBD mediated affinity purification from HEK-293 cells incubated in EBSS for the indicated times and visualized with specific anti-RalA and anti-RalB antibodies. The normalized GTP-loaded index for RalA and RalB was calculated as Ral(GTP)/Total Ral to generate the scatterplot.
B-F: RalB regulates Beclin1/exocyst subcomplex interactions. The indicated proteins were expressed in HEK-293 cells and immunoprecipitated with antibodies directed to the appropriate tag. Immunoprecipitates were analyzed for coprecipitation with Flag-Beclin1, Flag RalB(23V), and endogenous Sec8 as shown. Whole cell lysate (WCL), Immunoprecipitation (IP).
G: Nutrient status specifies distinct endogenous RalB/effector interactions. Endogenous RalB was immunoprecipitated with anti-RalB antibody from HEK-293 cells incubated in DMEM or EBSS for 90 minutes as indicated and analyzed for coprecipitation of Exo84, Sec5, ATG14L, UVRAG, and ULK1.
H: 48 hours post-transfection, HEK-293 cells were incubated in DMEM or EBSS for 90 minutes as indicated. Flag-RalB immunoprecipitates were examined for coprecipitation of HA-Sec5. The indicated normalized IP / input ratio was calculated by dividing immunoprecipitated Sec5 by total expressed Sec5, then normalizing the calculated values to DMEM condition.
I: Ral-inhibition induces accumulation of Exo84/Rubicon interactions. Co-expression, co-IPs with the indicated proteins were performed as in (B-F).
The RalB/Exo84 effector pathway mobilizes VPS34 activity
To further probe the relationship of Exo84 versus Sec5 complexes to mobilization of autophagosomes, we examined the consequence of nutrient depletion or Ral activation on recruitment of VPS34 to exocyst/Beclin1 complexes. Beclin1 has been heavily implicated as a positive regulatory cofactor of the VPS34 lipid kinase, which is thought to be a biochemical trigger for initiation of autophagosome isolation membrane assembly and elongation (Vergne et al., 2009; Zeng et al., 2006). Like the Exo84/Beclin1 relationship, we found that nutrient depletion resulted in accumulation of Exo84/VPS34 complexes (Figure 6A). Again, as we had seen with Beclin1, the opposite relationship was observed with Sec5 (Figure 6B). Expression of active RalB in the absence of nutrient depletion mirrored these observations. Namely, RalB(G23V) drove assembly of Exo84/VPS34 complexes (Figure 6C) and drove disassembly of Sec5/VPS34 complexes in a manner dependent upon direct RalB/effector interactions (Figure 6D).
Figure 6. RalB expression drives assembly of Exo84/Vps34 and Exo84/ATG14L complexes.
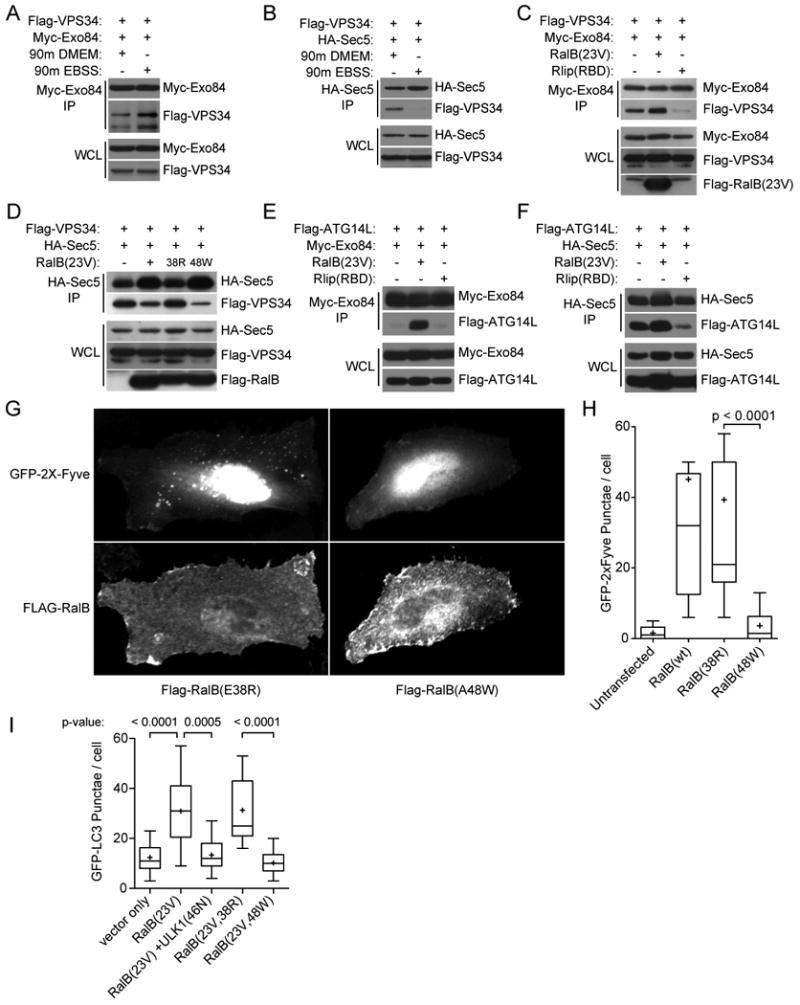
A-F: VPS34 and ATG14L/exocyst subcomplexes are regulated by nutrient limitation and RalB activation. HEK-293 cells expressing the indicated proteins were incubated in DMEM or EBSS for 90 minutes as indicated. Tagged exocyst subunits were immunoprecipitated and analyzed for coprecipitation with Flag-VPS34 and Flag-ATG14L where indicated.
G: RalB/Exo84 effector interactions mobilize VPS34 activity. Hela cells expressing GFP-2X-Fyve together with RalB partial loss of function mutants Flag-RalB(E38R) or Flag-RalB(A48W) are shown as indicated.
H: GFP-2X-Fyve punctae in cells treated as in (G) were quantitated. The distribution of GFP-2X-Fyve punctae/cell is displayed as box-and-whisker plots. P-values were calculated using the student's t-test.
I: RalB(G23V) and RalB(G23V,E38R) are sufficient to induce accumulation of GFP-LC3 punctae and kinase dead ULK1(K46N) blocks the increase observed with RalB(G23V) expression. Hela cells stably expressing GFP-LC3 were transfected with the indicated constructs then visualized by immunofluorescence of the indicated tags. The distribution of GFP-LC3 punctae/cell is displayed as box-and-whisker plots. P-values were calculated using the student's t-test.
The presence of ATG14L in complex with Beclin1 and VPS34 is thought to specify the participation of this complex in autophagy as opposed to other cell processes where VPS34 activity has been implicated (Itakura et al., 2008; Matsunaga et al., 2009; Sun et al., 2008; Zhong et al., 2009). We found that dynamic interactions of Exo84 and Sec5 with ATG14L were also remarkably similar to those observed with Beclin1. Specifically, under nutrient rich growth conditions, Exo84 association with ATG14L was induced by RalB(G23V) expression (Figure 6E) while preexisting Sec5/ATG14L complexes were inhibited in the presence of the dominant inhibitory peptide Rlip(RBD) (Figure 6F). Furthermore, RalB(E38R) but not RalB(A48W) expression was sufficient to drive accumulation of PI-(3)-P positive punctae, the product of active VPS34, as visualized by accumulation of the GFP-2X-FYVE probe (Figure 6G,H). Similar results were observed for the accumulation of GFP-LC3 punctae (Figure 6I). These observations suggest that induction of autophagy proceeds through assembly of Beclin1-ATG14L-VPS34 complexes on Exo84, while the interaction of these components with Sec5 may represent organization of inactive components in a preinitation complex and/or a signal termination complex. Importantly, the increased accumulation of GFP-LC3 observed upon RalB(G23V) expression was reversed by co-expression of kinase dead mutant ULK1(K46N), suggesting that RalB may act upstream of ULK1 to promote autophagy (Figure 6I).
Active ULK1 assembles on Exo84 upon induction of autophagy
ULK1 activation is the most apical positive inductive signal, among Atg proteins, yet identified for initiation of autophagy. We have found that RalB is activated upon nutrient starvation and that this correlates with the induction of RalB/ULK1 complexes (Figures 5A and 5G respectively). Remarkably, RalB(G23V) expression was sufficient to promote assembly of ULK1/Beclin1 complexes, which have not been described previously but which may represent a mechanistic link between ULK1 activation and the VPS34 vesicle nucleation complex (Figure 7A). Furthermore, either nutrient depletion or RalB(G23V) expression was sufficient to induce assembly of ULK1/Exo84 complexes (Figure 7B,F). Depletion of Exo84 eliminated the capacity of RalB(G23V) to induce ULK1/Beclin1 complex formation, indicating that Exo84 is required for this assembly event (Figure 7C). In contrast to ULK1/Exo84 interactions, we observed increased Sec5/ULK1 complex assembly when Ral signaling was blocked by expression of Rlip(RBD) (Figure 7D). Analysis of Beclin1 deletion constructs indicated that, unlike Exo84 and Sec5, ULK1 requires the evolutionarily conserved domain (aa 244-337) for Beclin1 association (Figure S3A). Importantly, while ULK1 was present in both Exo84 and Sec5 complexes under nutrient poor conditions (Figure 7E,F), only Exo84-associated ULK1 displayed significant catalytic activity (Figure 7F,G).
Figure 7. Active ULK1 associates with Exo84.
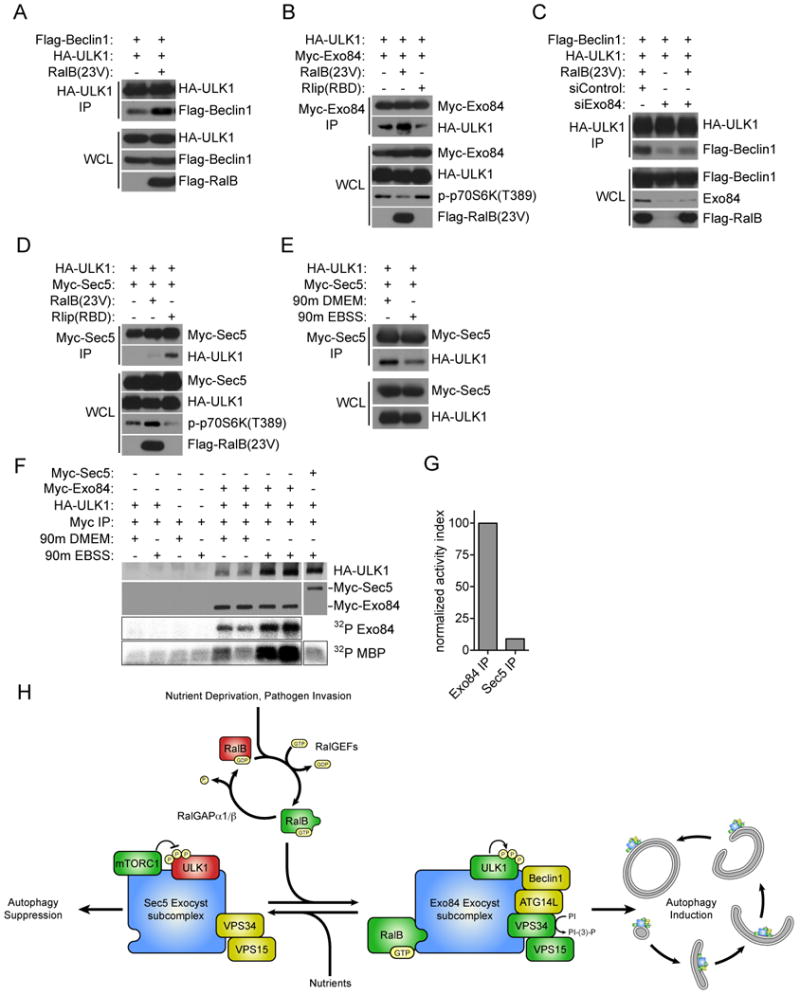
A: RalB induces ULK1/Beclin1 Complex formation. ULK1 immunoprecipitates were analyzed for coprecipitation with Flag-Beclin1 upon RalB(23V) expression as indicated.
B: ULK1/Exo84 complexes are regulated by RalB. The indicated proteins were expressed in HEK-293 cells. Myc-tagged Exo84 was immunoprecipitated and analyzed for coprecipitation with HA-ULK1.
C: RalB induced ULK1/Beclin1 complexes require Exo84. HEK-293 cells were first transfected with siControl or siExo84 siRNAs before the indicated proteins were expressed 24 hours later. ULK1 immunoprecipitates were analyzed for coprecipitation with Flag-Beclin1 upon RalB(23V) expression as indicated.
D: ULK1/Sec5 complexes accumulate upon Ral inhibition. Co-expression, co-IPs with the indicated proteins were performed as in (B).
E: ULK1/Sec5 complexes dissociate upon nutrient deprivation. Co-expression, co-IPs with the indicated proteins were performed as in (B) with the addition of 90 minute incubation in DMEM or EBSS as indicated.
F: Amino-acid starvation induces association of Exo84 with catalytically active ULK1. Exo84 and Sec5 complexes were assayed for coprecipitation with ULK1 and for associated protein kinase activity as indicated.
G: The normalized activity ratio for EBSS stimulated Exo84 and Sec5 precipitates was calculated by the division of MBP 32P incorporation by the HA-ULK1 signal coprecipitated from (F).
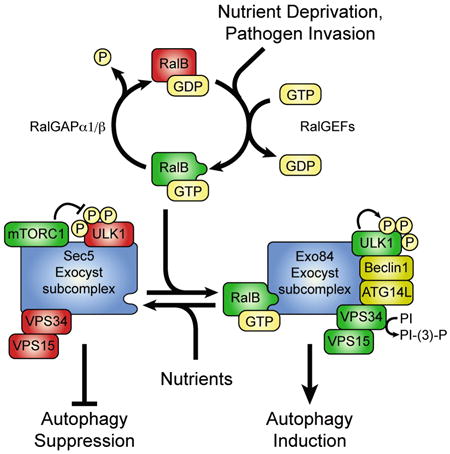
H: Working model of RalB/exocyst dependent mobilization of autophagy.
See also Figure S3.
ULK1 and mTORC1 have been reported to inhibit each other by reciprocal phosphorylation (Hosokawa et al., 2009a; Lee et al., 2007). Consistent with catalytically active Exo84/ULK1 complexes, expression of RalB(G23V) and Exo84 was sufficient to inhibit base-line mTORC1 activity as observed by reduced accumulation of phospho-threonine 389 on p70S6K (Figure 7B). In contrast, expression of RalB(G23V) and Sec5 resulted in increased accumulation of phospho-p70S6K (Figure 7D). Of note, endogenous mTORC1 was present in Sec5 but not Exo84 immunoprecipitates (Figure S3B). The assembly of ULK1 with Exo84 and disassembly of Beclin1 from Sec5 are responsive to mTOR inhibition, but only upon chronic exposure to rapamycin, suggesting this is a consequence of the indirect effects that mimic nutrient starvation (Figure S3C,D). These combined observations indicate that the RalB/Exo84 effector relationship engages autophagy through direct modulation of a ULK1/Beclin1 initiation complex.
Discussion
Our findings are consistent with a model in which the Ras-like G-protein RalB acts as a regulatory switch to promote autophagosome biogenesis, in response to inductive signaling events, by mobilizing assembly of ULK1/Beclin1/VPS34 autophagy initiation complexes (Figure 7H). RalB is activated during the autophagic response, is localized to sites of incipient autophagosome formation, and is necessary and sufficient for induction of autophagic flux. In response to RalB activation, the direct RalB effector Exo84 is engaged as an essential assembly platform for catalytically active autophagy induction (ULK1/FIP200) and vesicle initiation (Beclin1/ATG14L,VPS34) complexes.
In all cases examined, we found that dynamic assembly of active autophagosome biogenesis machinery on Exo84 was coordinated with disassembly of this same machinery from Sec5. Both Exo84 and Sec5 are Ral family G-protein effectors and subunits of the exocsyt, a heterooctomeric secretory vesicle trafficking complex (Lipschutz and Mostov, 2002). Previous work has shown that distinct Exo84 and Sec5 subcomplexes are directly engaged by Ral signaling to mobilize exocyst holocomplex formation in support of the dynamic vesicle targeting and tethering events required for stimulus-dependent exocytosis (Moskalenko et al., 2002; Moskalenko et al., 2003). Remarkably, the Sec5/autophagy protein disassembly event and the Exo84/autophagy protein assembly event, described here, both require direct interaction with active RalB proteins. Thus induction of autophagy through RalB activation triggers dynamic autophagy protein reassembly events centered on two independent exocyst subunits. The tethering of autophagosome biogenesis machinery to distinct exocyst subcomplexes may provide appropriate spatial and temporal resolution of localized autophagic triggers. Consistent with this, we find that Sec5 and Exo84 accumulate in discrete cellular compartments that segregate with localization of Beclin1 pre (Sec5) and post (Exo84) induction of autophagy. It will be of great interest to determine if these locations represent the source location and assembly sites, respectively, of the membrane proteins required for autophagosome isolation membrane construction. Known exocyst subunit-autonomous mechanisms specifying subcellular localization patterns include interactions with organelle-specific proteins and membrane-selective phosphoinositides (He and Guo, 2009).
Recent observations indicate that detection of conserved pathogen-associated molecular patterns (PAMPs) by Toll-like receptors will mobilize autophagy together with activation of canonical innate-immune pathway activation (Delgado et al., 2008; Shi and Kehrl, 2010). Within this context of host defense surveillance and response systems, we have previously shown that RalB can engage Sec5 to activate the innate immunity signaling kinase TBK1 and the subsequent IRF3 transcription factor-dependent interferon response (Chien et al., 2006). Our observations here indicate that RalB can separately engage Exo84 to facilitate activation of the autophagy kinase ULK1 and induction of autophagosome biogenesis. Together, this suggests that RalB and the exocyst represent a regulatory hub, through bifurcating activation of TBK1 and Beclin1/VPS34, that helps engage concomitant activation of the gene expression and organelle biogenesis responses supporting systemic pathogen recognition and clearance. The required coordination of such time and location-specified responses may account for the adaptation of exocyst function to support of signal transduction cascades in metazoans.
The RalA and RalB G-proteins are signal propagation molecules coupled to mitogenic, trophic and cytokine signaling signaling systems (Bodemann and White, 2008; Feig, 2003). This connectivity potentially provides appropriate functional coupling of autophagic responses to diverse cellular milieus. Ral activation occurs through engagement of one or more of a family of 5 Ral-specific guanyl nucleotide exchange factors that can selectively couple to RalA or RalB through mechanisms that remain to be determined (Bodemann and White, 2008). Of note, RalA has recently been shown to promote mTORC1 activation, potentially through PLD1 and phosphatidic acid-dependent mTORC1/2 assembly (Maehama et al., 2008; Toschi et al., 2009; Voss et al., 1999). This regulatory relationship could be directly antagonistic to autophagosome biosynthesis given the capacity of mTORC1 to restrain ULK1 activity through direct inhibitory phosphorylation events (Hosokawa et al., 2009a; Jung et al., 2009). Whether these are independent or interconnected regulatory arms remains to be determined. However, the RalB GDP/GTP cycle and its effector relationships comprise a regulatory mechanism that can directly control dynamic transition between metabolic states supporting cell growth versus cell maintenance.
Materials and Methods
Plasmids and Antibodies
Detailed information on plasmid and antibody origins, immunofluorescence protocols, image capture procedures, and image quantitation are described in supplemental methods.
Yeast Two-Hybrid
The coding sequence for full-length human SEC3 (GenBank gi:7023219) was cloned into pB27 as a C-terminal fusion to LexA and used as a bait to screen at saturation a high-complexity random-primed human placenta cDNA library as previously described (Fromont-Racine et al., 1997).
Immunoprecipitation and kinase assays
Immunoisolation of tagged or native proteins was performed using standard procedures from non-denaturing cell extracts (20 mM Tris-HCl pH 7.4, 137mM NaCl, 1% Triton-X-100, 0.5% Sodium Deoxycholate, 10mM MgCl2, 2mM EGTA). Kinase assays were carried out in (25 mM MOPS pH 7.5, 1mM EGTA, 0.1mM Sodium Vanadate, 15mM MgCl2, 5 mM β-glycerol phosphate). See supplemental methods for extended details.
Salmonella typhimurium Infection
Exposure to GFP-expressing Salmonella typhimurium (obtained from Mary O'Riordan, University of Michigan) was performed as described (Radtke et al., 2007).
Supplementary Material
Acknowledgments
We thank Noboru Mishuzima for GFP-Atg5 and GFP-LC3, Zhenyu Yue for Flag-ATG14L and Flag-RUBICON, and Noriko Okazaki for HA-ULK1 and HA-ULK1(K46N). We thank members of the White and Levine labs for productive discussions. We thank Melanie Cobb for helpful discussion and assistance with kinase assays. This work was supported by grants from the National Institutes of Health (CA71443 and CA129451 to MW, andCA84254 and CA109618 to BL), ARC4845 to JC, and the Welch Foundation (I-1414 to MW). BB was supported by DOD Award Number W81XWH-06-1-0749. RR was supported by T32GM008203. YO was supported by CPRIT RP101496.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amar N, Lustig G, Ichimura Y, Ohsumi Y, Elazar Z. Two newly identified sites in the ubiquitin-like protein Atg8 are essential for autophagy. EMBO Rep. 2006;7:635–642. doi: 10.1038/sj.embor.7400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva M, Rosse C, Langevin J, Chien YC, Gho M, Gonzy-Treboul G, Voegeling-Lemaire S, Aresta S, Lepesant JA, Bellaiche Y, et al. The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol Cell Biol. 2006;26:8953–8963. doi: 10.1128/MCB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvanakantham R, Li J, Tan TT, Ng ML. Human Sec3 protein is a novel transcriptional and translational repressor of flavivirus. Cell Microbiol. 2010;12:453–472. doi: 10.1111/j.1462-5822.2009.01407.x. [DOI] [PubMed] [Google Scholar]
- Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- Cascone I, Selimoglu R, Ozdemir C, Del Nery E, Yeaman C, White M, Camonis J. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–2387. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chen XW, Leto D, Chiang SH, Wang Q, Saltiel AR. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev Cell. 2007;13:391–404. doi: 10.1016/j.devcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frische EW, Pellis-van Berkel W, van Haaften G, Cuppen E, Plasterk RH, Tijsterman M, Bos JL, Zwartkruis FJ. RAP-1 and the RAL-1/exocyst pathway coordinate hypodermal cell organization in Caenorhabditis elegans. EMBO J. 2007;26:5083–5092. doi: 10.1038/sj.emboj.7601922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindstaff KK, Yeaman C, Anandasabapathy N, Hsu SC, Rodriguez-Boulan E, Scheller RH, Nelson WJ. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 1998;93:731–740. doi: 10.1016/s0092-8674(00)81435-x. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, Ito M, Watarai H, Hazelett CC, Yeaman C, et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol. 2009;11:1427–1432. doi: 10.1038/ncb1990. [DOI] [PubMed] [Google Scholar]
- He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009a;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009b;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 2005;24:2064–2074. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Lalli G, Hall A. Ral GTPases regulate neurite branching through GAP-43 and the exocyst complex. J Cell Biol. 2005;171:857–869. doi: 10.1083/jcb.200507061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, Kim JM, Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–365. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipschutz JH, Mostov KE. Exocytosis: the many masters of the exocyst. Curr Biol. 2002;12:R212–214. doi: 10.1016/s0960-9822(02)00753-4. [DOI] [PubMed] [Google Scholar]
- Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283:35053–35059. doi: 10.1074/jbc.M805822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res. 2000;85:1–12. doi: 10.1016/s0169-328x(00)00218-7. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MX. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiczka KS, Yeaman C. Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J Cell Sci. 2008;121:2880–2891. doi: 10.1242/jcs.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss M, Weernink PA, Haupenthal S, Moller U, Cool RH, Bauer B, Camonis JH, Jakobs KH, Schmidt M. Phospholipase D stimulation by receptor tyrosine kinases mediated by protein kinase C and a Ras/Ral signaling cascade. J Biol Chem. 1999;274:34691–34698. doi: 10.1074/jbc.274.49.34691. [DOI] [PubMed] [Google Scholar]
- Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


