Abstract
U2449 is one of many invariant residues in the central loop of domain V of 23S rRNA, a region that constitutes part of the peptidyltransferase center of the ribosome. In Escherichia coli, this U is post-transcriptionally modified to dihydrouridine (D) and is the only D modification found in E.coli rRNAs. To analyze the role of this base and its modification in ribosomal function, all three base substitutions were constructed on a plasmid copy of the rrnB operon and assayed for their ability to support cell growth in a strain of E.coli lacking chromosomal rrn operons. Both purine substitution mutations were not viable. However, growth and antibiotic sensitivity of cells expressing only the mutant D2449C rRNA was indistinguishable from wild type. We conclude that while a pyrimidine is required at position 2449 for proper ribosomal function, the D modification is dispensable.
INTRODUCTION
The regions of large and small subunit rRNAs that comprise the functional centers of the ribosome contain many invariant residues, and at least some of these residues are post-transcriptionally modified in all organisms (1,2). The localization of such modifications to conserved, functionally important regions of rRNA has given rise to the notion that these modifications must somehow be important for ribosome assembly and/or function (3). In agreement with this idea, in vitro reconstitution of Escherichia coli 50S subunits from the individual RNA and protein components was found to require the presence of one or more modifications contained within an ∼80 nt segment of 23S rRNA (positions 2445–2523; 4). However, reconstitution of 50S subunits and peptidyltransferase activity from Thermus aquaticus (5) or Bacillus stearothermophilus (6) ribosomal proteins and in vitro transcribed rRNA was accomplished without the need for any modifications, albeit with lower activity than that obtained with fully modified rRNAs. These data may indicate a difference between mesophilic and thermophilic organisms with respect to their requirement for rRNA modifications, and suggest that post-transcriptional modifications may be dispensable for catalytic function.
Only a minority of the genes encoding rRNA methylases have been identified (7,8) and disruption of many of these methylase genes has little effect on cell growth or ribosome function (2). However, in yeast mitochondrial ribosomes, loss of G2251 modification leads to defects in subunit assembly (9). Lack of pseudouridylation of several residues in the 1916 loop in domain IV of 23S rRNA in E.coli has substantial effects on cell growth (10) while lack of ribose modification of U2552 affects subunit–subunit interactions (11).
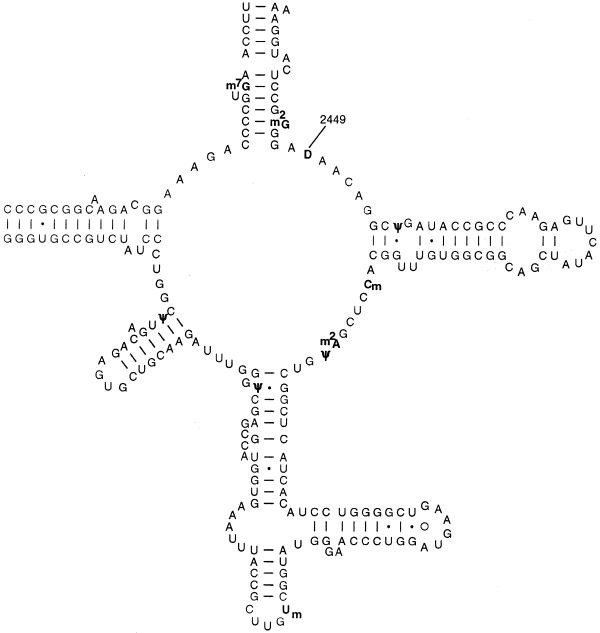
The central loop of domain V of 23S rRNA is the site of peptidyltransferase activity and this region of rRNA is also rich in post-transcriptional modifications (Fig. 1). In addition, this region of rRNA is one of the sites of ribosomal interaction with antibiotics that inhibit peptidyltransferase activity and growth of peptide chains (12). In E.coli, the invariant U2449 is modified to dihydrouridine (D) and, in contrast to its abundance in tRNAs, this is the only D modification found in E.coli rRNAs (13). Studies with tRNAs have suggested that D allows conformational flexibility of RNA molecules (14). This fact, together with the importance of the 2449 region of 23S rRNA in antibiotic binding and peptidyltransferase function prompted us to investigate the importance of the D modification at this position in E.coli 23S rRNA. In the absence of any knowledge concerning the location and nature of the rRNA-specific dihydrouridine synthetase, our approach has been to construct all three base substitution mutations at position 2449 on a plasmid-borne copy of the rrnB operon and attempt to express these mutant rRNAs in a strain of E.coli lacking all chromosomal rrn operons. Our findings were that while D2449A and D2449G transversion mutants were not viable under these conditions, the D2449C transition mutant was without any obvious phenotype. Our conclusions are that despite its extreme conservation, a U at position 2449 is not essential for ribosomal function. Moreover, since D2449 could be replaced with another (unmodified) pyrimidine without apparent consequence, we also conclude that the D modification at 2449 is dispensable.
Figure 1.
Sequence and secondary structure of the central loop region of domain V of E.coli 23S rRNA. The 10 post-transcriptional modifications are indicated in bold.
MATERIALS AND METHODS
Bacterial strains and plasmids
Plasmids pMO10 and pKK3535 are derived from plasmids pSC101 and pBR322, respectively, and contain the intact rrnB operon under the control of the native P1P2 promoters (15). In plasmid pLK35, the rrnB operon is transcribed from the inducible λPL promoter and, in the presence of the thermolabile λcI repressor, transcription of the rrnB operon is induced by a temperature shift to 42°C. Plasmid pts1192U was constructed by ligating the BamHI fragment containing the intact rrnB operon (and carrying the spectinomycin resistance C1192U mutation) from plasmid pKK1192U (16) to BamHI-cleaved pMAK705 (17). Strains MC242 (λ– Hfr relA1 spoT1 metB1) or MC140 [F– Δ(lac-pro) thi– recA– srl–] transformed with pLG857 (encoding the temperature-sensitive λcI857 repressor) were used as hosts for pLK35-derived plasmids. The Δ7 prrn strain MC227 (ΔrrnE ΔrrnB ΔrrnA ΔrrnH ΔrrnG::lacZ ΔrrnC::cat ΔrrnD::cat ΔrecA/pTRNA66, pts1192U; 18) was constructed by replacing the rrnC-containing plasmid pHKrrnC in strain AVS69009 (ΔrrnE ΔrrnB ΔrrnA ΔrrnH ΔrrnG::lacZ ΔrrnC::cat ΔrrnD::cat ΔrecA/pTRNA66, pHKrrnC; 19) with pts1192U, selecting for spectinomycin resistance and screening for neomycin-sensitive transformants. Strain TA542 (ΔrrnE ΔrrnB ΔrrnA ΔrrnH ΔrrnG::cat ΔrrnC::cat ΔrrnD::cat ΔrecA56/pTRNA66 pHKrrnC) was used as a host for pKK3535 and its mutant derivatives. The neomycin-resistant rrnB-containing plasmid, pHKrrnC, in strain TA542 was replaced with ampicillin-resistant, pKK3535-derived plasmids by transforming TA542 with the ampicillin-resistant plasmids and growing the transformants in the absence of selection for pHKrrnC. Loss of pHKrrnC was monitored by the appearance of neomycin-sensitive clones after three to four cycles of overnight growth and dilution into fresh medium in the absence of neomycin selection. Plasmid prrnS12 was constructed by inserting the rrnB operon from plasmid pKK3535 and the wild-type rpsL gene from plasmid pNO1523 (20) into plasmid pLG339 (21). MC230 is ΔrrnE ΔrrnB ΔrrnH ΔrrnG::cat ΔrrnA ΔrrnD::cat ΔrrnC::cat recA56/pts1192U p70. MC250 is a derivative of MC230 containing the rpsL121 streptomycin resistance mutation and carrying plasmid prrnS12 as the sole source of rRNA genes.
The pSG and pBRlac series of lacZ mutant plasmids contain nonsense or frameshift mutations in the 5′-end of the lacZ coding region (22,23).
Mutagenesis
Site-directed mutagenesis was performed as described by Kunkel et al. (24), using an M13 clone carrying the EcoRI–BamHI fragment of plasmid pLK35, encoding the 3′ half of 23S rRNA as a template. Mutant M13 clones were digested with SphI and BamHI and ligated to SphI/BamHI/I-CeuI-treated pLK35. DNA sequencing of the intact plasmids confirmed the presence of the desired mutations. The D2449C mutation was transferred to plasmid pKK3535 by fragment exchanges using BglII and CelII. The same mutation was constructed in plasmid pMO10 by ligating the BamHI fragment from pKK2449C with BamHI-treated pLG339 (21).
Isolation and analysis of rRNA
Total RNA was extracted from detergent-treated cells as described (23). Ribosomes were extracted from logarithmically growing cells following a 150 min temperature shift to induce transcription of mutant rRNA. 30S and 50S subunits were separated from 70S ribosomes and polysomes on sucrose gradients as described (25). The amounts of plasmid-encoded rRNA in each gradient fraction were determined by primer extension as described by Sigmund et al. (26) using an oligonucleotide complementary to bases 2470–2450. The bands corresponding to plasmid- and chromosomally-encoded rRNA extension products were quantitated using a Fuji phosphorimager. Growth rates and β-galactosidase assays were carried out as described previously (23,27).
RESULTS
Construction and expression of mutations at position 2449 in 23S rRNA
All three base substitution mutations at position D2449 were constructed by site-directed mutagenesis. Despite repeated attempts, only the D2449C mutation could be expressed from the strong constitutive P1P2 promoters in plasmid pKK3535. Accordingly, mutant rRNAs were expressed from the inducible λ PL promoter in plasmid pLK35 in strains also expressing the λ temperature-sensitive repressor. In this expression system, ∼50–60% of the total rRNA is plasmid-encoded upon induction of transcription from PL (Table 1; 15,23). After induction of mutant rRNA synthesis, cells expressing the D2449G rRNA displayed a slightly prolonged doubling time while expression of the D2449A mutant rRNA increased the doubling time substantially (Table 1). In contrast, expression of the D2449C mutant had no effect on growth rate. Taken together, the inability to express the D2449G and D2449A mutant rRNAs from the P1P2 promoters, and the slower growth rates after induction from λ PL, suggested that the purine base substitutions at D2449 might be deleterious to ribosome function.
Table 1. Effects of 2449 mutations on growth rate, the accuracy of decoding and distribution of plasmid encoded rRNA in 50S subunits, 70S ribosomes and polysomes.
| rRNA plasmid | Doubling time (min) | Readthrough and frameshifting levels in lacZ mutants | % distribution of mutant rRNA in gradient fractions | ||||||
| |
|
pSG12-6 (UAG) |
pSG34-11 (UGA) |
pSG12DP (–1 FS) |
pSGlac7 (+1 FS) |
PSG163 (UAG) |
50S |
70S |
Polysomes |
| pLK35 wt |
43 ± 4 |
13 ± 1 |
27 ± 3 |
79 ± 5 |
27 ± 1 |
21 ± 1 |
ND |
ND |
ND |
| pLK2449A |
59 ± 3 |
18 ± 1 |
49 ± 2 |
152 ± 7 |
34 ± 1 |
46 ± 3 |
66 ± 5 |
57 ± 6 |
50 ± 3 |
| pLK2449C |
41 ± 3 |
14 ± 1 |
29 ± 2 |
86 ± 4 |
28 ± 2 |
21 ± 1 |
73 ± 1 |
64 ± 7 |
66 ± 8 |
| pLK2449G | 48 ± 3 | 17 ± 1 | 34 ± 2 | 92 ± 10 | 25 ± 1 | 28 ± 1 | 70 ± 3 | 62 ± 6 | 48 ± 1 |
All plasmids for growth rate determinations and ribosome preparations were maintained in strain MC242 co-transformed with pLG857, encoding the temperature-sensitive λcI857 repressor. Growth rates were measured following induction of plasmid-encoded rRNA transcription at 42°C. Ribosomes were harvested from cells grown at 42°C for 150 min. The relative proportions of plasmid and chromosomally encoded rRNAs were determined by primer extension assays on RNA from three independent ribosome preparations. ND, not determined. Values for stop codon readthrough and frameshifting are expressed in Miller units of β-galactosidase (29). β-Galactosidase activities were measured after induction of plasmid-encoded rRNA transcription at 42°C for 150 min. Each value represents the mean ± SE of three to five independent measurements.
The recent development of an E.coli strain that lacks all seven chromosomal rrn operons and expresses plasmid encoded rRNA exclusively (Δ7 prrn; 18) has facilitated the analysis of rRNA mutations. Construction of strains expressing only mutant rRNA involves transformation of Δ7 prrn with the relevant mutant rrn plasmid and subsequent displacement of the resident wild-type rrn plasmid. Strain TA548 was transformed with pLK35 and each of the D2449 mutant derivatives. Growth of these transformants on medium containing ampicillin but lacking neomycin led to loss of the resident rrn plasmid, pHKrrnC, in strains expressing wild-type or 2449C rRNAs. However, after three cycles of overnight growth and dilution into fresh medium, no neomycin-sensitive transformants were obtained with strains expressing 2449G or 2449A rRNAs, suggesting that these mutant rRNAs were unable to supply the total protein synthetic needs of the cell.
The plasmid displacement system described above relies on passive loss of the resident rrn plasmid in the absence of antibiotic selection. To facilitate displacement of the resident rrn plasmid by mutant derivatives and to allow recovery of deleterious rRNA mutants, we have developed a system that actively selects for displacement of the resident rrn plasmid. This system relies on the observation that resistance to streptomycin caused by mutations in rpsL is recessive (28). Derivatives of the Δ7 prrn strain, such as MC250, carrying the rpsL121 mutation on the chromosome are sensitive to streptomycin because they also contain plasmid prrnS12 carrying an intact rrnB operon and a wild-type rpsL gene. Resistance to streptomycin can be recovered when prrnS12 is displaced from this strain by another rrn plasmid that does not contain the rpsL gene. Strain MC250 was, therefore, transformed with pLK35 and each of the mutant D2449 derivatives and ampicillin-resistant transformants were streaked on plates containing ampicillin and streptomycin. MC250 cells transformed with either the wild-type or D2449C mutant plasmid were able to grow on streptomycin-containing plates and these transformants were subsequently shown to have lost the prrnS12-associated neomycin resistance. In contrast, the pLK2449A and pLK2449G transformants remained streptomycin sensitive (Fig. 2). This indicated that both purine substitutions at position 2449 had deleterious effects on ribosome function whereas the pyrimidine substitution was viable. While we have not been able to analyze unmodified U2449 rRNAs, we believe that D2449C ribosomes are functionally equivalent to such unmodified ribosomes and thus conclude that the D modification itself is not essential for ribosome function.
Figure 2.
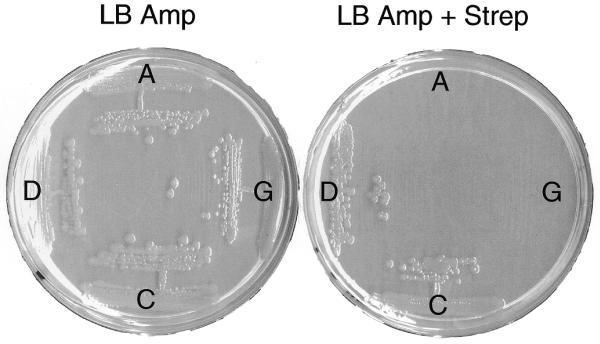
Displacement of the resident rrn plasmid, prrnS12, from MC250 by pLK35 (D, wild type at position 2449) and each of the D2449 mutant deriviatives (A, C and G). The MC250 transformants were streaked on LB ampicillin (left) and on LB ampicillin + streptomycin plates (right). Loss of prrnS12 allows expression of rpsL121-associated streptomycin resistance and growth on streptomycin. Streptomycin-resistant colonies were subsequently shown to have lost the prrnS12-associated neomycin resistance.
Lethal mutations at position 2449
The inability to construct strains expressing the D2449A or D2449G mutant rRNAs exclusively necessitated their further analysis in an inducible expression system in a strain carrying intact chromosomal rrn operons. Our previous work had shown that several mutations in the peptidyltransferase center, including mutations at G2447, Ψ2504 (M.O’Connor and A.E.Dahlberg, unpublished data) and G2458 (29) affected the accuracy of translation. The effects of the D2449 mutations on stop codon readthrough and frameshifting were examined using a series of lacZ reporter gene constructs. Cells expressing the D2449A mutation showed a 1.3–2.2-fold increase in the levels of stop codon readthrough and frameshifting (Table 1). Some very modest, but reproducible increases in readthrough and frameshifting were also observed with the less deleterious D2449G mutation, whereas the D2449C mutant was indistinguishable from wild type. Apart from some under-representation of D2449G and D2449A in the polysome fractions, ribosomes isolated from induced cells showed normal levels of subunits, 70S ribosomes and polysomes on sucrose gradients (data not shown) and the mutant rRNA was well-represented in all ribosome fractions (Table 1). These results showed that while both of the deleterious purine substitution mutations decreased the accuracy of decoding, none of the D2449 mutations had dramatic effects on 50S subunit assembly, 70S ribosome formation or entry into the polysome pool.
Characterization of the D2449C mutation
The effects of the D2449C substitution on cell growth and translational fidelity were examined in Δ7 prrn strains that expressed the mutant rRNA exclusively from the native, growth-regulated P1P2 promoters in plasmid pKK3535 rather than from the λ PL promoter in the pLK35 plasmids used above. Successful replacement of the resident wild-type pts1192U rrn plasmid by pKK2449C in the Δ7 prrn strain MC227 was assessed both by loss of pts1192U-associated spectinomycin resistance and by primer extension of total RNA (26; data not shown). No difference in growth rate was observed between strains expressing only wild-type or the D2229C rRNAs under any of the conditions used (Table 2). Moreover, both wild-type and mutant strains grew at equivalent rates on solid, minimal and rich media at 25, 30, 37 and 42°C (data not shown). The influence of the D2449C mutation on recovery from heat stress was addressed by incubating mutant or wild-type cells for varying lengths of time at 52°C and measuring the number of colony forming units in the cultures before and after heat treatment. After 25 min incubation at high temperature, both wild-type and mutant strains displayed the same 100-fold loss of viability. A further 25 min incubation at 52°C led to an additional 2-fold decrease in the viability of both cultures. The effect of the D2449C mutation on low temperature survival was assessed by incubating diluted, mutant and wild-type cultures at 15°C and measuring the increases in OD600 and the numbers of viable cells in the cultures during a 60-h period after transfer to 15°C. No difference in low temperature survival and recovery was observed between the wild-type and D2449C mutant strains (data not shown). Analysis of ribosomes on sucrose gradients revealed no differences between mutant and wild-type in ribosome assembly or subunit–subunit interactions (data not shown). When assayed under conditions where only mutant rRNA was expressed, the D2449 mutation had no effect on stop codon readthrough or frameshifting (Table 2). Many mutations in the central loop of domain V of 23S rRNA have been associated with altered sensitivity to antibiotics that inhibit peptide bond formation or growth of peptide chains (12). We were unable to examine any effects of the D2449C mutation on chloramphenicol resistance since the Δ7 prrn strain contains multiple chloramphenicol acetyltransferase (CAT) cassettes inserted in chromosomal rrn operons. However, the D2449C alteration did not affect sensitivity of the strain to the antibiotics tylosin, spiramycin, oleandomycin, erythromycin, clindamycin, tetracycline, paromomycin, kasugamycin or fusidic acid (data not shown). Thus, in summary, it appeared that substitution of the D modification by a C at position 2449 had no detectable effect on cell growth, heat or cold shock survival, translational fidelity or antibiotic sensitivity.
Table 2. Growth rates and decoding fidelity of strains expressing wild-type or D2449C rRNA exclusively.
| rRNA mutant | Doubling times (min) | Readthrough and frameshifting levels in lacZ mutants | |||||
| |
30°C |
37°C |
42°C |
p240 |
p415 |
p12-6 |
plac7 |
| Wild-type |
74 ± 4 |
60 ± 2 |
80 ± 6 |
11 ± 1 |
392 ± 27 |
38 ± 3 |
41 ± 7 |
| D2449C | 82 ± 4 | 64 ± 4 | 76 ± 3 | 9 ± 1 | 384 ± 30 | 38 ± 5 | 36 ± 7 |
Growth rates were determined for pKK3535 and pKK2449C-containing derivatives of the Δ7prrn strain, TA542, where all rRNA is plasmid encoded. β-Galactosidase measurements were performed on TA542 derivatives expressing wild-type or D2449C rRNA from plasmids pMO10 and pMO2449C, respectively; lacZ constructs were carried on pBR322-derived plasmids. Values for stop codon readthrough and frameshifting are expressed in Miller units of β-galactosidase (29). All values represent the mean ± SE of three to five independent determinations.
DISCUSSION
Substantial biochemical data as well as the recently solved crystal structure of the 50S ribosomal subunit indicate that the central loop of domain V of 23S rRNA forms part of the peptidyltransferase center of the ribosome (30,31). In keeping with its central role in protein synthesis, many of the rRNA residues in this region of large subunit rRNAs are invariant. The clustering of modified bases in the central loop has given rise to several models in which modified bases play essential roles in translation (32). The observation that D in tRNAs promotes conformational flexibility has led to the suggestion that its presence in a functional center of 23S rRNA may be necessary to accommodate the dynamic, conformational changes that must occur in this region of the ribosome (14). The enzyme(s) responsible for D modification at position 2449 in E.coli has not yet been identified, preventing a direct analysis of unmodified, U2449 ribosomes. However, our observation that the D2449C mutation is without apparent phenotype indicates that D modification is not essential for ribosome function, and thus the role of this modification remains unclear. Consistent with our data is the finding that although the rRNA modifications of Aeromonas hydrophilia are very similar to those found in E.coli, D is conspicuously absent (33). Analysis of the published rRNA sequences indicates that a U is always found at position 2449 in large subunit rRNAs. However, at least in the context of the E.coli ribosome, a C appears to function as well as D and thus neither the modification nor the uridine residue itself appears to be essential for ribosome function. The possibility remains that the D modification aids in ribosome assembly or ribosome function under particular conditions not revealed by the laboratory growth conditions utilized here.
A majority of the mutations that have been made in the central loop of domain V of 23S rRNA have deleterious effects on ribosome function (34–36). While effects on peptidyltransferase activity and antibiotic sensitivity have been documented for many of these deleterious mutations, a more surprising finding is that several of the mutations that have been linked with peptide bond formation also affect the fidelity of decoding, an activity associated with the small ribosomal subunit. Such a phenotype is observed with the D2449A and D2449G mutations studied here. Similar effects on decoding have also been observed with mutations at the adjacent positions G2447 and Ψ2504 (M.O’Connor and A.E.Dahlberg, unpublished data) as well as at several positions in helix 89 (29). These findings highlight the interconnectedness of the central ribosomal activities of decoding, translocation and peptide bond formation. However, the effects of the D2449A and D2449G mutations on decoding are very modest in comparison to other (viable) ribosomal mutations (37) and are thus unlikely to account for the lethality of these base substitutions in the Δ7 prrn strain. The distribution of D2449A and D2449G rRNAs in the subunit, 70S ribosome and polysome (Table 1) indicates that the mutant RNAs are fully assembled into 50S particles, competent to interact with 30S subunits and initiate translation. However, their inability to support the total protein synthetic needs of the cell indicates that one or more of the 50S-associated elongation functions is compromised in the mutant ribosomes. The recently published crystal structure of the 50S ribosomal subunit of Haloarcula marismortuii indicated that residues at positions 2444 and 2446 participate in a base triple interaction with U2449 (31). The phenotypic effects of purine substitutions at position 2449 may thus reflect their inability to participate in an equivalent base triple interaction in the E.coli ribosome. In addition, the crystal structure of the catalytic center of the 50S subunit as well as an anomalous pKa associated with A2451 has been interpreted to indicate that A2451 may play an essential role in peptide bond formation (31,38). This raises the possibility that the defects associated with the D2449A and D2449G mutants may be due to alterations in the efficiency of the peptidyltransferase reaction.
Finally, we have developed a method that facilitates replacement of the resident rrn plasmid in Δ7 prrn strains with other mutant rrn plasmids. This method permits recovery of deleterious rRNA mutations and should allow rapid screening and analysis of rRNA mutations.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Drs James McCloskey, Jeff Kowalak and Joseph Dalluge for suggesting this project and sharing their data with us. We thank Steven Gregory for his comments on the manuscript, Anton Vila-Sanjurjo for assistance with the figures and George Q. Pennabble for his bravura badinage. This work was supported by grants #GMS19756 (A.E.D.) and #GMS24751 (C.L.S.) from the National Institutes of Health.
References
- 1.Maden B.E. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 2.Ofengand J. and Rudd,K.E. (2000) Bacterial, Archaeal, and organellar rRNA pseudouridines and methylated nucleosides and their enzymes. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function,Antibiotics, and Cellular Interactions. American Society for Microbiology, Washington, DC, pp. 175–189.
- 3.Brimacombe R., Mitchell,P., Osswald,M., Stade,K. and Bochkariov,D. (1993) Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J., 7, 161–167. [DOI] [PubMed] [Google Scholar]
- 4.Green R. and Noller,H.F. (1996) In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA, 2, 1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 5.Khaitovich P., Tenson,T., Kloss,P. and Mankin,A.S. (1999) Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry, 38, 1780–1788. [DOI] [PubMed] [Google Scholar]
- 6.Green R. and Noller,H.F. (1999) Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry, 38, 1772–1779. [DOI] [PubMed] [Google Scholar]
- 7.Bjork G.R. (1995) Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog. Nucleic Acid Res. Mol. Biol., 50, 263–338. [DOI] [PubMed] [Google Scholar]
- 8.Raychaudhuri S., Niu,L., Conrad,J., Lane,B.G. and Ofengand,J. (1999) Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J. Biol. Chem., 274, 18880–18886. [DOI] [PubMed] [Google Scholar]
- 9.Sirum-Connolly K. and Mason,T.L. (1993) Functional requirement of a site-specific ribose methylation in ribosomalRNA. Science, 262, 1886–1889. [DOI] [PubMed] [Google Scholar]
- 10. Raychaudhuri S., Conrad,J., Hall,B.G. and Ofengand,J. (1998) A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA, 11, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldas T., Binet,E., Bouloc,P. and Richarme,G. (2000) Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun., 271, 714–718. [DOI] [PubMed] [Google Scholar]
- 12.Porse B.T. and Garrett,R.A. (1999) Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23 S rRNA and the synergism of their inhibitory mechanisms. J. Mol. Biol., 286, 375–387. [DOI] [PubMed] [Google Scholar]
- 13.Kowalak J.A., Bruenger,E. and McCloskey,J.A. (1995) Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J. Biol. Chem., 270, 17758–17764. [DOI] [PubMed] [Google Scholar]
- 14.Dalluge J.J., Hashizume,T., Sopchik,A.E., McCloskey,J.A. and Davis,D.R. (1996) Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res., 24, 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory S.T., Brunelli,C.A., Lodmell,J.S., O’Connor,M. and Dahlberg,A.E. (1998) Genetic selection of rRNA mutations. Methods Mol. Biol., 77, 271–281. [DOI] [PubMed] [Google Scholar]
- 16.Makosky P.C. and Dahlberg,A.E. (1987) Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie, 69, 885–889. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton C.M., Aldea,M., Washburn,B.K., Babitzke,P. and Kushner,S.R. (1989) New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol ., 171, 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asai,T., Zaporojets,D., Squires,C. and Squires,C.L. (1999) An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA, 96, 1971–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vila-Sanjurjo A., Squires,C.L. and Dahlberg,A.E. (1999) Isolation of kasugamycin resistant mutants in the 16 S ribosomal RNA of Escherichia coli. J. Mol. Biol., 293, 1–8. [DOI] [PubMed] [Google Scholar]
- 20.Dean D. (1981) A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gene, 15, 99–102. [DOI] [PubMed] [Google Scholar]
- 21.Stoker N.G., Fairweather,N.F. and Spratt,B.G. (1982) Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene, 18, 335–341. [DOI] [PubMed] [Google Scholar]
- 22.O’Connor M., Wills,N.M., Bossi,L., Gesteland,R.F. and Atkins,J.F. (1993) Functional tRNAs with altered 3′ ends. EMBO J., 12, 2559–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor M. and Dahlberg,A.E. (1993) Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc. Natl Acad. Sci. USA, 90, 9214–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel T.A., Bebenek,K. and McClary,J. (1991) Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol., 204, 125–139. [DOI] [PubMed] [Google Scholar]
- 25.Tapprich,W.E., Goss,D.J. and Dahlberg,A.E. (1989) Mutation at position 791 in Escherichia coli 16S ribosomal RNA affects processes involved in the initiation of protein synthesis. Proc. Natl Acad. Sci. USA, 86, 4927–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigmund C.D., Ettayebi,M., Borden,A. and Morgan,E.A. (1988) Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol., 164, 673–690. [DOI] [PubMed] [Google Scholar]
- 27.Miller J.H. (1991) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Breckenridge L. and Gorini,L. (1969) The dominance of streptomycin sensitivity re-examined. Proc. Natl Acad. Sci. USA, 62, 979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connor M. and Dahlberg,A.E. (1995) The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol., 254, 838–847. [DOI] [PubMed] [Google Scholar]
- 30.Green R. and Noller,H.F. (1997) Ribosomes and translation. Annu. Rev. Biochem., 66, 679–716. [DOI] [PubMed] [Google Scholar]
- 31.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 32.Lane B.G., Ofengand,J. and Gray,M.W. (1992) Pseudouridine in the large-subunit (23 S-like) ribosomal RNA. The site of peptidyl transfer in the ribosome? FEBS Lett., 302, 1–4. [DOI] [PubMed] [Google Scholar]
- 33.Dalluge J.J. (1996) PhD dissertation. University of Utah, Salt Lake City, UT.
- 34.Vester B. and Garrett,R.A. (1988) The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer centre of Escherichia coli 23S ribosomal RNA. EMBO J., 7, 3577–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porse B.T. and Garrett,R.A. (1995) Mapping important nucleotides in the peptidyl transferase centre of 23 S rRNA using a random mutagenesis approach. J. Mol. Biol., 249, 1–10. [DOI] [PubMed] [Google Scholar]
- 36.Triman K.L. (1999) Mutational analysis of 23S ribosomal RNA structure and function in Escherichia coli. Adv. Genet., 41, 157–195. [DOI] [PubMed] [Google Scholar]
- 37.Andersson D.I., Bohman,K., Isaksson,L.A. and Kurland,C.G. (1982) Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet., 187, 467–472. [DOI] [PubMed] [Google Scholar]
- 38.Muth G.W., Ortoleva-Donnelly,L. and Strobel,S.A. (2000) A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science, 289, 947–950. [DOI] [PubMed] [Google Scholar]