Abstract
The mammalian retina contains more than 50 distinct neuronal types, which are broadly classified into several major classes: photoreceptor, bipolar, horizontal, amacrine, and ganglion cells. Although some of the developmental mechanisms involved in the differentiation of retinal ganglion cells (RGCs) are beginning to be understood, there is little information regarding the genetic and molecular determinants of the distinct morphologies of the 15 – 20 mammalian RGC cell types. Previous work has shown that the transcription factor Brn3b/Pou4f2 plays a major role in the development and survival of many RGCs. The roles of the closely related family members, Brn3a/Pou4f1 and Brn3c/Pou4f3 in RGC development are less clear. Using a genetically-directed method for sparse cell labeling and sparse conditional gene ablation in mice, we describe here the sets of RGC types in which each of the three Brn3/Pou4f transcription factors are expressed and the consequences of ablating these factors on the development of RGC morphologies.
Keywords: Mouse, Retina Development, Retinal Ganglion Cell, Brn3 Transcription Factors, Dendritic Arbor
1. Introduction
Visual information undergoes a first level of processing in the retina, and the features extracted from the stimulus are conveyed to the brain through parallel channels subserved by 15–20 distinct retinal ganglion cell (RGC) types (Masland, 2001; Ramon y Cajal, 1972; Wassle, 2004 and references within). RGC types can be distinguished by their physiological properties (Barlow et al., 1964; Levick, 1967; Cleland and Levick, 1974a, 1974b; Roska and Werblin, 2001; Troy and Shou, 2002), target nuclei in the brain (Pu & Amthor, 1990a, 1990b; Rodieck and Watanabe, 1993; Simpson, 1984), patterns of dendritic arborization in the retina (Ramon y Cajal, 1972; Boycott and Wassle, 1974; Levick, 1975; Amthor et al., 1984; Rockhill et al., 2002) and, in a few cases, molecular markers (Siegert et al., 2009; Kim et al., 2010).
RGC typography depends on the classification criteria applied and species studied (Masland and Martin, 2007). Combining information about central brain projections, physiological light responses, and dendritic morphology is highly advanced in cat and primate retinas and has created a detailed picture of RGC types (Rodieck and Watanabe, 1993; Dacey et al., 2003). In the mouse, classification schemes based solely on morphological (Sun et al., 2002; Badea and Nathans, 2004; Kong et al., 2005; Coombs et al., 2006; Volgyi et al., 2009), or physiological (Carcieri et al., 2003; Pang et al., 2003; Zeck and Masland, 2007) criteria have recently been complemented by the discovery of molecular markers for individual RGC cell types (Hattar et al., 2002; Huberman et al., 2008; Kim et al., 2008; Yonehara et al., 2008; Huberman et al., 2009; Siegert et al., 2009; Kim et al., 2010). RGC type-specific expression of fluorescent, immunohistochemical, or enzymatic reporters has further advanced the integration of dendritic morphology, physiology, and projection patterns (Berson et al., 2002; Hattar et al., 2002; Huberman et al., 2008; Kim et al., 2008; Yonehara et al., 2008; Do et al., 2009; Huberman et al., 2009; Yonehara et al., 2009). Finally, genetic manipulations targeted to specific RGC types permit type-specific cell deletion or modification and an analysis of the resulting effect on visual reflexes and behaviors (Guler et al., 2008; Badea et al., 2009a). These results underscore the utility of understanding the molecular genetics and development of RGC types in the mouse retina.
Studying the morphology of RGC dendritic arbors is of interest not only as an aid in cell type classification, but because dendritic morphology relates directly to RGC response properties. In particular, the area of the arbor is closely correlated with the size of the RGC receptive field and the level of dendritic lamination in the inner plexiform layer determines the type of amacrine and bipolar cells from which the RGC receives its input (Boycott and Wassle, 1974; Famiglietti and Kolb, 1976; Amthor et al., 1984; Kim et al., 2008). At present, the molecular mechanisms that specify the area, shape, and stratification level(s) of RGC dendritic arbors are still poorly understood (Yamagata et al., 2002; Fuerst et al., 2008; Yamagata and Sanes, 2008; Badea et al., 2009a; Fuerst et al., 2009).
One family of transcription factors that is expressed in the retina exclusively in RGCs is the Pou4f/Brn3 family of transcription factors (Xiang et al., 1995; Erkman et al., 1996; Gan et al., 1996; Xiang et al., 1996; Xiang et al., 1997; Badea et al., 2009a). The three members of the Brn3 family - Brn3a, Brn3b and Brn3c - are present in partially overlapping subpopulations of RGCs. They are also expressed in primary sensory neurons in the dorsal root and trigeminal ganglia, and in auditory and vestibular hair cells (Brn3c) or their synaptic partners in the vestibular system (Brn3a and Brn3b) (Xiang et al., 1995; Erkman et al., 1996; Gan et al., 1996; Xiang et al., 1996; Xiang et al., 1997). In the retina, targeted deletion of Brn3b results in the loss of 70 – 80% of RGCs, with many of the surviving RGCs exhibiting severe axonal defects (Erkman et al., 1996; Gan et al., 1996; Erkman et al., 2000; Badea et al., 2009a). Deletion of Brn3a specifically in the retina results in a characteristic change in the distribution of RGC dendritic morphologies (Badea et al., 2009a). Deletion of Brn3c does not have a major effect on RGC survival, but the Brn3b and Brn3c double mutant shows an increase in the severity of RGC loss compared to the Brn3b mutant (Wang et al., 2002). Thus, each Brn3 gene plays a distinctive role in RGC development.
Several questions remain open regarding the role of the Brn3 transcription factors in RGC development and type specification. Which RGC types express each of the Brn3 transcription factors? Does Brn3c have a role in RGC development independent of its effects on cell survival in combination with Brn3b? What are the molecular mechanisms by which the Brn3 proteins regulate RGC cell type specification?
To begin to address these questions, we have developed a strategy in which Cre-mediated recombination of a conditional Brn3 allele simultaneously excises the Brn3 coding region and activates a previously silent histochemical reporter (alkaline phosphatase; AP) by bringing the AP coding region under the control of the Brn3 promoter (Badea et al., 2009a and references within). Using this technique under conditions that produce a sparse collection of recombined cells, one can observe and quantify the morphologies of individual RGCs expressing the targeted Brn3 gene. In addition, by comparing mouse lines in which the conditional Brn3 allele is paired either with a wild type (WT) allele or with a conventional null allele, one can compare the morphologies of individual RGcs that are either homozygous mutant or heterozygous, respectively, for the Brn3 gene being manipulated.
In the present study we define the morphological subtypes of RGCs that express Brn3a, Brn3b, and Brn3c, and for each of the Brn3 genes we quantify a series of parameters that characterize the dendritic morphology in wild type compared to mutant RGCs.
2. Materials and Methods
2.1 Mouse strains and crosses
Mouse lines carrying alleles Brn3a−, Brn3b−, Brn3c−, Brn3aCKOAP, Brn3bCKOAP, and R26rtTACreER were previously described (Gan et al., 1996; Xiang et al., 1996; Xiang et al., 1997; Badea et al., 2009a, 2009b). The Brn3cCKOAP allele (T. Badea, J. Williams, P. Smallwood, and J. Nathans, manuscript in preparation) is a conditional knock-in AP reporter allele analogous to the Brn3aCKOAP and Brn3bCKOAP alleles described in Badea et al. (2009a).
To obtain sparse Cre-mediated recombination in RGCs, the following crosses were set up: (1) R26rtTACreER/+rtTACreER/+;Brn3a+/− male × Brn3aCKOAP/CKOAP female, (2) R26rtTACreER/+;Brn3b+/− male × Brn3bCKOAP/CKOAP female, and (3) R26rtTACreER/+;Brn3c+/− male × Brn3cCKOAP/CKOAP female. Conception date was determined by examining the copulation plug, and pregnant females were moved to cages with food pellets containing Doxycyline (1.75 mg/gram) at gestational day 2–4. At gestational day 11 – 13, 100 – 250 µg of 4-hydroxytamoxifen in sunflower seed oil vehicle was delivered by intraperitoneal injection, and the doxycycline diet was continued until birth (Table 1). A detailed description of this protocol can be found in Badea et al. (2009b).
Table 1.
Doxycycline and 4-HT treatment
| Brn3 locus genotype | Mouse tag# | Doxycycline regime | 4 HT administration |
|---|---|---|---|
| Brn3aCKOAP/+ | 8664 | E4-P0 = 1 | E11 = 150 microgr |
| 8640 | E4-P0 = food | E13 = 100 microgr | |
| 9015, 9018 | E3-P2 = food | E12 = 200 microgr | |
| 9030 | E3-P5 = food | E12 = 250 microgr | |
| Brn3aCKOAP/− | 8641, 8643 | E4-P0 = food | E13 = 100 microgr |
| 9026, 9031 | E3-P5 = food | E12 = 250 microgr | |
| Brn3bCKOAP/+ | 8740 | E4-P0 = food | E12 = 200 |
| 8753 | E4-P0 = food | E11 = 100 | |
| Brn3bCKOAP/− | 8741 | E4-P0 = food | E12 = 200 |
| 8754 | E4-P0 = food | E11 = 100 | |
| Brn3cCKOAP/+ | 8881 | E5-P0 = food | E14 = 400 microgr |
| 8933 | E3-P0 = food | E13 = 250 microgr | |
| 9058, 9171 | E2-P0 = food | E12 = 200 microgr | |
| Brn3cCKOAP/− | 8934, 8938 | E3-P0 = food | E13 = 250 microgr |
For each mouse both retinas were analyzed.
2.2 Histochemistry
At 4–8 weeks after birth, mice of the six desired genotypes (R26rtTACreER/+;Brn3aCKOAP/+, R26rtTACreER/+;Brn3aCKOAP/−, R26rtTACreER/+;Brn3bCKOAP/+, R26rtTACreER/+;Brn3bCKOAP/−, R26rtTACreER/+;Brn3cCKOAP/+, and R26rtTACreER/+;Brn3cCKOAP/−) were anesthetized with ketamine and xylazine, and fixed with 4% paraformaldehyde in PBS by intracardiac perfusion. Retinas were dissected, flat mounted, and histochemically stained for AP as previously described (Badea et al., 2003). Retina flat mounts were dehydrated in an ethanol series and cleared and mounted in benzyl benzoate: benzyl alcohol (2:1).
2.3 Image collection and analysis
Montages of the whole retina were generated at 5X magnification using a Zeiss Z1 Imager microscope equipped with a motorized xyz stage. Individual RGCs were imaged under DIC/Nomarsky transmitted light, and z-stacks were collected with a 20X objective (3.125 pixels/µm) at 1 µm vertical (z dimension) intervals. For each RGC dendritic arbor, the area and the inner (vitreal) and outer (scleral) limits of the stratification depth within the inner plexiform layer (IPL) were determined (Badea et al., 2003; Badea et al., 2009a, 2009b; Fig. 1H,I). Scatter plots were prepared to assess the relationships among the measured parameters using MatLab and Excel. Individual examples of distinct morphological types were reconstructed (i.e. traced) using Neuromantic freeware (www.rdg.ac.uk/neuromantic/).
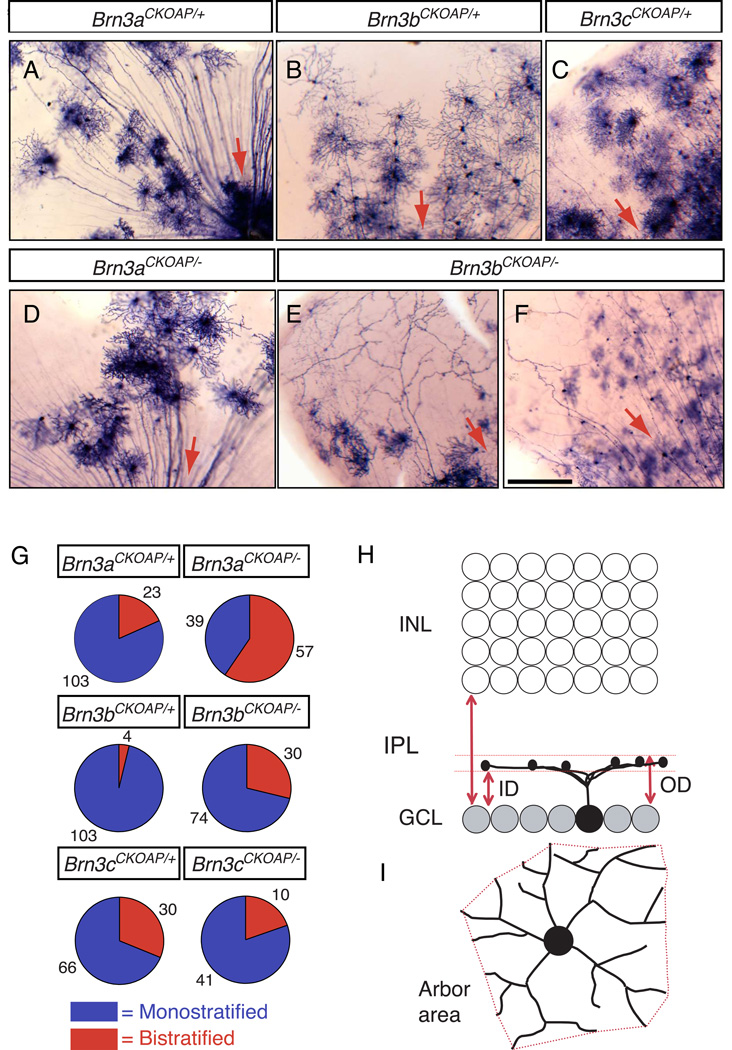
Fig. 1. Mouse RGCs visualized by sparse recombination of conditional AP reporter knock-in alleles at the Brn3a, Brn3b, and Brn3c loci.
(A–F) Flat mount preparations of retinas from (A) R26rtTACreER/+;Brn3aCKOAP/+, (B) R26rtTACreER/+;Brn3bCKOAP/+, (C) R26rtT-CreER/+;Brn3cCKOAP/+, (D) R26rtTACreER/+;Brn3aCKOAP/−, and (E,F) R26rtTACreER/+;Brn3bCKOAP/− mice, histochemically stained for AP activity. Red arrows indicate the direction of the optic disc. (G) Pie charts showing the ratios of monostratified (blue): bistratified (red) RGCs from retinas of the indicated genotypes. The number of cells of each type is indicated. (H,I), Morphological parameters for the analysis of RGC dendritic arbors. In (H), the inner distance (ID) and outer distance (OD), normalized to the IPL thickness, together define the level of arbor stratification and thickness. ID and OD equal 0 at the level of the ganglion cell layer, and they equal 1 at the level of the INL. In (I), the arbor area is calculated as the area of the polygon (in red) with the shortest perimeter that connects the tips of the RGC dendrites when projected in the plane of the retina. Scale bar in F, 100 µm.
3. Results
3.1 Genetically-directed sparse labeling of RGCs
We used a Cre recombinase-estrogen receptor ligand binding domain fusion protein (CreER) to effect recombination of a pair of loxP target sites in each Brn3 conditional allele to visualize the dendritic arbors of RGCs expressing each Brn3 transcription factor (Badea et al., 2003; Badea and Nathans, 2004). For improved control over recombination efficiency at early developmental time points, we used a recently generated R26rtTACreER knock-in allele, in which Cre recombinase activity is under two levels of pharmacologic control: expression of CreER is regulated at the transcriptional level by the doxycycline-dependent activation of the reverse tetracycline trans-activator (rtTA), and nuclear import of CreER is controlled, in a dose-dependent manner, by 4–hydroxytamoxifen (4-HT) (Badea et al., 2009b).
To specifically label RGCs expressing Brn3a, Brn3b, or Brn3c, we used the previously described Brn3aCKOAP and Brn3bCKOAP conditional knock-in AP reporter alleles, and an analogous, newly generated Brn3cCKOAP allele (Badea, Williams, Smallwood, and Nathans, manuscript in preparation). Using an optimized doxycycline and 4-HT delivery protocol (Badea et al., 2009b), we induced sparse recombination at each of the three Brn3CKOAP alleles, resulting in excision of the coresponding Brn3 coding region and its replacement with AP (Brn3AP). As noted in the Introduction, the neurons that have undergone Cre-mediated recombination become heterozygous for the targeted transcription factor if the Brn3 allele on the homologous chromosome is WT (starting genotype Brn3CKOAP/+; final genotype Brn3AP/+), or they become null for the targeted transcription factor if the Brn3 allele on the homologous chromosome is a conventional null mutation (starting genotype Brn3CKOAP/−; final genotype Brn3AP/−). In either case, the surrounding cells remain phenotypically WT. Retinas from mice with each of the six relevant genetic backgrounds (R26rtTACreER/+;Brn3aCKOAP/+, R26rtTACreER/+;Brn3aCKOAP/−, R26rtTACreER/+;Brn3bCKOAP/+, R26rtTACreER/+;Brn3bCKOAP/−, R26rtTACreER/+;Brn3cCKOAP/+, R26rtTACreER/+;Brn3cCKOAP/−) were histochemically stained in flat mount preparations for AP activity (Fig. 1A–F). Since all of the data presented in this study used the R26rtTACreER allele as a source of Cre recombinase, we will refer hereafter only to the Brn3 alleles in describing the genotype of each line, e.g. Brn3aCKOAP/+ instead of R26rtTACreER/+;Brn3aCKOAP/+. Panels A–C in Fig. 1 show examples of retina flat mounts in which the AP+ RGCs are heterozygous for the indicated Brn3 transcription factor, whereas panels D–F in Fig. 1 show AP+ RGCs that lack the indicated transcription factor. Many aberrant AP+ neurites are seen following loss of Brn3b (Fig. 1E,F), consistent with our earlier experiments in which recombination was driven by a Pax6-αCre transgene (Badea et al., 2009a). In all of the figures the labels refer to the genotype of the mouse (e.g. Brn3bCKOAP/−) rather than the genotype of the individual AP-expressing RGCs (e.g. Brn3bAP/−).
3.2 General characteristics of RGCs: morphologic and parametric analyses
A total of 580 RGCs were analyzed. They were obtained from 38 retinas (Table 1) and were distributed approximately equally among the six genotypes. Individual RGC axons and dendrites were imaged using DIC/Nomarski optics under transmitted light, and successive optical sections where collected at 1 µm vertical (z dimension) intervals. To classify RGCs based on dendrite morphology, cells were first separated into monostratified or bistratified types based on the absence or presence of two well-separated, flat dendritic arbors. Fig. 1G and Table 2 show that the proportion of monostratified and bistratified types varies among WT RGCs that express the different Brn3 genes, with Brn3b-expressing RGCs showing the lowest proportion of bistratified cells and Brn3c-expressing RGCs showing the highest proportion. The effects of deleting each Brn3 gene on the ratio of monostratified to bistratified RGCs are described in section 3.7. For each dendritic arbor, the inner (vitreal) and outer (scleral) limits – abbreviated as ID (inner distance) and OD (outer distance), respectively - were measured (Fig. 1H). ID and OD were normalized to the total thickness of the IPL at the same (x,y) position. Finally, the arbor area was measured by constructing a bounding polygon with vertices defined by the most distal points along the dendritic tree (Fig. 1I).
Table 2.
AP-expressing monostratified and bistratified RGCs
| monostratified | bistratified | total | monostratified % | Bistratified % | |
|---|---|---|---|---|---|
| Brn3aAP/+ | 103 | 23 | 126 | 82 | 18 |
| Brn3aAP/− | 39 | 57 | 96 | 41 | 59 |
| Brn3bAP/+ | 103 | 4 | 107 | 96 | 4 |
| Brn3bAP/− | 74 | 30 | 104 | 71 | 29 |
| Brn3cAP/+ | 66 | 30 | 96 | 69 | 31 |
| Brn3cAP/− | 41 | 10 | 51 | 80 | 20 |
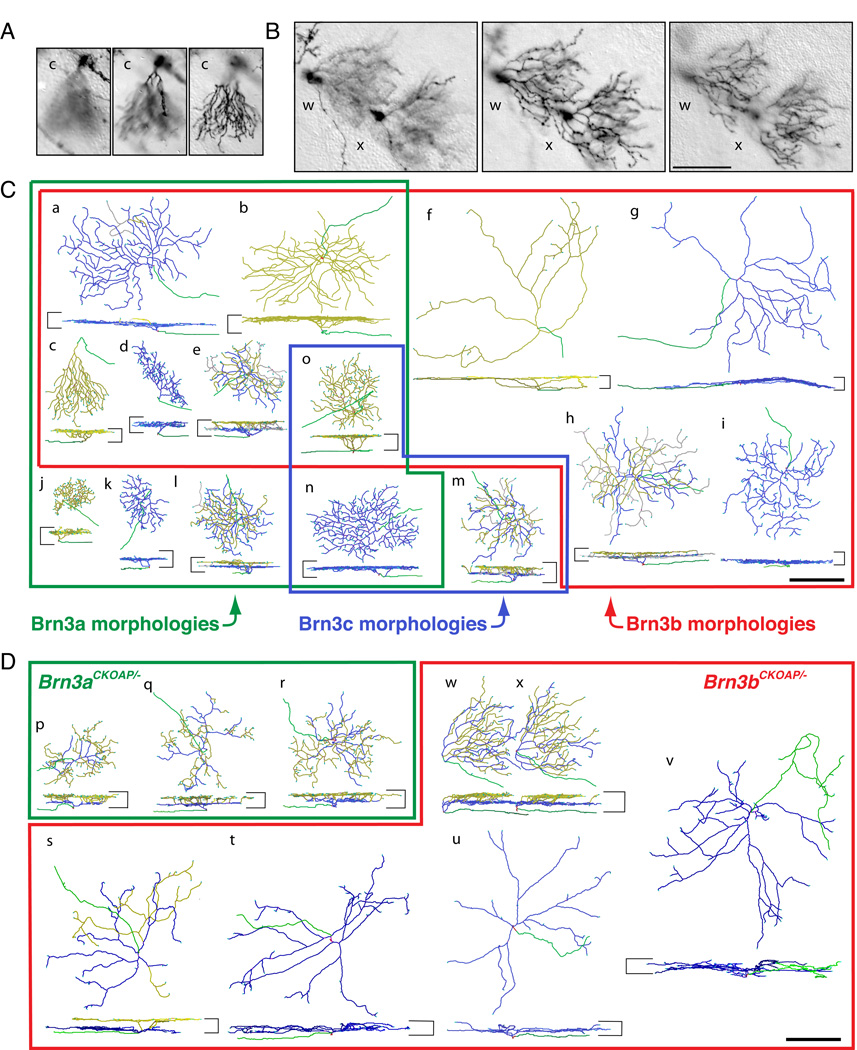
Fig. 2 shows examples of dendritic arbors imaged at various z-axis focal planes (panels A and B), together with a series of reconstructed Brn3AP/+ RGCs (panel C) and Brn3AP/− RGCs (panel D). Each reconstructed RGC in Fig. 2C and D is shown en face with respect to the plane of the retina (upper image) and from the side (lower image), and is assigned a lower case letter (a through x). The images in panels A and B correspond, respectively, to reconstructed cell c in panel C and cells w and x in panel D. In panel C, the colored borders and their associated labels indicate which RGC morphologies are characteristic of Brn3aAP/+, Brn3bAP/+, or Brn3cAP/+ RGCs; for example, the morphology exemplified by the RGC designated n in Fig. 2C is characteristic of Brn3aAP/+ and Brn3cAP/+ RGCs. The genotypes of each RGC in Fig. 2C are indicated in the scatter plots in Figs. 3 and 4, in which area and dendritic stratification parameters are cross-correlated for each RGC in the monostratified and bistratified data sets, respectively. Fig. 2D shows a series of reconstructed Brn3aAP/− and Brn3bAP/− RGCs; their corresponding data points are also indicated in the scatter plots in Figs. 3 and 4. In the text that follows, we will refer to each of these cells by its letter designation; as each cell is discussed, the reader is encouraged to consult both the reconstructed cell image (Fig. 2) and the scatter plots (Figs. 3 and 4).
Fig. 2. Examples of the morphologies of WT and mutant RGCs expressing each of the three Brn3AP alleles.
(A,B) Sequential z-dimension DIC images of one Brn3bAP/+ (A) and two Brn3bAP/− (B) RGCs in flat mounted retinas. The planes of the optical sections are at the nerve fiber layer (left panels), at ~0.3–0.4 IPL depth (central panels) and at the boundary of the IPL and INL (right panels). The images in (A) and (B) correspond, respectively, to reconstructed cell c in panel (C) and reconstructed cells w and x in panel (D). For cell c, the dendrites stratify adjacent to the INL, whereas for cells w and x there are two distinct arbors (coded blue and yellow in (D)), one stratifying at an IPL depth of ~0.35 and the second stratifying adjacent to the INL. (C,D) Reconstructions of individual Brn3AP/+ (C) and Brn3AP/− (D) RGCs obtained from DIC z-stacks similar to the ones sampled in panels (A–B). For each cell, en face (top) and vertical views (bottom) are shown. The boundaries of the IPL are shown as horizontal bars on the sides of each vertical projection. Axons are green, vitreal (“ON”) dendrites are blue, and scleral (“OFF”) dendrites are yellow. “Recurrent” dendrites, defined as originating in the scleral plexus and ending in the vitreal plexus, are gray (present in cells a, e, h, l, and m). (C) Distinct RGC morphological types representative of the Brn3aAP/+, Brn3bAP/+ and Brn3cAP/+ data sets are enclosed in green, red and blue outlines, respectively. Types j, k, and l are unique to Brn3aAP/+ RGCs; types f, g, h and i to Brn3bAP/+ RGCs; and type m to Brn3cAP/+ RGCs. Types a–e are observed among both Brn3aAP/+ and Brn3bAP/+ RGCs; type n in Brn3aAP/+ and Brn3cAP/+ RGCs; and type o in all three Brn3AP/+ RGCs. The specific examples shown are derived from Brn3aCKOAP/+ (a, b, e, j, k, l, n), Brn3bCKOAP/+ (c, d, f, g, h, i, o) and Brn3cCKOAP/+ (m) retinas. (D) Examples of RGC morphologies observed in the Brn3aCKOAP/− and Brn3bCKOAP/− retinas are enclosed in green and red outlines, respectively. Examples of bistratified Brn3aAP/− RGCs (p, q, r), bistratified Brn3bAP/− RGCs (s, w, x) and monostratified Brn3bAP/− RGCs (t, u, v) are shown. Scale bars in B, C and D, 100 µm.
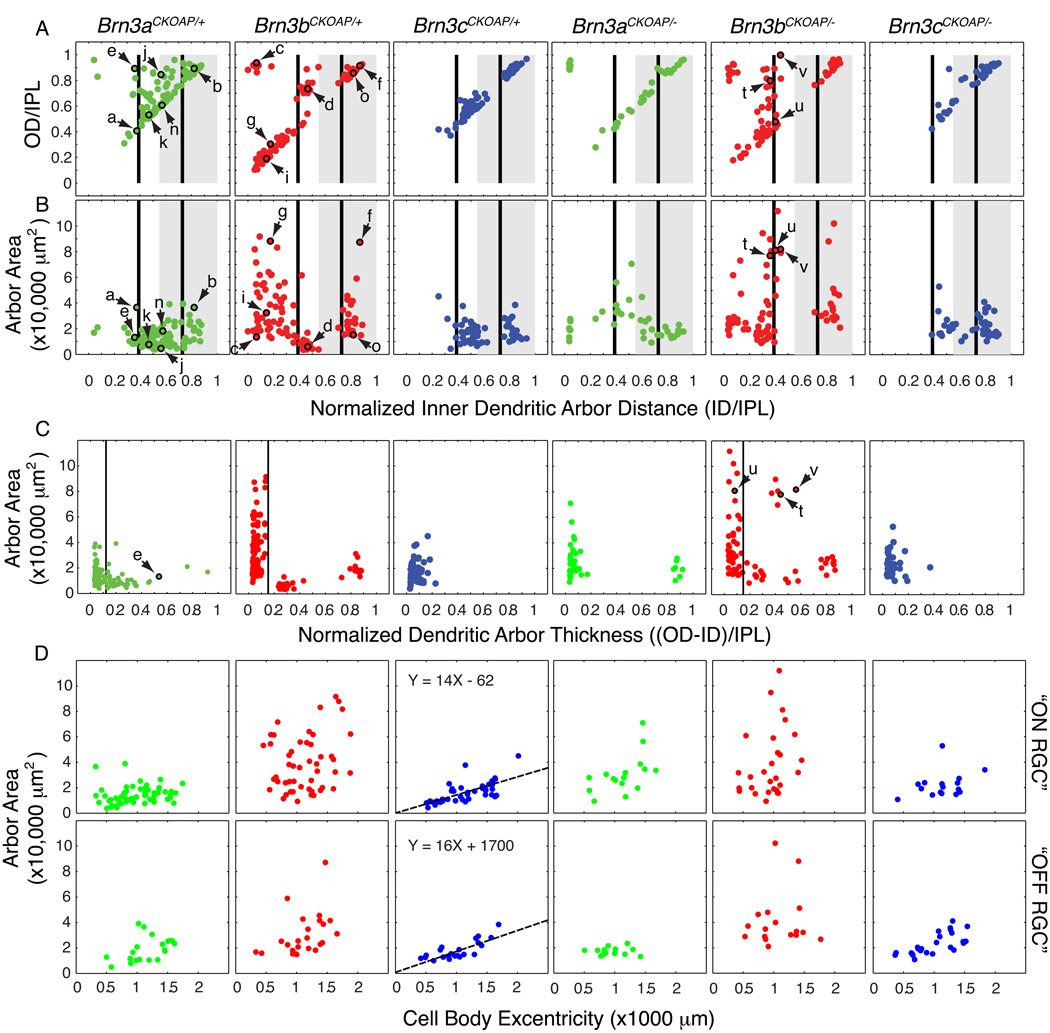
Fig. 3. Morphological parameters for monostratified Brn3AP/+ and Brn3AP/− RGCs.
Scatter plots in each vertical column refer to RGCs with the genotypes indicated at the top. Arrows and lower case letters identify the cells shown in Fig. 2. (A,B) Scatter plots of normalized inner distance (ID/IPL) vs. normalized outer distance (OD/IPL) and ID/IPL vs. dendritic arbor area. These parameters are defined in Fig. 1 H and I. The black vertical bars at 0.39 and 0.73 ID/IPL (referred to in the text as ~0.4 and ~0.7, respectively) represent the stratification levels of the “ON” (0.39) and “OFF” (0.73) bands of calretinin, calbindin, and choline acetyltransferase immunostaining, as measured by Haverkamp and Wassle (2000), Morgan et al. (2006), and Badea et al. (2009a). The grey rectangle spanning 0.55–1.0 ID/IPL represents the “OFF” sublamina of the IPL, as determined by the stratification level of the central band of calretinin/calbindin immunoreactivity (Haverkamp and Wassle, 2000; Badea et al 2009a) and of the axon arbor terminals of mGluR6-expressing, “ON” bipolar cells (Morgan et al 2006). (C) Scatter plots for the dendritic arbor thickness, (OD-ID)/IPL vs. arbor area. The dendritic arbor thickness represents the difference of the outer distance (OD) and inner distance (ID), as defined in Fig. 1H, normalized to the thickness of the IPL. (D) Dendritic arbor areas increase with eccentricity for flat monostratified Brn3cAP/+ and Brn3cAP/− RGCs in the mouse retina. Scatter plots of eccentricity (distance of the cell body from the optic nerve head) vs. area for flat monostratified RGCs. Flat monostratified RGCs include all monostratified Brn3cAP/+ RGCs, as well as Brn3aAP/+ and Brn3bAP/+ RGCs with normalized dendritic arbor thicknesses <0.12 and <0.15, respectively; these cutoffs are based on the positions of troughs in the arbor thickness distributions in panel C. For Brn3aAP/+ and Brn3bAP/+ RGCs, “ON” or “OFF” types were defined as having an ID/IPL ratio under or above 0.55, respectively. For Brn3cAP/+ RGCs, cells close to the 0.55 ON/OFF border were grouped with the RGCs that clustered nearby in parameter space in panels A and B. There is a linear correlation between eccentricity and dendritic arbor area for both “ON” and “OFF” Brn3cAP/+ and Brn3cAP/− RGCs, and the best fitting straight line is shown.
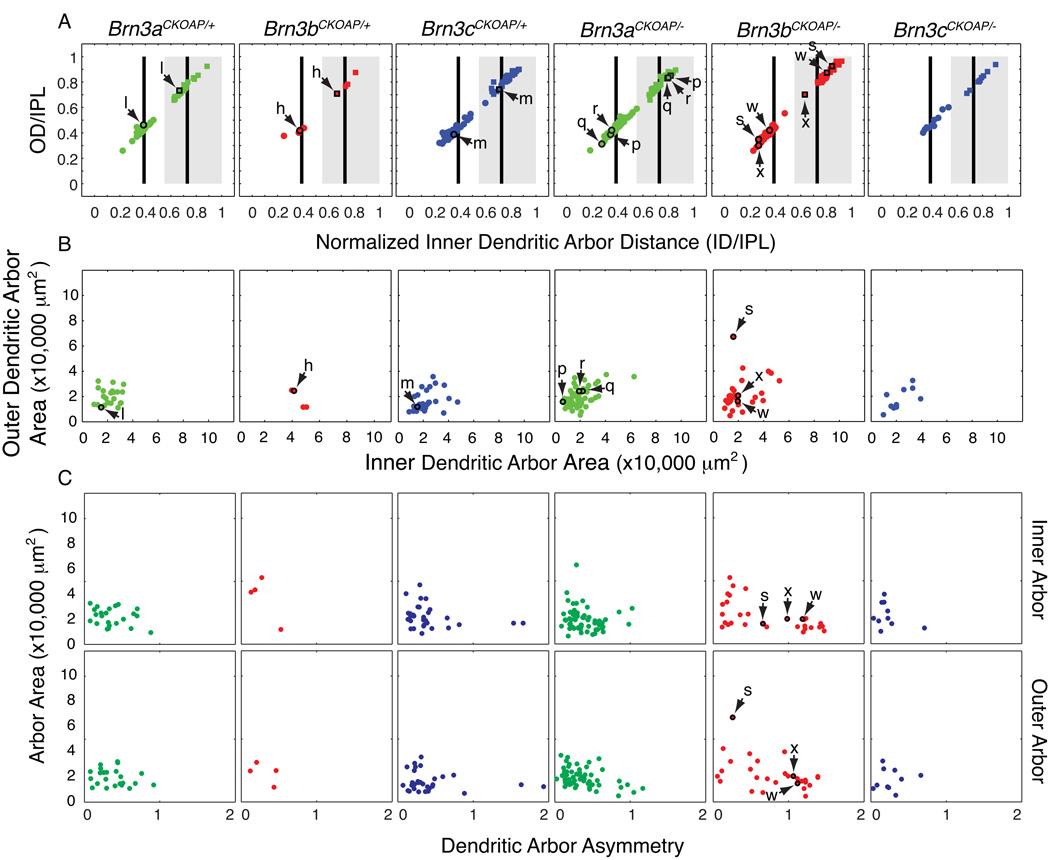
Fig. 4. Morphological parameters for bistratified Brn3AP/+ and Brn3AP/− RGCs.
Scatter plots in each vertical column refer to RGCs with the genotypes indicated at the top. Arrows and lower case letters identify the cells shown in Fig. 2. (A,B) Scatter plots of normalized inner distance (ID/IPL) vs. normalized outer distance (OD/IPL) and inner dendritic arbor area vs. outer dendritic arbor area. These parameters are defined in Fig. 1 H and I. For black vertical bars and gray rectangle landmarks in panel A, see Fig. 3 legend. In (A) stratification parameters for the inner dendritic arbors (circles) and outer dendritic arbors (squares) are plotted for each cell, e.g. inner and outer dendritic arbors for cell l, in the Brn3aCKOAP/+ scatter plot are plotted as a circle and a square, highlighted by a black outline, and indicated by a pair of arrows. (C) Scatter plots of dendritic arbor asymmetry vs. arbor area, for inner (top) and outer (bottom) arbors. The asymmetry parameter is defined as the distance from the xy coordinates of the cell body to the center of mass of the bounding polygon of the dendritic arbor, normalized by the average radius of the bounding polygon.
In general, the quantitative relationships between dendritic arbor area and arbor stratification level show a narrower range of variation within the set of WT bistratified RGCs compared to the WT monostratified RGCs, with the monostratified cells segregating into many distinct clusters across an extended hyper-volume of parametric space (compare Fig. 3A and B vs. Fig. 4A and B). Among monostratified Brn3bAP/+ and Brn3cAP/+ RGCs, the level of dendritic arborization, as measured by the normalized ID parameter, clusters into two relatively discrete groups when plotted against arbor area (Fig. 3B). Among the monostratified Brn3bAP/+ RGCs, these two groups further divide into four subgroups when ID is plotted against OD (Fig. 3A). By contrast, monostratified Brn3aAP/+ RGCs show a more continuous distribution across parametric space (Fig. 3A–C).
The thickness of a dendritic arbor in the z-dimension can be determined by subtracting ID from OD (Fig. 3C). This parameter, which can also be inferred from the plots of ID vs OD (Fig. 3A), shows a distinctive distribution for each Brn3 subset (Fig. 3C). For monostratified Brn3aAP/+ and Brn3bAP/+ RGCs, arbor areas show an inverse correlation with arbor thickness; in other words, arbors with large areas tend to be relatively thin in the z-dimension, whereas arbors with small areas tend to be relatively thick in the z-dimension. Using the (OD-ID)/IPL thickness parameter, monostratified Brn3bAP/+ RGC dendritic arbors can be subdivided in three distinct sets, with the thinnest set encompassing all of the arbors with large areas (Fig. 3C).
In sections 3.3 – 3.5 below we present a more detailed description of individual RGC types.
3.3 RGCs with small or asymmetric arbors
A distinct morphologic type of RGC has dendritic arbors that are sector shaped when viewed in the plane of the retina, slope from their origin at the cell body through the full width of the IPL, and end in a flat arbor apposed to the INL (e.g., cell c). This cell morphology, present among both Brn3aAP/+ and Brn3bAP/+ RGCs, has been previously described in unbiased morphologic surveys (Sun et al., 2002 - type C6; Badea and Nathans, 2004 - monostratified RGC cluster 6), and has recently been shown to express the molecular marker JamB and to respond selectively to upward motion (Kim et al., 2008; Kim et al., 2010).
A second subset of monostratified RGCs exhibits small, thick, more complex dendritic arbors, and is found only among Brn3aAP/+ and Brn3bAP/+ RGCs. RGCs of this general category may belong to several distinct types that are not well resolved with the parameters used here. One morphology belonging to this group exhibits an arbor thickness ((OD-ID)/IPL) of 0.4 – 0.6, is common among Brn3aAP/+ and Brn3bAP/+ RGCs, and spans from a normalized ID of ~0.35 to the INL (e.g., cell e). These cells could reasonably be classified as bistratified as their arbors, although not cleanly segregated into two planes, have higher dendritic densities at their inner and outer edges. The remainder of this group consists of RGCs with very dense dendritic arbors. The ones observed among Brn3bAP/+ RGCs (e.g., cell d) have small areas (mean area = 6,397 µm2, SD = 2,670 µm2) and are tightly clustered in parametric space (mean ID/IPL = 0.455; mean thickness = 0.29), suggesting a unique cell type, most likely an “ON” RGC. Similar Brn3aAP/+ RGCs have large arbor widths ((OD-ID)/IPL = 0.2 – 0.4) and small arbor areas (mean area = 9,436 µm2, SD = 6,418 µm2; e.g., cell j), but they span a broader range of stratification levels (ID/IPL = 0.34 – 0.74). Thus, the Brn3aAP/+ RGCs in this somewhat heterogeneous group very likely comprise both “ON” RGCs as well as “OFF” RGCs (e.g. cell j).
3.4 RGCs with flat, monostratified arbors
The majority of monostratified RGCs have relatively flat arbors. They distribute along the diagonal axis in the ID vs OD plots (Fig. 3A; i.e., ID and OD have similar values), and to the left side of the OD-ID vs area plots (Fig. 3C). All monostratified Brn3cAP/+ RGCs fall into this category (Fig. 3C).
Based on ID, OD, and area parameters Brn3cAP/+ RGCs cluster in two distinct groups (Fig. 3A, B). [In the descriptions that follows, we make use of the laminar positions of starburst amacrine cell processes at normalized distances from the inner border of the IPL of ~0.4 and ~0.7; these are indicated in Fig. 3A and B and Fig. 4A by vertical lines; see Haverkamp and Wassle (2000); Morgan et al. (2006); Badea et al. (2009a), for examples of choline acetyltransferase, calretinin and calbindin immunostaining. These landmarks, together with a central band of calretinin immunostaining midway between the starburst amacrine bands, define the junction between the ON and OFF laminae at a normalized distance of ~0.55 from the inner border of the IPL (Euler et al., 1996; Haverkamp and Wassle, 2000; Ghosh et al., 2004; Morgan et al., 2006). Although we have not determined the response properties of the RGCs described here, in the text that follows we take the liberty of provisionally referring to those RGCs with arbors vitreal to the ON/OFF junction at ~0.55 as “ON” RGCs and those RGCs with arbors scleral to that junction as “OFF” RGCs.] The “OFF” Brn3cAP/+ RGC population has dendritic arbors stratifying between the outer starburst amacrine band and the inner nuclear layer (INL; ID mean=0.83, SD=0.04; OD mean=0.89, SD=0.037), whereas the majority of the dendritic arbors of the “ON” Brn3cAP/+ RGC population are confined between the inner starburst amacrine band and the ON/OFF boundary (ID mean=0.48, SD=0.074; OD mean=0.57, SD=0.075). RGCs with similar morphologies to the “OFF” Brn3cAP/+ RGC population can be seen among both Brn3aAP/+ and Brn3bAP/+ RGCs (e.g., cell o), which represents the only cell morphology common to all three Brn3 transcription factors. The “ON” Brn3cAP/+ RGC morphological type, which is easily recognized in the flat mount view because of its highly ramified and homogeneously distributed dendrite pattern, is also present among Brn3aAP/+ RGCs (e.g., cell n), but not among Brn3bAP/+ RGCs.
Flat monostratified Brn3bAP/+ “OFF” RGCs constitute a more complex set, but they can be subdivided based on arbor stratification levels and areas. The “OFF” Brn3bAP/+ RGCs stratify between the outer starburst amacrine lamina and the INL with a broad distribution of area values. In addition to morphologies similar to the “OFF” Brn3cAP/+ RGCs, (e.g., cell o), two larger cells are also seen. Cell f is reminiscent of the melanopsin expressing “OFF” RGC morphology (Hattar et al., 2002; Berson et al., 2010; Ecker et al., 2010), with an extremely large area (87,246 µm2 for this example) and a sparse dendritic arbor. A second group of large “OFF” RGCs, with dense and straight dendrites is present among both Brn3aAP/+ and Brn3bAP/+ RGCs (e.g., cell b), but cannot be separated as a distinct cluster from the smaller o type cells based only on the parameters used here.
“ON” RGCs with flat dendritic arbors most likely represent a mixture of cell types in both the Brn3aAP/+ and Brn3bAP/+ RGC sets. Rather than attempting a classification of the different subtypes, we provide below a general description of their morphology, along with some representative examples. The dendritic arbors of “ON” Brn3bAP/+ RGCs (minimum area = 9,088 µm2, maximum area = 91,566 µm2, mean = 38,879 µm2) are, on average, much larger than their Brn3aAP/+ (minimum area = 3,823 µm2, maximum area = 38,877 µm2, mean = 14,479 µm2) and Brn3cAP/+ (minimum area = 4211.4 µm2, maximum area = 45,075 µm2, mean = 16,256 µm2) counterparts. Many “ON” Brn3bAP/+ RGCs are distinctive in that their dendrites stratify close to the ganglion cell layer (GCL), with ID/IPL generally less than 0.4, whereas the dendrites of “ON” Brn3aAP/+ RGCs have a minimal ID/IPL ratio of 0.27, and generally stratify at, or scleral to, the starburst amacrine landmark at 0.4 (Fig. 3A,B). Cells g and i represent examples of flat “ON” Brn3bAP/+ RGCs with large dendritic arbors stratifying in close proximity to the GCL. These morphologies, like the large “OFF” cell f, are unique to Brn3bAP/+ RGCs. Based on their areas and stratification levels, cells f and g most likely correspond to the M1 and M2 morphologies, respectively, of intrinsically photosensitive RGCs (Hattar et al., 2002; Berson et al., 2010; Ecker et al., 2010).
One particular type of flat, “ON” dendritic arbor, shared by Brn3aAP/+ and Brn3bAP/+ RGCs is exemplified by cell a. The arbor stratifies at the 0.4 IPL starburst amacrine cell landmark, is extremely flat, and, in some examples exhibits isolated recurrent dendrites looping into the “OFF” layer before joining the principal arbor. This morphology is characteristic of the ON direction selective (ON-DS) RGC, which has been extensively characterized both physiologically and morphologically in the mouse (Sun et al., 2006; Yonehara et al., 2008; Yonehara et al., 2009). Brn3aAP/+ RGCs also include flat “ON” RGCs with small areas (e.g. cell k) that are not seen among Brn3bAP/+ or Brn3cAP/+ RGCs.
3.5 RGCs with bistratified arbors
Bistratified RGCs with stratification levels characteristic of ON-OFF direction selective cells are commonly observed among Brn3aAP/+ and Brn3cAP/+ RGCs but rarely observed among Brn3bAP/+ RGCs (e.g., cells h, l, and m; see also Figs. 1G and 4). The few Brn3bAP/+ bistratified RGCs analyzed here have, on average, larger inner dendritic arbor areas than Brn3aAP/+ and Brn3cAP/+ RGCs (Fig. 4B). In comparing reconstructed cells l and m, a distinction between bistratified Brn3aAP/+ and Brn3cAP/+ RGCs becomes apparent. Whereas in cell l, all “ON” dendrites are derived from the cell body or other “ON” dendrites (drawn in blue), in cell m, about 50 % of “ON” stratifying dendrites derive from dendrites that had already descended into the “OFF” arbor and then looped back into the “ON” strata (drawn in gray). Although not quantified, this seems to be a consistent finding, and it might have implications for the “ON” vs. “OFF” input computations happening in ON-OFF DS RGCs.
3.6 Changes in arbor area with retinal eccentricity
Whereas average dendritic arbor areas of “ON” and “OFF” Brn3cAP/+ RGC populations are comparable (mean area of “ON” RGCs = 16,256 µm2, SD = 8,159 µm2; mean area of “OFF” RGCs = 18,067 µm2, SD = 7,351 µm2), both cell types display a roughly linear correlation between area and retinal eccentricity, as defined by the distance from the cell body to the optic disc (Fig. 3D; for “ON” RGCs, R = 0.694, P = 1.755 e−7; for “OFF” RGCs, R = 0.784, P = 1.6 e−5). The analogous Brn3aAP/+ and Brn3bAP/+ scatter plots in Fig. 3D suggest a similar trend, although the greater heterogeneity of these RGC populations would be expected to partially obscure any strong eccentricity trend that is limited to one or a few RGC subtypes. This is, to our knowledge, the first report of an eccentricity effect in dendritic arbor areas for mouse RGCs. Interestingly, the variation in area is quite large, comprising an average change of ~2-fold between 0.5 and 1.5 mm eccentricity.
3.7 Effect of deleting individual Brn3 genes on RGC morphology
What are the consequences of deleting Brn3 transcription factors on the morphologies of individual RGCs? Previously, we reported major axon arbor defects in Pax6αCre;Brn3bAP/− RGCs and the loss of specific dendritic arbor morphologies together with an increase in the proportion of bistratified RGCs in Pax6αCre;Brn3aAP/− retinas (Badea et al., 2009a). These phenotypes are independently confirmed in the current study (Figs. 1D–G, 2D, 3, and 4; Table 2). We also observe here a higher proportion of bistratified dendritic morphologies among Brn3bAP/− RGCs compared to Brn3bAP/+ RGCs (Fig. 1G; Table 2). The small decrease in the proportion of bistratified dendritic morphologies among Brn3cAP/− RGCs compared to Brn3cAP/+ RGCs (Fig. 2G) is not statistically significant and, indeed, no statistically significant differences in RGC morphologies were observed between Brn3cAP/+ and Brn3cAP/− samples with any of the parameters we measured (Figs. 3 and 4).
When Brn3a is removed from individual RGCs by sparse, R26rtTACreER-induced recombination, surviving Brn3aAP/− cells encompass four basic morphologies: bistratified RGCs (e.g., cells p–r), flat “ON” or “OFF” RGCs (mostly costratifying with the 0.4 and 0.7 starburst amacrine cell landmarks), and sector cells. These data are in good agreement with our previous analysis of RGC morphologies in Pax6αCre; Brn3aAP/− retinas, and they support our previous suggestion that Brn3a may be necessary for the establishment of RGCs with small and thick dendritic arbors (cells d, e, and j; Badea et al 2009a). Interestingly, among bistratified Brn3aAP/− RGCs, many exhibit relatively simplified “ON” dendritic arbors, that expand into isolated and separated “OFF” arbors (compare cells p–r with cells e and l).
Many of the sparsely recombined Brn3bAP/− RGCs observed in this study exhibit the same abnormal features observed in Pax6αCre;Brn3bAP/− retinas, such as bifurcated axons penetrating in the IPL (Figs. 1E and F, and cell v), and small dendritic arbors arising from axonless cell bodies in the INL. The finding of amacrine or amacrine-like Brn3bAP/− (but not Brn3bAP/+) neurons is consistent with a role for Brn3b in suppressing amacrine cell fates (Qiu et al., 2008). Stratification levels and areas of monostratified Brn3bAP/− dendritic arbors overlap substantially with those of Brn3bAP/+ (Fig. 3) except for the presence of a subset of Brn3bAP/− RGCs with wide and thick arbors (e.g. cells t–v). These cells resemble the large “ON” morphologies seen both among Brn3bAP/+ and Brn3bAP/− RGCs, except that individual Brn3bAP/− arbors encompass multiple IPL strata (compare cells g and u). In some cases, these arbors resemble those of large bistratified Melanopsin cells (e.g., cell s; Berson et al., 2010).
As noted above, a striking, and previously unreported, observation is the expansion in the fraction of bistratified RGCs among Brn3bAP/− RGCs compared to Brn3bAP/+ RGCs (Figs. 1G and 4). The majority of these bistratified neurons have inner arbors stratifying proximal to the 0.4 IPL landmark and outer arbors stratifying distal the 0.7 IPL landmark. Thus, the two dendritic arbor strata are further apart than is typical for ON-OFF DS bistratified RGCs (compare cells l and m with cell s) and do not coincide with the ChAT IPL bands. Moreover, in a subset of these RGCs, the cell bodies are offset from the dendritic arbor in a manner reminiscent of the monostratified sector cells (cells c, w, and x). To quantify this phenomenon, we computed the relative offset of the cell body in relation to the dendritic arbors for all inner and outer arbors of bistratified RGCs (Fig. 4C). The asymmetry parameter – defined as the distance between the cell body and the center of mass of the bounding polygon of either the inner or outer dendritic arbor, divided by the mean radius of the bounding polygon - is 0 for a perfectly symmetrical cell with a centered cell body, 1 for a cell body placed on the circumference of a circular bounding polygon, and greater than one for a cell body positioned beyond the circumference of a circular bounding polygon. Fig. 4C shows the extended distribution of asymmetry parameters calculated for Brn3bAP/− dendritic arbors, with a large fraction having values larger than 1.
4. Discussion
4.1 Patterns of Brn3 transcription factor expression and the division of RGCs into subtypes
How many types of RGCs are there in the mouse retina? Depending on the imaging approach, the parameters measured, and the clustering algorithms applied, estimates for morphologic types vary widely: 17 in Sun et al. 2002, 11 in Badea and Nathans, 2004, 11 in Kong et al., 2005, 14 in Coombs et al., 2006, and 22 in Volgyi et al., 2009. In the present study, at least 15 distinct RGC morphologies are visualized by using sparse recombination in the three Brn3CKOAP/+ mouse lines.
These estimates represent a lower bound for a full classification system, as each morphological class could encompass multiple functional classes. Well-documented examples of this situation are bistratified ON-OFF DS cells, which have similar morphological parameters, yet can be differentiated by their preference for four distinct directions of stimulus movement (Barlow et al., 1964; Weng et al., 2005). For ON-OFF DS cells, dendritic arbor reconstructions using genetically driven reporters have recently been described (Schubert et al., 2005b; Huberman et al., 2009). Interestingly, Huberman et al report transgenic labeling of ON-OFF DS cells selectively responsive to stimuli moving in one direction, raising the possibility that the four functional ON-OFF DS RGCs subtypes might express distinctive molecular markers. More subtle distinctions between bistratified RGCs based on the number of recurrent dendrites looping from the “OFF” arbor to the “ON” arbor, as seen in comparing Brn3aAP/+ RGC l and Brn3cAP/+ RGC m, may add additional categories of ON-OFF bistratified RGCs.
Monostratified ON-DS RGCs (e.g., cell a), although morphologically indistinguishable from each other, also comprise three distinct functional classes, responsive to moving stimuli in three directions, “up”, “forward” and “backward”. These cells provide the afferent signals for the visual component of the vestibulo-ocular reflex, are labeled in Brn3aCKOAP/+ and Brn3bCKOAP/+ but not Brn3cCKOAP/+ retinas, and project to three brainstem nuclei in the accessory optic system (Simpson, 1984; Yonehara et al., 2008; Badea et al., 2009a; Yonehara et al., 2009). The recent reports by Yonehara et al suggest that, within this morphological group, there is molecular diversity that correlates with functional diversity. By contrast, the sector cells present among Brn3aAP/+ and Brn3bAP/+ RGCs (e.g., cell c), appear to constitute a unique type, identifiable by physiology, morphology and JamB expression (Kim et al., 2008).
In our data set, cells with morphologies similar to melanopsin-expressing RGCs are seen exclusively in Brn3bCKOAP/+ retinas (cells f, g, and i; Hattar et al., 2002; Berson et al., 2010). This is consistent with the previous observation that Brn3bAP/+ RGCs project to the OPN and IGL, and show a weak projection to the SCN, all of which are targets of melanopsin RGCs, whereas Brn3aAP/+ and Brn3cAP/+ RGCs avoid many of these targets (Badea et al., 2009a; Badea, Williams, Smallwood, and Nathans, manuscript in preparation).
The physiologic and morphologic properties, as well as one genetic marker, have been described for mouse “OFF” alpha RGCs (Pang et al., 2003; Schubert et al., 2005a; Völgyi et al., 2005; Huberman et al., 2008). This type is likely present in both Brn3aAP/+ and Brn3bAP/+ RGC data sets, exemplified by cell b, based on dendritic area and stratification, and the relatively straight, acutely angled dendrites. The flat monostratified “OFF” RGC type (cell o), is the only morphological type shared by all Brn3AP/+ RGCs and represents the only “OFF” RGC labeled among Brn3cAP/+ RGCs.
Cells with small and thick dendritic arbors (cells d and j) have been previously reported (Sun et al., 2002; Badea and Nathans, 2004; Coombs et al., 2006; Volgyi et al., 2009). We speculate that they represent the mouse beta RGCs, since they come in both “ON” (cell d) and “OFF” (cell j) variants, and stratify close to the middle of the IPL. These cells likely correspond to the small thick arbor clusters G5, G8, G13 and G14 described by Volgyi 2009. We note that similar RGCs with small areas and similar stratification levels have been reported as bistratified in Schubert et al., 2005b. In the present study, it is difficult to discern two arbors.
Another morphology present among both Brn3aAP/+ and Brn3bAP/+ RGCs, which is difficult to match with previously described morphologies, is exemplified by cell e. It most likely is classified as a bistratified RGC in Coombs et al., 2006, and is a member of cluster 5 in Badea and Nathans, 2004. In all of the examples of this RGC type in our data set and in the literature, the “OFF” arbor is sparser than the “ON” arbor, and “OFF” dendrites ascend vertically from the “ON” plexus to the inner edge of the INL. Although the physiological properties of this cell have not been reported in the mouse retina, bistratified RGCs with a similarly large distance between the two arbors have been described as blue – yellow color opponent cells in the primate (Dacey and Lee, 1994; Crook et al., 2009), and as “uniformity detectors” in rabbit (Famiglietti, 2009). Bistratified cells similar to cell e have been shown not to be color opponent in guinea pigs, which, like mice, have some cones that co-express the short- and medium-wavelength pigments (Applebury et al., 2000; Yin et al., 2009).
Brn3cAP/+ RGCs can be divided in three distinct morphologies with tightly clustered morphological parameters: an “ON” RGC (cell n), an “OFF” RGC (cell o), and a bistratified, most likely an “ON-OFF”, RGC (cell m). The dendritic arbor areas of all three cells exhibit a linear increase with eccentricity, implying a central-to-peripheral decrease in visual acuity for these three channels.
Brn3aAP/+ and Brn3bAP/+ RGCs each encompass at least 10 distinct cell morphologies, consistent with immunohistochemical data showing that each transcription factor is expressed in ~75% of all RGCs (Badea et al. 2009a). In the present study, five distinct cell morphologies (cells a–e) were observed to be shared among Brn3aAP/+ and Brn3bAP/+ RGCs, one (cell n) is shared between Brn3aAP/+ and Brn3cAP/+ RGCs, and one (cell o) is shared by all three Brn3AP/+ RGCs. This pattern is consistent with the hypothesis that the Brn3 transcription factors constitute a combinatorial code for RGC cell type specification. This pattern also holds for other molecular markers identified thus far. For example, the photopigment Melanopsin is expressed in several distinct RGC cell types (Berson et al., 2010; Ecker et al., 2010); the secreted Ig domain molecule Spig1 is expressed in one type of ON-DS RGC in most of the retina, but in virtually all RGCs in the dorsal arc as well as in some amacrine cells (Yonehara et al., 2008); the Ig domain adhesion molecule JamB is transiently expressed in sector-shaped, upward direction selective RGCs, but also in a population of amacrine cells (Kim et al., 2008); and a calretinin-GFP bacterial artificial chromosome (BAC) transgene is expressed in transient OFF alpha RGCs as well as in a subset of amacrine cells (Huberman et al., 2008). Thus, genetic labeling and/or manipulation of only a single neuronal cell type will likely require intersectional strategies for gene regulation, as have been used in mice (Badea et al., 2009b) and Drosophila (Potter et al., 2010).
Do labeled RGCs in Brn3CKOAP/+ retinas cover the entire spectrum of RGC types in the mouse? For at least one cell population, the answer is no. As previously reported, the SCN is not targeted by axons from Brn3aAP/+ and Brn3cAP/+ RGCs and only weakly innervated in its latero-caudal aspect by Brn3bAP/+ RGCs (Badea et al., 2009a; Badea, Williams, Smallwood, and Nathans, manuscript in preparation). Thus, at least one melanopsin-expressing, SCN-projecting cell population is Brn3 negative. Another RGC morphology that appears to be absent from the Brn3 collection is the parvalbumin-5 “approach detecting” RGC described by Münch et al. (2009).
4.2 The role of Brn3 transcription factors in RGC specification
What are the roles of Brn3 transcription factors in RGC development? Previous reports indicated that Brn3b plays a role in RGC axon development, whereas Brn3a is involved in differentiation of dendritic arbors (Erkman et al., 1996; Gan et al., 1996; Erkman et al., 2000; Wang et al., 2000; Badea et al., 2009a). The current study confirms and extends these findings. First, we observe axon branching defects in individual, sparsely labeled Brn3bAP/− RGCs, as well as small, axonless amacrine-like Brn3bAP/− cells in the INL. Since more than 99 % of RGCs in these retinas are phenotypically WT (i.e. they have not undergone Cre-mediated recombination), these findings indicate that the aberrant morphologies are not caused by a defect at the optic nerve, in cues for axon path-finding, or in any other non-cell autonomous processes, but rather are the result of cell-autonomous defects secondary to Brn3b ablation. Second, among Brn3bAP/− RGCs with novel morphologies, there is a population with large arbor areas and dendrites that populate multiple IPL laminae (e.g., cells t and v). In general, RGC dendritic arbor area is anticorrelated with arbor thickness, so that RGCs that integrate signals over a large retinal area usually occupy a narrow stratum of the IPL, thereby receiving input from relatively limited bipolar and amacrine cell populations. By contrast, a significant fraction of RGCs with small dendritic arbor areas collect information from several IPL strata, and could therefore receive synaptic inputs from a broader population of bipolar and amacrine cell types. Finally, a class of Brn3bAP/− RGCs with abnormal dendritic arbor morphologies is seen in the expanded population of AP-expressing bistratified RGCs. Roughly half of these bistratified Brn3bAP/− RGCs exhibit extremely asymmetric dendritic arbors, reminiscent of the sector/JamB population of monostratified RGCs (cells w, x). These asymmetries could plausibly arise from abnormal cell body movements among otherwise normal bistratified RGCs, or they could derive from sector RGCs that have developed a second dendritic arbor.
Acknowledgements
Supported by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amthor FR, Oyster CW, Takahashi ES. Morphology of on-off direction-selective ganglion cells in the rabbit retina. Brain Res. 1984;298(1):187–190. doi: 10.1016/0006-8993(84)91167-3. [DOI] [PubMed] [Google Scholar]
- Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009a;61:852–864. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Hua ZL, Smallwood PM, Williams J, Rotolo T, Ye X, Nathans J. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One. 2009b;4:e7859. doi: 10.1371/journal.pone.0007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow HB, Hill RM, Levick WR. Retinal ganglion cells responding selectively to direction and speed of image motion in the rabbit. J Physiol. 1964;173:377–407. doi: 10.1113/jphysiol.1964.sp007463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- Berson DM, Castrucci AM, Provencio I. Morphology and mosaics of melanopsin-expressing retinal ganglion cell types in mice. J Comp Neurol. 2010;518:2405–2422. doi: 10.1002/cne.22381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott BB, Wassle H. The morphological types of ganglion cells of the domestic cat's retina. J Physiol. 1974;240:397–419. doi: 10.1113/jphysiol.1974.sp010616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcieri SM, Jacobs AL, Nirenberg S. Classification of retinal ganglion cells: a statistical approach. J Neurophysiol. 2003;90:1704–1713. doi: 10.1152/jn.00127.2003. [DOI] [PubMed] [Google Scholar]
- Cleland BG, Levick WR. Brisk and sluggish concentrically organized ganglion cells in the cat's retina. J Physiol. 1974a;240:421–456. doi: 10.1113/jphysiol.1974.sp010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland BG, Levick WR. Properties of rarely encountered types of ganglion cells in the cat's retina and an overall classification. J Physiol. 1974b;240:457–492. doi: 10.1113/jphysiol.1974.sp010618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs J, van der List D, Wang G, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience. 2006;140:123–136. doi: 10.1016/j.neuroscience.2006.02.079. [DOI] [PubMed] [Google Scholar]
- Crook JD, Davenport CM, Peterson BB, Packer OS, Detwiler PB, Dacey DM. Parallel ON and OFF cone bipolar inputs establish spatially coextensive receptive field structure of blue-yellow ganglion cells in primate retina. J Neurosci. 2009;29:8372–8387. doi: 10.1523/JNEUROSCI.1218-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacey DM, Lee BB. The 'blue-on' opponent pathway in primate retina originates from a distinct bistratified ganglion cell type. Nature. 1994;367:731–735. doi: 10.1038/367731a0. [DOI] [PubMed] [Google Scholar]
- Dacey DM, Peterson BB, Robinson FR, Gamlin PD. Fireworks in the primate retina: in vitro photodynamics reveals diverse LGN-projecting ganglion cell types. Neuron. 2003;37:15–27. doi: 10.1016/s0896-6273(02)01143-1. [DOI] [PubMed] [Google Scholar]
- Do MT, Kang SH, Xue T, Zhong H, Liao H, Bergles DE, Yau K. Photon capture and signalling by melanopsin retinal ganglion cells. Nature. 2009;457:281–287. doi: 10.1038/nature07682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JL, Dumitrescu ON, Wong KY, Alam NM, Chen SK, LeGates T, Renna JM, Prusky JT, Berson DM, Hattar S. Melanopsin-expressing retinal ganglioncell photoreceptors: cellular diversity and role in pattern vision. Neuron. 2010 Jul;67(1):49–60. doi: 10.1016/j.neuron.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O'Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, O'Connell SM, Krones AI, Kirby MA, Rapaport DH, Bermingham JR, O'Leary DD, Rosenfeld MG. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000;28:779–792. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- Euler T, Schneider H, Wassle H. Glutamate responses of bipolar cells in a slice preparation of the rat retina. J Neurosci. 1996;16:2934–2944. doi: 10.1523/JNEUROSCI.16-09-02934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiglietti EV, Kolb H. Structural basis for ON-and OFF-center responses in retinal ganglion cells. Science. 1976;194:193–195. doi: 10.1126/science.959847. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV. Bistratified ganglion cells of rabbit retina: Neural architecture for contrast-independent visual responses. Vis Neurosci. 2009:1–19. doi: 10.1017/S0952523808080929. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64:484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wassle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- Guler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao H, Barnard AR, Cahill H, Badea TC, Zhao H, Hankins MW, Berson DM, Lucas RJ, Yau K, Hattar S. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, Barres BA. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, Barres BA. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature. 2008;452:478–482. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- Kim I, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30:1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Fish DR, Rockhill RL, Masland RH. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. J Comp Neurol. 2005;489:293–310. doi: 10.1002/cne.20631. [DOI] [PubMed] [Google Scholar]
- Levick WR. Receptive fields and trigger features of ganglion cells in the visual streak of the rabbits retina. J Physiol. 1967;188:285–307. doi: 10.1113/jphysiol.1967.sp008140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levick WR. Form and function of cat retinal ganglion cells. Nature. 1975;254:659–662. doi: 10.1038/254659a0. [DOI] [PubMed] [Google Scholar]
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001;11:431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- Masland RH, Martin PR. The unsolved mystery of vision. Curr Biol. 2007;17:R577–R582. doi: 10.1016/j.cub.2007.05.040. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Dhingra A, Vardi N, Wong RO. Axons and dendrites originate from neuroepithelial-like processes of retinal bipolar cells. Nat Neurosci. 2006;9:85–92. doi: 10.1038/nn1615. [DOI] [PubMed] [Google Scholar]
- Münch TA, da Silveira RA, Siegert S, Viney TJ, Awatramani GB, Roska B. Approach sensitivity in the retina processed by a multifunctional neural circuit. Nat Neurosci. 2009;12:1308–1316. doi: 10.1038/nn.2389. [DOI] [PubMed] [Google Scholar]
- Pang J, Gao F, Wu SM. Light-evoked excitatory and inhibitory synaptic inputs to ON and OFF alpha ganglion cells in the mouse retina. J Neurosci. 2003;23:6063–6073. doi: 10.1523/JNEUROSCI.23-14-06063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu ML, Amthor FR. Dendritic morphologies of retinal ganglion cells projecting to the lateral geniculate nucleus in the rabbit. J Comp Neurol. 1990a;302:675–693. doi: 10.1002/cne.903020320. [DOI] [PubMed] [Google Scholar]
- Pu ML, Amthor FR. Dendritic morphologies of retinal ganglion cells projecting to the nucleus of the optic tract in the rabbit. J Comp Neurol. 1990b;302:657–674. doi: 10.1002/cne.903020319. [DOI] [PubMed] [Google Scholar]
- Qiu F, Jiang H, Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J Neurosci. 2008;28:3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y, Cajal S. The structure of the retina. Springfield, Ill.: C.C. Thomas; 1972. [Google Scholar]
- Rockhill RL, Daly FJ, MacNeil MA, Brown SP, Masland RH. The diversity of ganglion cells in a mammalian retina. J Neurosci. 2002;22:3831–3843. doi: 10.1523/JNEUROSCI.22-09-03831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW, Watanabe M. Survey of the morphology of macaque retinal ganglion cells that project to the pretectum, superior colliculus, and parvicellular laminae of the lateral geniculate nucleus. J Comp Neurol. 1993;338:289–303. doi: 10.1002/cne.903380211. [DOI] [PubMed] [Google Scholar]
- Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–587. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- Schubert T, Degen J, Willecke K, Hormuzdi SG, Monyer H, Weiler R. Connexin36 mediates gap junctional coupling of alpha-ganglion cells in mouse retina. J Comp Neurol. 2005a;485:191–201. doi: 10.1002/cne.20510. [DOI] [PubMed] [Google Scholar]
- Schubert T, Maxeiner S, Kruger O, Willecke K, Weiler R. Connexin45 mediates gap junctional coupling of bistratified ganglion cells in the mouse retina. J Comp Neurol. 2005b;490:29–39. doi: 10.1002/cne.20621. [DOI] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Simpson JI. The accessory optic system. Annu Rev Neurosci. 1984;7:13–41. doi: 10.1146/annurev.ne.07.030184.000305. [DOI] [PubMed] [Google Scholar]
- Sun W, Li N, He S. Large-scale morphological survey of mouse retinal ganglion cells. J Comp Neurol. 2002;451:115–126. doi: 10.1002/cne.10323. [DOI] [PubMed] [Google Scholar]
- Sun W, Deng Q, Levick WR, He S. ON direction-selective ganglion cells in the mouse retina. J Physiol. 2006;576:197–202. doi: 10.1113/jphysiol.2006.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy JB, Shou T. The receptive fields of cat retinal ganglion cells in physiological and pathological states: where we are after half a century of research. Prog Retin Eye Res. 2002;21:263–302. doi: 10.1016/s1350-9462(02)00002-2. [DOI] [PubMed] [Google Scholar]
- Volgyi B, Chheda S, Bloomfield SA. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. J Comp Neurol. 2009;512:664–687. doi: 10.1002/cne.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Gan L, Martin SE, Klein WH. Abnormal polarization and axon outgrowth in retinal ganglion cells lacking the POU-domain transcription factor Brn-3b. Mol Cell Neurosci. 2000;16:141–156. doi: 10.1006/mcne.2000.0860. [DOI] [PubMed] [Google Scholar]
- Wang SW, Mu X, Bowers WJ, Kim D, Plas DJ, Crair MC, Federoff HJ, Gan L, Klein WH. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002;129:467–477. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- Wassle H. Parallel processing in the mammalian retina. Nat Rev Neurosci. 2004;5:747–757. doi: 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]
- Weng S, Sun W, He S. Identification of ON-OFF direction-selective ganglion cells in the mouse retina. J Physiol. 2005;562:915–923. doi: 10.1113/jphysiol.2004.076695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Li D, Chen ZY, Zhou L, O'Malley BW, Jr, Klein W, Nathans J. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Zhou L, Klein WH, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci U S A. 1996;93:11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yin L, Smith RG, Sterling P, Brainard DH. Physiology and morphology of color-opponent ganglion cells in a retina expressing a dual gradient of S and M opsins. J Neurosci. 2009;29:2706–2724. doi: 10.1523/JNEUROSCI.5471-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Shintani T, Suzuki R, Sakuta H, Takeuchi Y, Nakamura-Yonehara K, Noda M. Expression of SPIG1 reveals development of a retinal ganglion cell subtype projecting to the medial terminal nucleus in the mouse. PLoS ONE. 2008;3:e1533. doi: 10.1371/journal.pone.0001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, Noda M. Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PLoS One. 2009;4:e4320. doi: 10.1371/journal.pone.0004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeck GM, Masland RH. Spike train signatures of retinal ganglion cell types. Eur J Neurosci. 2007;26:367–380. doi: 10.1111/j.1460-9568.2007.05670.x. [DOI] [PubMed] [Google Scholar]