We report that San, an acetyltransferase required for sister chromatid cohesion, also acetylates β-tubulin at lysine 252. The acetylation happens only on free tubulin heterodimers, and it delays the incorporation of modified tubulins into microtubules in vivo.
Abstract
Dynamic instability is a critical property of microtubules (MTs). By regulating the rate of tubulin polymerization and depolymerization, cells organize the MT cytoskeleton to accommodate their specific functions. Among many processes, posttranslational modifications of tubulin are implicated in regulating MT functions. Here we report a novel tubulin acetylation catalyzed by acetyltransferase San at lysine 252 (K252) of β-tubulin. This acetylation, which is also detected in vivo, is added to soluble tubulin heterodimers but not tubulins in MTs. The acetylation-mimicking K252A/Q mutants were incorporated into the MT cytoskeleton in HeLa cells without causing any obvious MT defect. However, after cold-induced catastrophe, MT regrowth is accelerated in San-siRNA cells while the incorporation of acetylation-mimicking mutant tubulins is severely impeded. K252 of β-tubulin localizes at the interface of α-/β-tubulins and interacts with the phosphate group of the α-tubulin-bound GTP. We propose that the acetylation slows down tubulin incorporation into MTs by neutralizing the positive charge on K252 and allowing tubulin heterodimers to adopt a conformation that disfavors tubulin incorporation.
INTRODUCTION
Tubulin, the building block of microtubules (MTs), is regulated by various MT-associated proteins and many posttranslational modifications (Westermann and Weber, 2003; Gaertig and Wloga, 2008; Hammond et al., 2008). These modifications are thought to be important for fine-tuning the property of tubulin and MTs to suit their diverse functions. Tubulin acetylation was first documented in 1981 (McKeithan and Rosenbaum, 1981), and the best characterized example is lysine 40 (K40) acetylation of α-tubulin (LeDizet and Piperno, 1987). Although this modification always associates with stable MTs, its exact role remains elusive. Nonetheless, it has been directly and indirectly implicated in many biological processes, such as primary cilium disassembly (Pugacheva et al., 2007), cell migration (Hubbert et al., 2002), and autophagy (Iwata et al., 2005). The acetylation of tubulin is not limited to K40 of α-tubulin, as additional acetylations on both α- and β-tubulins have been revealed in a recent proteomic study (Choudhary et al., 2009). Although the biological significance of these modifications remains to be determined, it suggests that tubulins and MTs may be regulated by multiple acetylation events.
The enzymes that catalyze K40 acetylation and deacetylation have been identified. The elongation factor and MEC-17/αTAT1 are K40 acetyltransferases (Creppe et al., 2009; Akella et al., 2010; Shida et al., 2010), and HDAC6 and SirT2 are the deacetylases (Hubbert et al., 2002; North et al., 2003). Interestingly, these enzymes also modify other proteins and are involved in other biological processes unrelated to tubulins (Valenzuela-Fernandez et al., 2008). We have been working on an acetyltransferase called San, which is required for sister chromatid cohesion in Drosophila and human cells (Williams et al., 2003; Hou et al., 2007). In addition, San is also a subunit of the N-terminal acetyltransferase NatA complex, which is conserved from yeast to human (Evjenth et al., 2009). San by itself, however, has the acetyltransferase activity, and its function in sister chromatid cohesion does not require other subunits of the NatA complex (Hou et al., 2007).
Here we report our unexpected discovery that San is also a tubulin acetyltransferase (TAT). It modifies tubulin heterodimers but not polymerized MTs in vitro. San is not responsible for K40 acetylation of α-tubulin. Instead, it catalyzes a novel acetylation at K252 of β-tubulin. Depleting up to 80% of San from HeLa cells significantly increases the rate of MT regrowth after MT catastrophe, although no gross defect in MT cytoskeleton is observed. In contrast, two acetylation-mimicking mutants are incorporated into MTs approximately 20-fold more slowly in the same MT regrowth assay. Therefore, we conclude that K252 acetylation of β-tubulin negatively regulates MT regrowth rate by slowing down the incorporation of the acetylated tubulins.
RESULTS
San acetylates tubulin
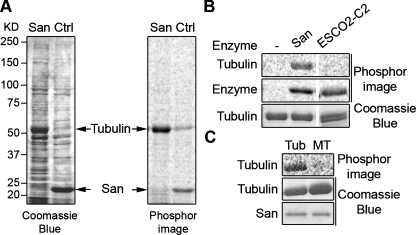
To identify San substrates, purified recombinant San was immobilized on sepharose beads, which were next mixed with cytoplasmic and nucleoplasmic fractions prepared from 293T cells (Mendez and Stillman, 2000). After three washes, the beads were incubated at 37°C for 1 h with 14C-labeled acetyl-CoA to radiolabel potential substrates. On SDS–PAGE, approximately eight bands were both enriched and acetylated by San (Supplemental Figure S1). After increasing the sodium chloride concentration in the wash buffer to 100 mM, a ∼50 kDa band, which likely represented a higher affinity interacting protein, exhibited the most prominent signal on the Coomassie Blue–stained gel and the phosphorimage (Figure 1A). The band was sliced out, and mass spectrometry analysis identified it as α- or β-tubulin. To directly validate tubulin as a San substrate, we performed the in vitro acetylation assay using commercial bovine tubulin. As shown in Figure 1B, tubulin was acetylated by San, but not by acetyltransferase ESCO2 (Hou and Zou, 2005), indicating that San specifically acetylates tubulin. Interestingly, San did not acetylate Taxol-stabilized MTs (Figure 1C), suggesting that the targeted residue is inaccessible in MTs or that San does not interact with MTs. Taken together, San is a TAT that specifically modifies soluble tubulin heterodimers.
FIGURE 1:
San acetylates tubulin heterodimers but not MTs. (A) Tubulin is a San substrate. HeLa cell lysate was incubated with either San-conjugated NHS beads (San) or empty beads (Ctrl), and proteins bound to the beads were analyzed in the in vitro San acetylation assay. For the empty beads control, recombinant San was added after the pull-down experiment. After resolving the proteins by SDS–PAGE, the gel was stained with Coomassie Blue (left panel) and exposed to phosphorimage (right panel). The 52 kDa protein band was identified as tubulin by MS. In the San beads sample, San proteins were covalently linked to the beads and therefore not visible on the gel. (B) Tubulins were acetylated by San but not ESCO2. Bovine tubulins were subjected to in vitro acetylation with buffer (–), San, or ESCO2-C2 (Hou and Zou, 2005). The tubulin doublet shown in the third lane was due to partial separation of α- and β-tubulin on the gel. The self-acetylations of San and ESCO2-C2 were used as controls for enzymatic activity. (C) Tubulins but not MTs were acetylated by San. MTs made of bovine tubulins were either depolymerized to tubulins by incubation on ice (Tub) or stabilized by Taxol (MT). Both preparations were subjected to in vitro acetylation with San.
San slows tubulin regrowth after cold-induced catastrophe
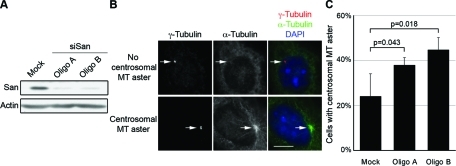
To investigate how the San-catalyzed tubulin acetylation affects MT functions in vivo, we first overexpressed San in HeLa cells. No obvious MT abnormality was detected when wild-type San was overexpressed (Supplemental Figure S2). It is possible that the activity of San is tightly regulated. We also knocked down San expression to approximately 20% of the physiological level in HeLa cells by using two siRNA oligonucleotides (Figure 2A). Again, no obvious defects in MT cytoskeleton were detected in interphase cells (Supplemental Figure S3). In mitosis the spindle was disorganized, but it could have been caused by premature sister chromatid separation as we and others reported previously (Hou et al., 2007; Dai et al., 2009). Next we investigated whether MT dynamics are affected by San knockdown. To this end, we performed a cold-induced MT depolymerization and regrowth assay to analyze the rate of MT depolymerization and polymerization. We did not observe any significant difference in terms of MT depolymerization when cells were incubated at 4°C. After cells were shifted back to 37°C, however, the rate of MT regrowth from the centrosome was noticeably faster in San-siRNA cells. The difference was quantified as the percentage of cells with centrosomal MT asters after 1.5 min of incubation at 37°C (Figure 2B). Centrosomal MT asters appeared in approximately 25% of the mock-treated cells, but in approximately 40% of the San-siRNA cells (Figure 2C). At 2.5 min, near 100% of cells with or without siRNA transfection contained centrosomal MT asters. Taken together, these results indicate that San negatively affects the rate of MT polymerization/nucleation.
FIGURE 2:
Centrosomal MT regrowth is accelerated in San-siRNA cells. (A) San expression level in RNAi cells. HeLa cells were transfected with mock or two siRNA oligonucleotides (oligo A and B) targeting San, and the expression levels were analyzed by immunoblot. Actin was probed as the loading control. (B) Cells with or without centrosomal MT aster in the MT regrowth assay. To enrich San-depleted interphase cells, HeLa cells were treated with 2 mM thymidine for 20 h prior to harvesting. Cells were put on ice to depolymerize MTs and then incubated in warm medium for 1.5 min to allow regrowth of MTs from the centrosome. Cells were stained to visualize centrosomes (γ-tubulin, red and marked by arrows), MTs (α-tubulin, green), and DNA (DAPI, blue). Scale bar, 10 μm. (C) Quantification of cells with centrosomal MT asters. For each sample, at least 50 cells were counted, and the percentage of cells with centrosomal MT aster was determined from three independent experiments. Error bars indicate standard deviation. The p value was calculated using Student’s t test.
San does not acetylate α-tubulin at K40
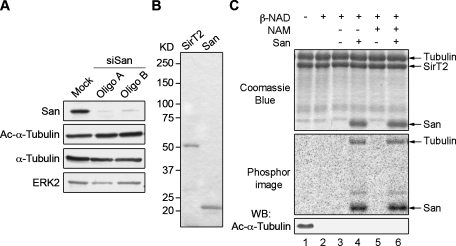
To investigate whether the faster MT reassembly in San-siRNA cells is related to the reduced level of San-catalyzed tubulin acetylation, we mapped the acetylation site so that mutations could be introduced to block and/or mimic the acetylation. First, we investigated whether the acetylation is at K40 of α-tubulin. To this end, we compared the Ac-K40 level in normal and San-siRNA cells. Using an Ac-K40 specific monoclonal antibody, we did not detect any change upon San depletion (Figure 3A). Therefore, it is unlikely that San is the major acetyltransferase that catalyzes the K40 acetylation. To directly test whether the K40 acetylation can be catalyzed by San, we removed the K40 acetylation from bovine tubulin using recombinant SirT2 (Figure 3B), one of the two Ac-K40 deacetylases (North et al., 2003). After incubation with SirT2, the bovine tubulin became free of Ac-K40 (Figure 3C, lane 2). Next the deacetylated tubulin was reacetylated using San in the presence of 14C-acetyl-CoA and nicotinamide (NAM), a SirT2 inhibitor. We detected marked increases in total tubulin acetylation, measured by 14C incorporation. The level of Ac-K40, however, measured by immunoblot using the Ac-K40 antibody, remained undetectable (Figure 3C, lane 6). This finding indicates that San catalyzes a different acetylation from the K40 acetylation. Furthermore, the level of 14C incorporation was indistinguishable from that in the absence of NAM (Figure 3C, lane 4), suggesting that, unlike Ac-K40, the acetylation catalyzed by San is resistant to SirT2. We conclude that San does not acetylate K40 of α-tubulin.
FIGURE 3:
San does not acetylate α-tubulin K40. (A) K40 acetylation level was unchanged in San-siRNA cells. Protein lysates from San-siRNA HeLa cells were analyzed by immunoblot. ERK2 was probed as the loading control. (B) Recombinant SirT2 and San. To examine the purity of recombinant proteins, 0.8 mg of His6-SirT2 and 1 mg of His6-San were subjected to SDS–PAGE and stained with Coomassie Blue. (C) San-catalyzed acetylation on tubulin was not at α-tubulin K40. Bovine tubulins were incubated with His6-SirT2 and β-NAD to deacetylate K40 of α-tubulin (lane 2). These deacetylated tubulins were then subjected to in vitro acetylation in the presence or absence of San and NAM (lanes 3–6). Proteins were analyzed by Coomassie Blue staining, phosphorimaging, and immunoblot.
San acetylates β-tubulin at K252
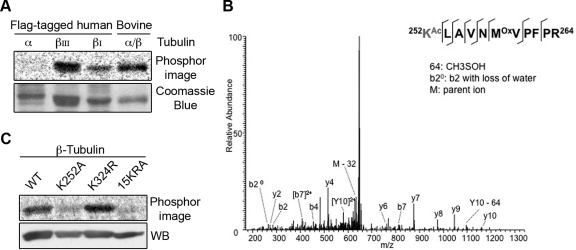
Having demonstrated that San-catalyzed tubulin acetylation was previously uncharacterized, we investigated which tubulin subunit was acetylated. To this end, we cloned the ubiquitous isotype of α-tubulin and the ubiquitous β-tubulin isotype I (βI) and added a flag tag at their C termini. The tagged tubulins were expressed in 293T cells, immunopurified on anti-flag beads, and eluted with flag peptides. Next, the purified tubulins were incubated with recombinant San in the presence of 14C-acetyl-CoA. As shown in Figure 4A, San acetylated only β-tubulin. The acetylation was also detected on the neuronal-specific isotype βIII, suggesting that the acetylation may not be isotype-specific. Taken together, we conclude that San is a β-TAT.
FIGURE 4:
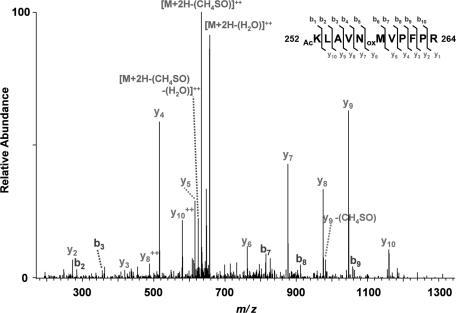
San acetylates β-tubulin at K252. (A) San acetylated β- but not α-tubulin. Flag-tagged tubulins were expressed in 293T cells, purified on anti-flag affinity gel, and analyzed by the in vitro acetylation assay. Bovine tubulin serves as a positive control. (B) Identification of K252 acetylation on βI-tubulin in vitro. MS/MS spectrum of m/z 664.7 identifies K252 as being acetylated. The labels b and y designate the N- and C-terminal fragments, respectively, of the peptide produced by breakage at the peptide bond in the mass spectrometer. The number represents the number of N- or C-terminal residues present in the peptide fragment. KAc and Mox represent acetyl-lysine and oxidized methionine residues, respectively. (C) β-tubulin K252A mutant was not efficiently acetylated by San in vitro. Wild-type and mutant β-tubulins were expressed in 293T cells, purified on anti-flag affinity gel, eluted by TEV protease, and analyzed by the in vitro acetylation assay. Immunoblot with β-tubulin antibody demonstrated equal loading.
To map the acetylation site on β-tubulin, we prepared recombinant βI-tubulin-flag from 293T cells. After incubation with San for 2 h, βI-tubulin-flag was separated from San by SDS–PAGE, sliced out of the gel, and analyzed by mass spectrometry. Acetylation at K252 was detected on several peptides (Figure 4B). In addition, acetylation at lysine 324 (K324) was also suggested but with a much lower confidence. To investigate whether these acetylations were catalyzed by San, we constructed K-to-A or K-to-R mutations at these two sites and compared the acetylation of the mutants with that of the wild type. As shown in Figure 4C, the K324R mutation did not significantly reduce the level of acetylation, indicating that K324 is not the primary site modified by San. In contrast, the acetylation of K252A mutants was reduced to approximately 30% of that of the wild type. The residual labeling was presumably due to the Nα-acetyltransferase activity of San (Evjenth et al., 2009) because the signal was comparable with that of the 15KRA mutant, in which all 15 lysines are substituted with arginines or alanines. Taken together, these results indicate that β-tubulin is specifically acetylated at K252 by San.
To determine whether K252 is acetylated in vivo, we purified endogenous tubulin from hTert1-RPE cells and analyzed its modification by mass spectrometry. As shown in Figure 5, acetylation at K252 was detected on βI-tubulin, indicating that this site is indeed acetylated in vivo. In addition, we detected acetylation at K324 (Supplemental Figure S4A). As mentioned earlier in the text, however, San does not catalyze this acetylation in vitro. Finally, a third acetylation at K58 of β-tubulin was recently identified (Choudhary et al., 2009). We therefore also examined whether San is capable of modifying this site in vitro by analyzing the acetylation of the K58R mutant. As shown in Figure S4B, this mutation did not reduce the level of acetylation, indicating that this site is not acetylated by San. In summary, among the three β-tubulin lysines known to be acetylated in vivo, San acetylates only K252.
FIGURE 5:
K252 of βI-tubulin is acetylated in vivo. Tubulins purified from hTert1-RPE cells were separated by SDS–PAGE. Tubulin gel bands were digested with trypsin, and data were collected on an LTQ-Orbitrap XL. MS/MS spectra were searched with Mascot, and peptide spectral matches indicative of lysine acetylation were subjected to manual validation. Precursor ion mass accuracy was used as orthogonal validation. Oxidized methionine resides are indicated by oxM. Neutral losses of methane sulfenic acid (-CH3SOH, -64 Da) and water (-H2O, -18 Da) from precursor and fragment ions are indicated.
The acetylation-mimicking mutant tubulins form less stable heterodimers but are incorporated into MTs in vivo
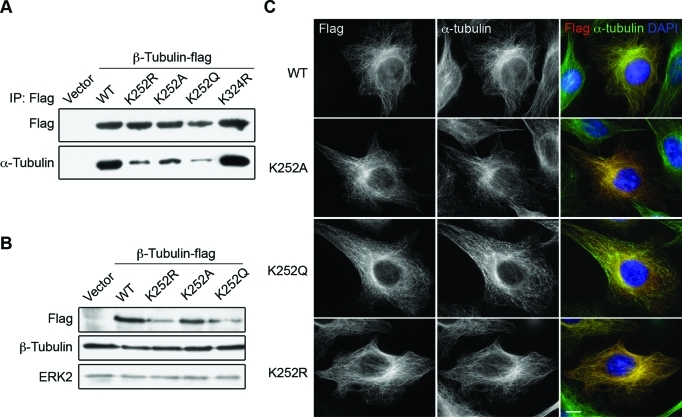
Because MT regrowth is accelerated in San-siRNA cells, we decided to investigate the incorporation rate of the β-tubulin K252A/Q and K252R mutants, which were routinely used to mimic the acetylated and unacetylated protein, respectively (Scroggins et al., 2007). First, we investigated whether the tubulin mutants were able to form heterodimers with α-tubulin. To this end, we expressed flag-tagged mutants in 293T cells and found that all three K252 mutants coimmunoprecipitated with less α-tubulin than the wild type did (Figure 6A), suggesting that the mutations result in less stable tubulin heterodimers. The β-tubulin mutants and the wild type, however, remained stable for at least 6 h in HeLa cells treated with cycloheximide (Supplemental Figure S5), indicating that the mutations do not significantly affect the short-term protein stability in vivo. We were unable to collect samples from later time points due to strong cytotoxicity, thus we could not rule out the possibility that the mutations affect protein stability in the long term. Nonetheless, the stability of the mutant tubulins suggests that they are not grossly misfolded. Consistent with this notion, all the K252 mutants were incorporated into MTs when expressed in HeLa cells (Figure 6C), indicating that they are able to form functional heterodimers in vivo. Expression of these exogenous β-tubulins did not cause any detectable change in total β-tubulin levels examined by Western blot (Figure 6B) and quantitative immunofluorescence staining in individual cells (Supplemental Figure S6). These findings indicate that the expression levels of the flag-tagged β-tubulins were much lower than that of the endogenous β-tubulin.
FIGURE 6:
β-tubulin K252 mutants are incorporated into MTs in HeLa cells. (A) β-tubulin K252 mutants pulled down less α-tubulin in vitro. Flag-tagged tubulins expressed in 293T cells were precipitated on anti-flag affinity gel, and the bound proteins were analyzed by immunoblot. (B) Protein level of β-tubulin mutants. Protein lysates from HeLa cells transfected with indicated plasmids were analyzed by immunoblot. ERK2 was probed as the loading control. (C) Cellular localization of tubulin mutants in HeLa cells. Cells transfected with plasmids carrying indicated β-tubulin constructs were stained for tubulin mutants (flag, red), MTs (α-tubulin, green), and DNA (DAPI, blue). Representative images are shown. Scale bar, 10 μm.
Cells expressing the acetylation-mimicking K252A and K252Q β-tubulins were indistinguishable from those expressing the wild-type control (Figure 6B and Supplemental Figure S6A), indicating that these mutants do not grossly affect MT cytoskeleton. The K252R mutant, which was designed to mimic unacetylatd tubulin, behaved differently, however. In approximately 50% of the transfected cells, this mutant was detected on seemingly normal MTs (Figure 6B and Supplemental Figure S6A). In the rest of the cells, the MT cytoskeleton became disorganized and/or disintegrated (Supplemental Figure S7A). The phenotype was also observed in mitotic cells where more than 50% of the mitotic spindles became multipolar and moved away from the center of the cell (Supplemental Figure S7B). Cells with abnormal MTs usually associated with higher expression level of the K252R mutant, but this was not always the case. Moreover, the phenotype does not correlate with the duration of mutant expression. Currently we do not understand why cells respond differently to the expression of the K252R mutant. We suspect that the mutant does not fully resemble the unacetylated tubulin. Because the arginine side chain is larger than that of unacetylated lysine, the substitution may result in an abnormal conformation even after the heterodimers are integrated into MTs. Incorporation of such aberrant tubulins may eventually lead to MT catastrophe in some (but not all) cells.
The acetylation-mimicking mutants are incorporated into regrowing MTs at a much slower rate
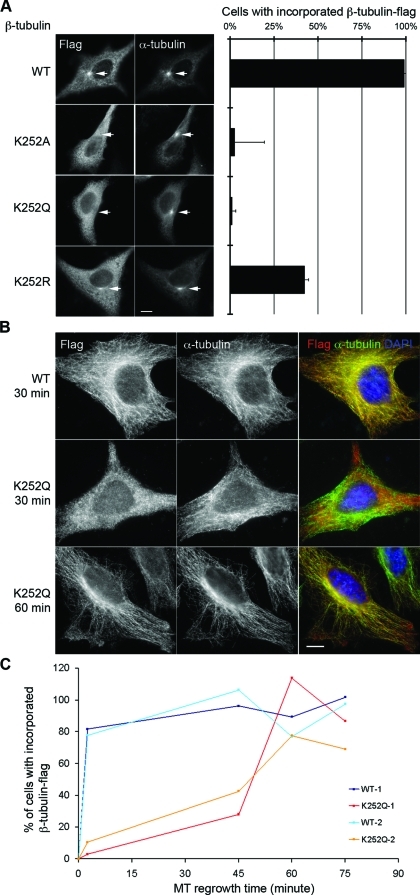
Because the acetylation-mimicking mutants could be incorporated into MTs, we next performed MT regrowth assays to analyze their incorporation rates. After expressing these flag-tagged tubulins, no defects were detected regarding the regrowth of overall centrosomal asters, which was detected in approximately 75% of the cells after shifting the culture to 37°C for 2.5 min (Supplemental Figure S8). This result was expected because the low expression level of the mutants was unlikely to dominant-negatively affect the overall MT dynamics. The K252R mutant was not incorporated faster than the wild type (Figure 7A), as one might predict on the basis of the increased MT regrowth rate in San-siRNA cells (Figure 2). One likely explanation is, as mentioned before, that this mutant does not fully mimic the unacetylated form. In contrast, the acetylation-mimicking K252A/Q mutants were more informative. These mutants were detected at the centrosomes in less than 2% of the expressing cells (Figure 7A). At this time point, the wild-type β-tubulin-flag was detected at the centrosomes in approximately 100% of the cells. After 30 min, the K252A and K252Q mutants were incorporated into MT cytoskeleton in 54% and 11% of the cells, respectively. At the 60-min time point, the mutants were incorporated in all the cells and became indistinguishable from the wild-type tubulin (Figure 7B). To better quantify the incorporation of flag-tagged tubulins over time, cells were extracted prior to fixation to remove soluble tubulins. The percentage of cells showing incorporated β-tubulin-flag was quantified and normalized to the percentage of cells expressing β-tubulin-flag. As shown in Figure 7C, whereas the wild-type β-tubulin-flag was incorporated in 80% of the cells at the 2.5-min time point, incorporation of the K252Q mutant, which was most delayed among all the mutants, was not detected until the 45-min time point. The K252Q mutant was incorporated in all the cells after 60 min of regrowth, consistent with the observation in cells without extraction. The much slower incorporation rate of the acetylation-mimicking mutants suggests that K252 acetylation of β-tubulin severely delays tubulin incorporation into the growing MTs. This observation is consistent with the accelerated MT regrowth in San-siRNA cells (Figure 2).
FIGURE 7:
Incorporation of the acetylation-mimicking β-tubulin mutants is delayed in the MT regrowth assay. (A) HeLa cells expressing indicated tubulin mutants were subjected to MT regrowth assay, fixed after 2.5 min of regrowth, and stained for tubulin mutants (flag) and MTs (α-tubulin). Representative images are shown. Arrows mark centrosomal MT asters. Scale bar, 10 μm. Right panel shows the quantification of flag-stained cells with flag-positive centrosomal MT asters. In each sample, at least 50 cells were counted, and the average percentage was determined from three independent experiments. Error bars indicate standard deviation. Between wild type and all three mutants, p < 0.001. (B) The K252Q mutant-expressing cells after 30 and 60 min of regrowth were stained for tubulin mutants (flag, red), MTσ (α-tubulin, green), and DNA (DAPI, blue). Representative images are shown. Scale bar, 10 μm. (C) Quantification of cells with incorporated β-tubulin-flag after extraction. For each data point, at least 150 cells were counted, and the percentage of cells with flag staining was normalized to the percentage of flag-stained cells without extraction. Data from two independent experiments are shown.
Because it took 60 min for the K252Q mutant to be incorporated in most of the cells, it is possible that only newly synthesized mutant tubulins are incorporated in the regrowth assay. In that case, the delay of mutant tubulin incorporation might only reflect the time required for cells to synthesize new proteins after a temperature shift. To address this issue, we added cycloheximide to the regrowth medium to block protein synthesis. As Supplemental Figure S9 shows, the K252Q mutant was incorporated in nearly all the cells after 60 min, regardless of cycloheximide treatment. This result indicates that the K252Q mutant tubulins are capable of polymerization after cold treatment and that the delayed incorporation is indeed a property of the mutant.
DISCUSSION
Previous studies have implicated San in N-terminal acetylation and sister chromatid cohesion in metazoans (Hou et al., 2007; Evjenth et al., 2009). We report here that San acetylates β-tubulin and negatively regulates tubulin incorporation. Unlike most tubulin modifications, this acetylation is added to free tubulin heterodimers but not to Taxol-stabilized MTs (Figure 1C). Such selectivity is similar to a previously described β-tubulin phosphorylation that also inhibits tubulin incorporation (Fourest-Lieuvin et al., 2006).
We mapped the acetylation site in vitro at K252 of β-tubulin. We propose that San is likely to catalyze this acetylation in vivo, as suggested by the following evidence. First, we detected the K252 acetylation in vivo (Figure 5). Second, depletion of San increases the rate of MT regrowth in vivo (Figure 2), suggesting that San negatively regulates MT assembly. Third, the K252A/Q mutants, which mimic the acetylated tubulins, are incorporated into MTs at a much slower rate (Figure 7), suggesting that the K252 acetylation impedes tubulin polymerization. A K252 acetylation specific antibody will provide the direct evidence addressing whether San acetylates K252 in vivo.
It remains unclear how exactly the acetylation affects tubulin property. In vitro acetylated tubulins failed to form MTs in a polymerization assay. This failure, however, may be due to the buffer condition as even unacetylated tubulins failed to polymerize after being mock incubated in the acetylation buffer. Nonetheless, studies of the mutant tubulins shed some light on the mechanism. We found that the K252 mutants form less stable heterodimers in vitro, yet are incorporated into MTs in vivo (Figure 6, A and C). This seemingly contradictory behavior has also been observed in the α-tubulin S140G mutant (Keays et al., 2007). It is possible that the mutant tubulin heterodimers are further stabilized by tubulin chaperones in vivo. Nonetheless, the reduced heterodimer stability in vitro suggests that the acetylation at K252 weakens the intradimer interaction. K252 locates at the interface between α- and β-tubulin heterodimer (Supplemental Figure S10). The residue is also close to the binding pocket of MT poison colchicine (Ravelli et al., 2004). Based on the structure of tubulin heterodimers, the positively charged K252 was proposed to neutralize the negative charge of the α-tubulin–bound GTP and stabilize the heterodimer (Lowe et al., 2001). Therefore, it is possible that the acetylation, by removing the charge at K252, weakens the intradimer interaction.
We further propose that San-catalyzed β-tubulin K252 acetylation impedes tubulin incorporation by abolishing the K252-GTP interaction and inducing a conformation of tubulin that disfavors MT polymerization. Tubulin heterodimers exist in at least two conformations. They adopt the straight conformation when they are polymerized into MTs (Nogales et al., 1998). In contrast, depolymerized tubulins adopt a curved conformation (Gigant et al., 2000; Wang and Nogales, 2005; Rice et al., 2008). The curved tubulins need to be straightened before being integrated into MTs and mechanisms that impede this conformational change negatively regulate MT polymerization. For example, MT destabilizing factor stathmin decreases MT polymerization by binding to and sequestering the curved tubulin heterodimers (Jourdain et al., 1997; Gigant et al., 2000). The acetylation at K252 may also induce a curved tubulin heterodimer. Such a model explains the delayed incorporation of the acetylation-mimicking K252A/Q mutants observed in the MT regrowth assay. It also explains the increased rate of MT regrowth in San-siRNA cells, where more tubulins are unacetylated and quickly adopt the straight conformation during polymerization.
Despite the fact that depletion of San accelerates the rate of MT regrowth, the exact physiological function of this novel tubulin modification may have to be determined in an animal model. In HeLa cells, we did not detect gross MT defects when San was depleted or when mutants were expressed. Because we can only deplete up to 80% of endogenous San, it is possible that the remaining 20% protein is able to maintain an overall healthy MT cytoskeleton. Alternatively, HeLa cells may not be the ideal system for analysis of tubulin mutants that exhibit subtle yet important differences. For example, HeLa cells expressing the aforementioned S140G mutant appear normal without defective MT organization, whereas mice heterozygous for this mutation exhibit abnormal neuronal migration and subsequent behavioral deficits (Keays et al., 2007). We speculate that manipulation of K252 acetylation may generate stronger phenotypes in neuronal cells, and further investigations using animal models will help reveal the physiological significance of the K252 acetylation.
MATERIALS AND METHODS
Antibodies and reagents
Antibodies to flag (Cat. No. F7425), Ac-K40 of α-tubulin (6–11 B-1), α-tubulin (DM1A), β-tubulin (Tub2.1), and γ-tubulin (Cat. No.T5192) were purchased from Sigma-Aldrich (St. Louis, MO). Antibody to ERK2 (C-14) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cy3 and Alexa Fluor 488–labeled secondary antibodies were purchased from Invitrogen (Carlsbad, CA). Taxol (paclitaxel), cytochalasin B, cycloheximide, β-nicotinamide adenine dinucleotide (β-NAD), and NAM were purchased from Sigma-Aldrich.
DNA constructs
Human San constructs in pET28 and pCS2 vectors were described previously (Hou et al., 2007). Human tubulin genes, including α (National Center for Biotechnology Information [NCBI] Reference Sequence: NM_006082), βI (GenBank: AB062393), and βIII (GenBank: BC000748) were amplified from a human thymus cDNA library (Clontech, Mountain View, CA) and subcloned into pCS2. These genes were fused with a carboxyl-terminal TEV cleavage site and a flag tag. Site-directed mutagenesis of βI-tubulin was carried out following the protocol of the QuickChange kit (Stratagene, La Jolla, CA). Human SirT2 gene (NCBI Reference Sequence: NM_012237) was amplified from the same cDNA library and subcloned into pET28.
Recombinant protein preparation
pET28 plasmids carrying San and SirT2 genes were transformed into Escherichia coli strain BL21 (DE3). Protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 4 h. His6-tagged proteins were purified using Ni-NTA resins (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. His6-San proteins were further purified on an S-100 gel filtration column (GE Healthcare, Piscataway, NJ) and eluted in acetylation buffer (50 mM HEPES, pH 8.0, 100 mM NaCl, 10% glycerol). His6-SirT2 proteins were dialyzed in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3). pCS2 plasmids carrying flag-tagged tubulin genes were transfected into 293T cells. The cells were harvested 48 h after transfection, washed in PBS, and lysed in IP buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 0.2% Nonidet P-40, 20 mM β-glycerol phosphate, 10% glycerol, 1 mM NaF, leupeptin/pepstatin/chymostatin at 1 μg/ml). After centrifugation at 100,000 × g for 30 min, the supernatant (S100) was incubated with anti-flag M2 affinity gel (Sigma-Aldrich) at 4°C for 3 h. The beads were washed three times in IP buffer followed by acetylation buffer, and the proteins were eluted by 1-h incubation at room temperature in acetylation buffer containing either flag peptide at 1 mg/ml or AcTev protease at 2 U/ml (Invitrogen). To examine the amount of α-tubulin on the beads, the proteins were eluted by Laemmli sample buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.2% bromophenol blue), resolved by SDS–PAGE, and analyzed by immunoblot.
Affinity purification of San substrates and identification of tubulin
His6-San proteins were covalently conjugated to NHS-activated Sepharose beads (GE Healthcare) according to the manufacturer’s instruction. HeLa cells were lysed by nitrogen cavitation (450 psi, on ice for 20 min) in acetylation buffer. After centrifugation at 50,000 × g for 30 min, 600 μl of the supernatant was incubated overnight at 4°C with 0.5 μl of San-conjugated or inactivated beads. Beads were washed three times in acetylation buffer, and proteins bound to the beads were subjected to the in vitro acetylation assay. For the empty beads control, His6-San was added after the pull-down experiment. After resolving the proteins by SDS–PAGE, the gel was stained by Coomassie Blue and exposed to a phosphorimage screen. A ∼50-kDa protein band extensively labeled by 14C was sliced out and sent for mass spectrometry. It was identified as an α-/β-tubulin heterodimer by the University of Texas Southwestern Protein Chemistry Technology Center using high-performance liquid chromatography/tandem mass spectrometry (HPLC/MS/MS).
Purification of endogenous tubulin for MS analysis
hTert1-RPE cells were harvested, washed twice in PBS plus cytochalasin B at 20 μg/ml, KHMD (78 mM KCl, 50 mM HEPES, pH 7.0, 4 mM MgCl2, 2 mM EGTA, 1 mM DTT, cytochalasin B at 20 μg/ml) once, and resuspended in KHMD plus protease inhibitors (leupeptin/pepstatin/chymostatin, 1μg/ml). Cells were Dounce-homogenized, and the extract was cleared by ultracentrifugation at 38,000 rpm for 15 min. All steps were carried out at 4°C unless otherwise noted. MT polymerization reactions were carried out in the presence of 10 μM Taxol at 33°C for 30 min. Polymerization reactions were layered onto a 50% wt/vol sucrose/KHMD cushion supplemented with 10 μM Taxol. Layered reactions were centrifuged at 39,000 rpm for 2 h in a TLS-55 (Beckman Coulter, Brea, CA) swinging bucket rotor. Pellets were washed twice in KHMD buffer, resuspended in Laemmli sample buffer, and resolved on 8% Nupage Bis-Tris gel (Invitrogen). Tubulin bands from Coomassie Blue–stained gel were excised and subjected to HPLC/MS/MS analysis.
Cell culture and siRNA depletions
HeLa and 293T cells were grown in DMEM with 10% fetal bovine serum. Plasmids and siRNA oligonucleotides were transfected using Effectene (Qiagen) and calcium phosphate, respectively. Cells were analyzed 72 h after transfection. The siRNA oligonucleotides A and B (sense/antisense), which were synthesized by GenePharma (Shanghai, China) are GCUACAAUGACAAGUUCUATT/UAGAACUUGUCAUUGUAGCTG and GCAAUGAGUCGGCAAUUGATT/UCAAUUGCCGACUCAUUGCTG, respectively.
In vitro acetylation by San
The assay was carried out in the acetylation buffer containing His6-San at 50 ng/ml, tubulin proteins at 200 ng/ml, and 100 μM 14C-acetyl-CoA (Perkin Elmer-Cetus, Waltham, MA). After a 1-h incubation at 37°C, Laemmli sample buffer was added to terminate the reaction. Acetylation was detected by exposing the SDS–PAGE gel to a phosphorimage screen. To compare the activity of San toward tubulins and MTs, bovine tubulins at 2.5 mg/ml (Cytoskeleton, Denver, CO) were polymerized in the presence of 1 mM GTP and 10% dimethyl sulfoxide at 37°C for 40 min. The mixture was divided into two aliquots, followed by centrifugation at 15,000 × g for 5 min at room temperature. One of the pellets was resuspended in cold BRB80 (80 mM PIPES, pH 6.8, 1 mM MgCl2, 5 mM EGTA) and incubated on ice for 10 min (tubulins). The other was resuspended in warm BRB80 plus 10 μM Taxol (MTs). Both preparations were then subjected to in vitro acetylation. To test whether San acetylates K40 on α-tubulin, bovine tubulins at 200 ng/ml were incubated with His6-SirT2 at 400 ng/ml and 1 mM β-NAD in acetylation buffer at 37°C for 4 h. These tubulins were next subjected to in vitro acetylation in the presence or absence of San and 5 mM NAM.
Protein turnover of β-tubulins
HeLa cells were transfected with flag-tagged β-tubulin constructs. After 48 h, cells were treated with cycloheximide at 100 ng/ml for up to 6 h and harvested at various time points with Laemmli sample buffer. The protein level of flag-tagged β-tubulin was analyzed by immunoblot using an anti-flag antibody.
MT regrowth assay
Cells seeded on coverslips were put on ice for 30 min to depolymerize MTs. Coverslips were then incubated in prewarmed medium at 37°C for the indicated time. After fixation by cold methanol, immunofluorescence staining was performed as previously described (Hou et al., 2007). To remove soluble tubulins, cells were extracted in extraction buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 5 mM EGTA, 0.5% Triton X-100) for 30 s prior to fixation. To block protein synthesis during the regrowth, cycloheximide at 100 ng/ml was added to prewarmed medium.
Epifluorescence microscopy
The analysis was performed on an Axio Observer Z1 microscope. (Carl Zeiss, Thornwood, NY). A Plan-Apochromat 63×/1.40 Oil DIC M27 objective was used, and images were acquired with an AxioCam MRm rev. 1–3 camera controlled by AxioVision software (Carl Zeiss, Thornwood, NY). Gamma adjustment and necessary cropping were performed in Photoshop CS2 (Adobe, San Jose, CA).
Supplementary Material
Acknowledgments
We thank Luke Rice for critical reading and helpful discussion of the manuscript. We also thank Zhijian (James) Chen for helping to map the in vivo acetylation sites on β-tubulin. This work was supported by a grant (R01CA126832) from the National Institutes of Health (NIH) to Y. Zhao and by a grant (R01GM081466) from the NIH and a grant (I-1594) from the Welch Foundation to H. Zou.
Abbreviations used:
- K40
lysine 40
- K252
lysine 252
- K324
lysine 324
- MT
microtubule
- NAM
nicotinamide
- TAT
tubulin acetyltransferase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0203) on December 22, 2010.
REFERENCES
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Creppe C, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of a-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- Dai J, Kateneva AV, Higgins JM. Studies of haspin-depleted cells reveal that spindle-pole integrity in mitosis requires chromosome cohesion. J Cell Sci. 2009;122:4168–4176. doi: 10.1242/jcs.054122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evjenth R, Hole K, Karlsen OA, Ziegler M, Arnesen T, Lillehaug JR. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J Biol Chem. 2009;284:31122–31129. doi: 10.1074/jbc.M109.001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, Job D. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol Biol Cell. 2006;17:1041–1050. doi: 10.1091/mbc.E05-07-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Wloga D. Ciliary tubulin and its post-translational modifications. Curr Top Dev Biol. 2008;85:83–113. doi: 10.1016/S0070-2153(08)00804-1. [DOI] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M. The 4 A X-ray structure of a tubulin:stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Chu CW, Kong X, Yokomori K, Zou H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J Cell Biol. 2007;177:587–597. doi: 10.1083/jcb.200701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Jourdain L, Curmi P, Sobel A, Pantaloni D, Carlier MF. Stathmin: a tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry. 1997;36:10817–10821. doi: 10.1021/bi971491b. [DOI] [PubMed] [Google Scholar]
- Keays DA, et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc Natl Acad Sci USA. 1987;84:5720–5724. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- McKeithan TW, Rosenbaum JL. Multiple forms of tubulin in the cytoskeletal and flagellar microtubules of Polytomella. J Cell Biol. 1981;91:352–360. doi: 10.1083/jcb.91.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci USA. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scroggins BT, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends in Cell Biol. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.