Here we address how Pax3 regulates stem cell maintenance and neurogenesis during caudal neural tube development. Pax3 acetylation at lysine residues K437 and K475 results in down-regulation of Hes1 and up-regulation of Neurog2 expression.
Abstract
Pax3 plays a role in regulating Hes1 and Neurog2 activity and thereby stem cell maintenance and neurogenesis. A mechanism for Pax3 regulation of these two opposing events, during caudal neural tube development, is examined in this study. Pax3 acetylation on C-terminal lysine residues K437 and K475 may be critical for proper regulation of Hes1 and Neurog2. Removal of these lysine residues increased Hes1 but decreased Neurog2 promoter activity. SIRT1 deacetylase may be a key component in regulating Pax3 acetylation. Chromatin immunoprecipitation assays showed that SIRT1 is associated with Hes1 and Neurog2 promoters during murine embryonic caudal neural tube development at E9.5, but not at E12.5. Overexpression of SIRT1 decreased Pax3 acetylation, Neurog2 and Brn3a positive staining. Conversely, siRNA-mediated silencing of SIRT1 increased these factors. These studies suggest that Pax3 acetylation results in decreased Hes1 and increased Neurog2 activity, thereby promoting sensory neuron differentiation.
INTRODUCTION
Pax3, a transcription factor and multifunctional regulatory protein, is expressed early in embryogenesis (Chalepakis and Gruss, 1995; Goulding et al., 1991, 1993; Epstein et al., 1991, 1993). In the nervous system, Pax3 is involved in neural tube closure (Li et al., 1999), neural crest development, and peripheral neuron differentiation (Goulding et al., 1991; Koblar et al., 1999). In muscle development, Pax3 ensures the survival of myogenic progenitor cells (Relaix et al., 2005) with Pax3-expressing progenitors giving rise to both skeletal and smooth muscle cells (Esner et al., 2006). Pax3 is also actively involved in adult regenerative myogenesis (Kuang et al., 2006).
Pax3 and its isoforms regulate downstream target genes (Mayanil et al., 2000, 2001, 2006; Lang et al., 2005; Wang et al., 2006; Nakazaki et al., 2008). In a previous paper, we demonstrated that Pax3 regulates two basic helix-loop-helix (bHLH) transcription factor genes, Hairy and enhancer of split homolog-1 (Hes1) and Neurogenin2 (Neurog2) (Nakazaki et al., 2008), by binding to cis-regulatory elements on their promoters. Hes1 and other Hes genes are important for neural stem cell maintenance (Shen et al., 2004; Theriault et al., 2005). Inactivation of Hes1 accelerates early neurogenesis and decreases the number of late-born neurons (Nakamura et al., 2000). Neurog2 plays a role in the acquisition of pan-neuronal properties (Scardigli et al., 2001; Sun et al., 2001; Lee and Paff, 2003), specification of neuronal subtypes (Ross et al., 2003, Schuurmans et al., 2004) sensory neurogenesis (Lo et al., 2002), and neural crest cell neurogenesis (Theriault et al., 2005).
In Splotch mutant mice, which do not express functional Pax3, neural crest cells undergo premature neurogenesis, potentially due to changes in regulation of bHLH transcription factors implicated in proliferation and differentiation (Nakazaki et al., 2008). Reeves et al. (1999) found that Pax3 expression is essential in maintaining the undifferentiated phenotype of immature ND7 cells, a rat dorsal root ganglion and mouse neuroblastoma hybrid cell line. In the absence of Pax3, these cells acquire many of the characteristics of mature neuronal cells. Overall, Pax3 appears to regulate stem cell maintenance and proliferation early in development, while initiating differentiation at a later time point. A question that arises from these observations is how Pax3 regulates two opposing bHLH transcription factors, Hes1 and Neurog2.
Many transcription factor functions are controlled by acetylation (Bannister and Miska, 2000; Das and Kundu, 2005). For instance, GATA-1 activity is regulated by p300-mediated acetylation (Boyes et al., 1998). Other well-characterized targets of non–histone protein acetylation include important cellular factors, such as p53, nuclear factor-κB (NF-κB), p65, CBP, p300, STAT3, tubulin, PC4, nuclear receptors, c-Myc, hypoxia-inducible factor (HIF)-1a, FoxO1, heat-shock protein (Hsp)-90, HMG, E2F, MyoD, Bcr–Abl, the FLT3 kinase, and c-Raf kinase (Glozak et al., 2005; Yang and Seto 2008). Pax3 acetylation could also potentially regulate Hes1 and Neurog2, such that Hes1 levels decrease and Neurog2 levels increase when neurogenesis begins.
In this communication, we investigated Pax3 acetylation and its potential role in neuronal differentiation. We also examined whether acetylated Pax3 is a substrate for SIRT1 deacetylase. SIRT1 expression patterns in developing mouse embryos (Sakamoto et al., 2004) suggest that it may be involved in neurogenesis (Libert et al., 2008). Comparable expression patterns of Hes1 and SIRT1 suggest that the two proteins coordinate the process of neural development. SIRT1 binds to HES1 and HEY2 and enhances their transcriptional repression activities (Takata and Ishikawa, 2003). Our results support the hypothesis that acetylated Pax3 decreases Hes1 activity and promotes differentiation by increasing Neurog2 activity. Two C-terminal lysine residues, K437 and K475, may be critical in this process. Furthermore, acetylated Pax3 is a substrate for SIRT1 deacetylase, which associates with chromatin structures on Hes1 and Neurog2 promoters.
RESULTS
Acetylated Pax3 is a substrate for SIRT1
Previously, we demonstrated an interaction between acetylated Pax3 and the histone deacetylase HDAC1. We also noted occupancy by acetylated proteins on some TGFβ2 regulatory elements used by Pax3 (Mayanil et al., 2006). In the present work, we took a closer look at the relationship between Pax3 and SIRT1 at Hes1 and Neurog2 promoters. SIRT1 is involved in stem cell maintenance and differentiation during embryonic development, (Kim et al., 2007; Han et al., 2008), and it associates with HES1 (Takata and Ishikawa, 2003). Because Pax3 regulates Hes1 and Neurog2 (Nakazaki et al., 2008), we surmised that an interaction between Pax3 and SIRT1 could be relevant in stem cell maintenance and differentiation. We hypothesized that 1) Pax3 is acetylated and acetylated Pax3 is a substrate for SIRT1 deacetylase and 2) acetylated Pax3 decreases stem cell proliferation by decreasing Hes1 activity and promotes differentiation by increasing Neurog2 activity.
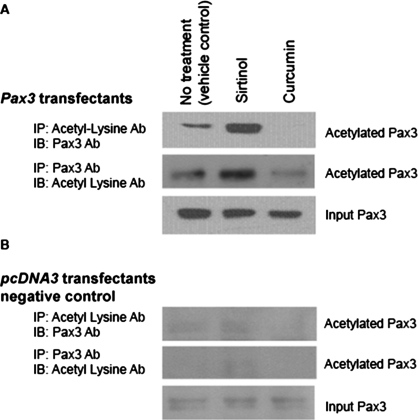
To examine Pax3 acetylation, DAOY cells transfected with Pax3 cDNA in pcDNA3 or pcDNA3 vector control were treated with sirtinol, a SIRT1 inhibitor (Araki et al., 2004) and curcumin (p300 acetyl transferase inhibitor) (Balasubramanyam et al., 2004). Cell lysates from treated cells were immunoprecipitated with anti–acetyl-lysine antibody and immunoblotted with Pax3 polyclonal antibody or vice versa (Figure 1, A and B). Immunoblot results showed low levels of acetylated Pax3 in nontreated controls and significant acetylated Pax3 in the presence of sirtinol but not in the presence of curcumin. These studies suggested that Pax3 is acetylated and a possible SIRT1 substrate.
FIGURE 1:
Pax3 acetylation. (A) DAOY cells transfected with Pax3-pcDNA3 expression plasmid were either not treated (vehicle control) or treated with sirtinol (100 μM) or curcumin (100 μM) for 4 h. Total cell lysates were immunoprecipitated with acetyl-lysine antibody and immunoblotted with Pax3 mAb, or immunoprecipitated with Pax3 mAb and immunoblotted with acetyl lysine polyclonal antibody. Pax3 input samples were immunoblotted to confirm the presence of Pax3. (B) DAOY cells transfected with pcDNA3 vector were used as negative Pax3 controls. These cells were treated as above. Minimal endogenous Pax3 levels, depicted in the input samples, were observed. Each experiment was performed in quadruplicate.
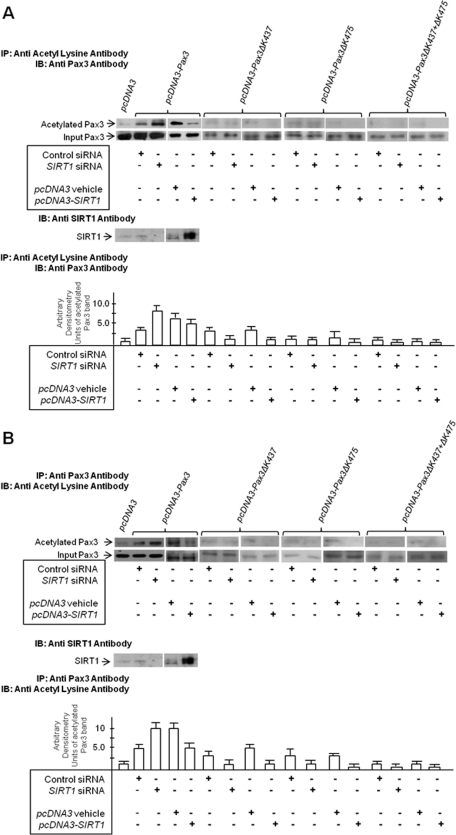
To confirm that acetylated Pax3 is a SIRT1 substrate, DAOY cells were cotransfected with pcDNA3-Pax3 expression construct and SIRT1 siRNA, control siRNA, SIRT1 expression plasmid, or pcDNA3 control plasmid (Figure 2, A and B). Cell extracts were prepared, immunoprecipitated with anti–acetyl-lysine antibody, and immunoblotted with Pax3 antibody (Figure 2A) or vice versa (Figure 2B). In the presence of SIRT1 siRNA, compared with control siRNA, Pax3 acetylation was more pronounced. When SIRT1 cDNA was cotransfected with Pax3 expression plasmid, Pax3 acetylation was reduced, compared with pcDNA3 control plasmid. This confirmed that Pax3 is acetylated and a substrate for SIRT1. SIRT1 cDNA did not completely abolish Pax3 acetylation; therefore, some acetylated lysine residues on Pax3 may not serve as SIRT1 substrates.
FIGURE 2:
Acetylation of C-terminal lysine residues, K437 and K475, is SIRT1-sensitive. DAOY cells transfected with the following expression plasmids: pcDNA3 vector control, Pax3-pcDNA3, pcDNA3-Pax3ΔK437, pcDNA3-Pax3ΔK475, or pcDNA3-Pax3ΔK437+ΔK475, were cotransfected with SIRT1 cDNA in pcDNA3, pcDNA3 vehicle, SIRT1 siRNA, or control scrambled siRNA. At 48 h posttransfection, total cell lysates were tested for the presence of SIRT1. Lysates were immunoprecipitated (IP) with acetyl-lysine polyclonal antibody and immunoblotted (IB) with Pax3 polyclonal antibody as shown in (A) or immunoprecipitated with Pax3 antibody and immunoblotted with acetyl lysine antibody as shown in (B). Bands were quantified by densitometry and the band intensity (ratio of immunoprecipitated band intensity to input Pax3 band intensity) expressed as arbitrary densitometry units. Results are presented as arbitrary densitometry units + SEM. Each experiment was conducted in quadruplicate and each data point in duplicate.
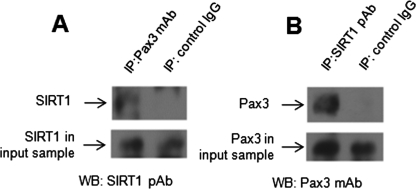
To show a direct interaction between Pax3 and SIRT1, coimmunoprecipitation assays were performed using ND7 cell lysates. Pax3 monoclonal antibody (mAb) was used for immunoprecipitation and SIRT1 polyclonal antibody for immunoblotting and vice versa. Mouse and rabbit immunoglobulin G (IgG) were used as negative controls. Figure 3 shows that control IgGs did not immunoprecipitate Pax3 or SIRT1. Pax3 mAb was able to immunoprecipitate SIRT1 (Figure 3A), and SIRT1 polyclonal antibody could immunoprecipitate Pax3 (Figure 3B). These studies show a direct interaction between Pax3 and SIRT1 in ND7 cells.
FIGURE 3:
Pax3 interacts directly with SIRT1. A total of 200 μg (total protein) from undifferentiated ND7 cell lysates was used for immunoprecipitation assays. (A) Anti-Pax3 mAb and mouse IgG control were used for immunoprecipitation. The immune complex was pulled down with Protein A-sepharose and immunoblotted using anti-SIRT1 rabbit polyclonal antibody (pAb). (B) Anti-SIRT1 rabbit pAb and rabbit IgG control were used for immunoprecipitation. The immune complex was pulled down with Protein A-sepharose and immunoblotted using anti-Pax3 mAb. For panels A and B, input samples were run in separate lanes to verify SIRT1 and Pax3 levels, respectively.
Acetylation of C-terminal lysine residues, K437 and K475, is SIRT1-sensitive
The C-terminal region (amino acids 298–481) of Pax3 (red brackets in Supplemental Figure S1) contains two lysine residues: K437 and K475 (underlined in Supplemental Figure S1). These lysine residues undergo ubiquitination and are responsible for Pax3 degradation (Boutet et al., 2007). Because the same lysine residues that get ubiquitinated may compete for acetylation (Benkirane et al., 2010), we hypothesized that these lysine residues are acetylated and their deacetylation is SIRT1-sensitive.
To ascertain whether K437 and K475 play a role in SIRT1-sensitive Pax3 acetylation, we cotransfected wild-type Pax3 (pcDNA3-Pax3) or Pax3 lysine deletion mutants (pcDNA3-Pax3ΔK437, pcDNA3-Pax3ΔK475, or pcDNA3-Pax3 ΔK437+ΔK475) in combination with SIRT1-siRNA or SIRT1-cDNA into DAOY cells. Appropriate scrambled siRNA-negative controls and control cDNA were also used. At 48 h post-transfection, the cells were lysed, immunoprecipitation assays were performed with anti–acetyl lysine rabbit polyclonal antibody, and immunoblots were done with anti-Pax3 mouse mAb and vice versa (Figure 2, A and B). The results showed that K437 and K475 single-deletion mutants were not immunoprecipitated with acetyl lysine antibody or anti-Pax3 antibody. Trace amounts of Pax3 were precipitated from control siRNA and SIRT1-siRNA cotransfected cells. With K437 and K475 double mutants, acetyl lysine antibody did not precipitate Pax3, nor did acetyl lysine antibody detect Pax3 immunoprecipitated with Pax3 antibody. The data support the conclusion that Pax3 lysine residues K437 and K475 are acetylated and that these residues can be deacetylated by SIRT1.
C-terminal lysine residues, K437 and K475, regulate Hes1 and Neurog2 promoter activity
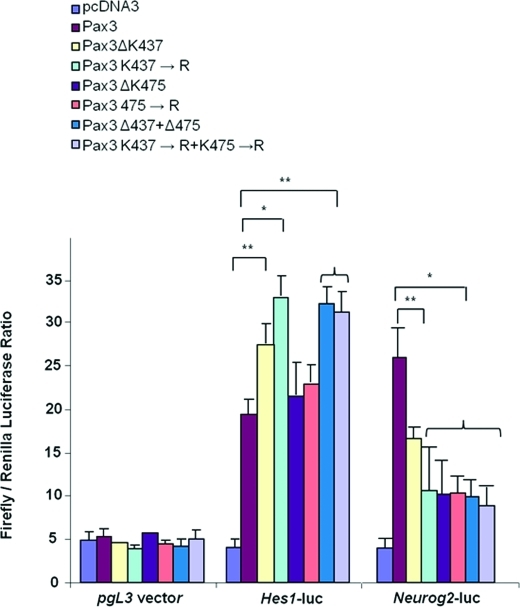
Because K437 and K475 undergo acetylation and are deacetylated by SIRT1, we hypothesized that they may be involved in regulating Hes1 and Neurog2 promoter activities. To test the hypothesis that K437 and K475 are important in Pax3 regulation of Hes1 and Neurog2, we cotransfected Hes1- and Neurog2-promoter-luciferase reporter constructs with pcDNA3 vector control, wild-type Pax3, Pax3 deletion mutants (Pax3ΔK437, Pax3ΔK475, Pax3ΔK437 + Pax3ΔK475), or Pax3 substitution mutants (Pax3K437→R; Pax3K475→R, Pax3K437→R+Pax3K475→R). Vector pgL3 was used as a negative control. The results showed that Hes1 and Neurog2 promoter activity significantly increased (p < 0.001) in the presence of wild-type Pax3, compared with pcDNA3 transfected control (Figure 4).
FIGURE 4:
C-terminal lysine residues K437 and K475 in Pax3 regulate Hes1 and Neurog2 promoter activity. Hes1-promoter and Neurog2-promoter activity in a transient cotransfection experiment using Hes1- and Neurog2-promoter luciferase and wild-type Pax3 expression construct or mutated Pax3-pcDNA3 expression constructs as follows: Pax3ΔK437, Pax3K437→R, Pax3ΔK475, Pax3K475→R, Pax3ΔK437+Δ475, Pax3K437→R+Pax3K475→R. Each experiment was done in quadruplicate and each data point in triplicate. *, p < 0.001; **, p < 0.05 (Student’s t test).
Compared with wild-type Pax3, Hes1 promoter activity increased significantly (p < 0.001) when K437 was deleted or substituted with arginine, but not when K475 was deleted or substituted with arginine. The double-lysine deletion or substitution mutants increased Hes1 promoter activity similar to levels seen with the single K437 deletion mutant (p < 0.001). For the Neurog2 promoter, K437 and K475 single or double deletions, or substitution mutants, decreased promoter activity compared with wild-type Pax3. These data suggest that K437 deacetylation increases Hes1 activity, whereas K437 and K475 deacetylation decreases Neurog2 activity.
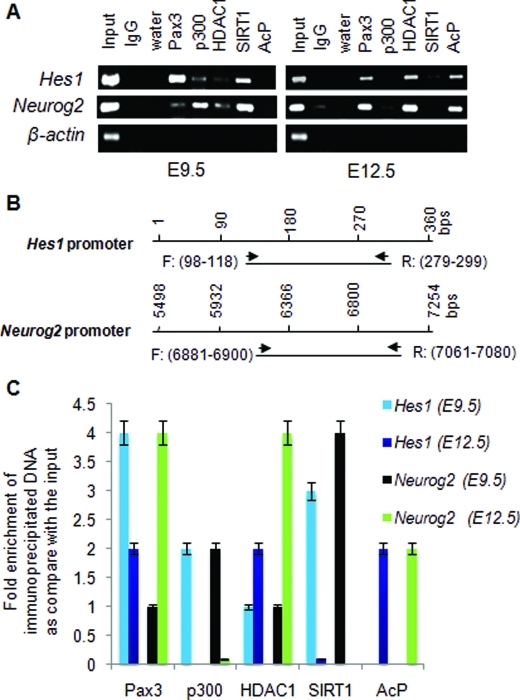
SIRT1 is associated with Hes1 and Neurog2 promoters in the developing caudal neural tube at E9.5
If deacetylation influences Hes1 and Neurog2 levels in one direction, acetylation could bring about opposite outcomes. We evaluated the association of deacetylases and acetylated proteins with chromatin structures on Hes1 and Neurog2 promoters. Chromatin immunoprecipitation assays were performed using mouse E9.5 and E12.5 caudal neural tubes. Hes1 expression between E8.5 and E10.5 is important for neural tube closure (Ishibashi et al., 1995; Nakazaki et al., 2008). Neurog2, a marker of sensory neurogenesis, is present at E12.5 (Simmons et al., 2001; Ribes et al., 2008). At E9.5, there was at least threefold higher Pax3 immunoprecipitable Hes1 than the Neurog2 (Figure 5A). Conversely, more Pax3 immunoprecipitable Neurog2 than Hes1 was observed at E12.5 (Figure 5A). At E9.5, both Hes1 and Neurog2 promoters were associated with SIRT1 deacetylase. At E12.5, both promoters were associated with acetylated proteins. Additionally, p300 was present at E9.5, but not E12.5, on both promoters. HDAC1 was nondifferentially associated with these promoters at either time point. These data indicate that 1) SIRT1 is associated with Hes1 and Neurog2 at E9.5 stage of neural tube development and 2) acetylated proteins are associated with Hes1 and Neurog2 promoters at E12.5. The presence of SIRT1 and the absence of acetylated proteins on the Hes1 promoter at E9.5 may increase stem cell proliferation and maintenance at this earlier time point, whereas these conditions on the Neurog2 promoter may decrease differentiation. The presence of acetylated proteins at a later time point, E12.5, may up-regulate Neurog2 and influence differentiation. Western blot data showing that Hes1 and SIRT1 expression decrease in ND7 cells upon differentiation (Supplemental Figure S2) support this conclusion.
FIGURE 5:
SIRT1 binds to Hes1 and Neurog2 promoters from E9.5, but not E12.5, mouse caudal neural tube. (A) ChIP assays were done with E9.5 and E12.5 mouse lumbar neural tube. ChIP compatible antibodies against Pax3, p300, HDAC1, SIRT1, and acetylated protein (AcP) were used to immunoprecipitate (IP) the protein–DNA complex. IgG was used as an IP-negative control. Murine β-actin primers were used as negative loading control. Amplified product was present only in the input and not in the control IgG or the immunoprecipitate. (B) The 200-bp amplified products using Hes1 and Neurog2 promoter primer sets are shown. Each ChIP experiment was performed in triplicate using one lumbar neural tube region per ChIP assay with a total of n = 4. (C) Immunoprecipitated DNA was subjected to quantitative PCR using murine primers for Hes1 and Neurog2 promoters. The data represent fold enrichment of immunoprecipitated DNA compared with input sample. Each ChIP experiment was performed in triplicate using one lumbar neural tube region per ChIP assay with a total n = 4.
Acetylated Pax3 binds to Hes1 and Neurog2 promoter
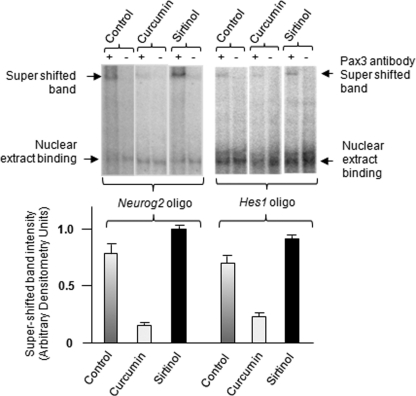
To confirm binding of acetylated Pax3 to Hes1 and Neurog2 promoters, we performed electromobility shift assay (EMSA) as described in Nakazaki et al. (2008), using ND7 cell nuclear extracts. ND7 cells (a generous gift from M. Calissano, Institute of Child Health London) were obtained by fusing neonatal rat (E12) dorsal root ganglion neurons with mouse neuroblastoma cells (Latchman and Polak, 1995). Figure 6 shows that Neurog2 and Hes1 promoter oligonucleotides (30mer) bind Pax3 (control) as evident by the supershifted band in the presence of Pax3 antibody. Supershifted band intensity was stronger in nuclear extracts from ND7 cells treated with sirtinol (SIRT1 inhibitor) and minimal in nuclear extracts from ND7 cells treated with curcumin (acetyltransferase inhibitor) (Figure 6). Pax3 binding seen in control cells may be due to a mixed population of acetylated and nonacetylated Pax3. These observations clearly demonstrate that acetylated Pax3 binds Neurog2 and Hes1 promoters.
FIGURE 6:
Identification of Neurog2 and Hes1 promoters as biological targets of acetylated Pax3. (A) EMSA binding reactions were performed with 32P-labeled double-stranded oligonucleotides, Neurog2 oligo: 5′- gacaccgtgctcggttccgggctgcgggga-3′ and Hes1oligo: 5′-cattggccgccagaccttgtgcctagcggc-3′, in the absence or presence of 100-fold molar excess of unlabeled oligonucleotide. Arrows indicate shifted and super-shifted bands. Nuclear extracts were from nontreated ND7 control cells in DMEM or from cells treated with curcumin (100 μM) or sirtinol (100 μM) for 4 h. Supershifted band intensity standardized to the control is shown in the histogram (average of three experiments ± SEM)
SIRT1 overexpression decreases sensory neurogenesis in ND7 cells
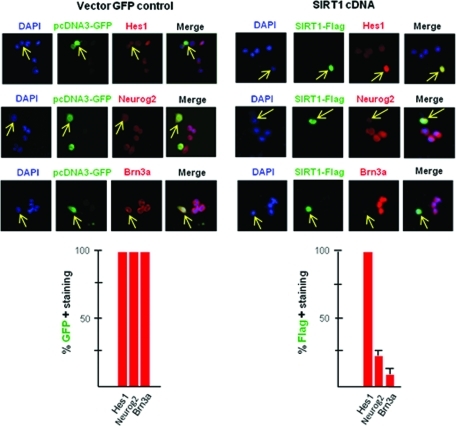
To further examine the role of SIRT1 in cell proliferation and sensory neurogenesis, SIRT1 was overexpressed in ND7 cells. Proliferating ND7 cells were cultured in DMEM containing 10% fetal bovine serum and 10% donor horse serum. These hybrid cells were transfected with pcDNA3-GFP (vector control), or SIRT1-flag-cDNA in pcDNA3-GFP (Figure 7), or scrambled negative control siRNA-FITC or SIRT1 siRNA (Supplemental Figure S3). At 48 h posttransfection, the cells were placed in growth media containing 20% serum and allowed to grow for 4 more days, after which they were immunostained for SIRT1, Hes1, Neurgo2, and Brn3a (Figure 7 and Supplemental Figure S3). SIRT1 overexpression increased Hes1 expression and decreased Neurog2 and Brn3a expression. Silencing SIRT1 with siRNA decreased Hes1 expression and increased Neurog2 and Brn3a immunostaining. This is in agreement with other studies showing an association between SIRT1 downregulation and neurogenesis (Prozorovski et al., 2008).
FIGURE 7:
Overexpression of SIRT1 decreases Neurog2 and Brn3a expression. ND7 cells were transfected with pcDNA3-GFP or SIRT1-flag-cDNA. The cells were fixed and immunostained for Hes1, Neurog2, and Brn3a (sensory neuron marker) and Flag to ascertain SIRT1 expression in SIRT1-flag-cDNA transfected cells. Yellow arrows show that pcDNA3-GFP transfection did not change Hes1, Neurog2, or Brn3a expression in transfected cells. SIRT1-flag-cDNA transfection increased Hes1 expression and decreased Neurog2 and Brn3a expression in transfected cells. The histograms show an average ± SEM of three separate experiments.
SIRT1 overexpression increases Hes1 and decreases Neurog2 expression in chick embryos
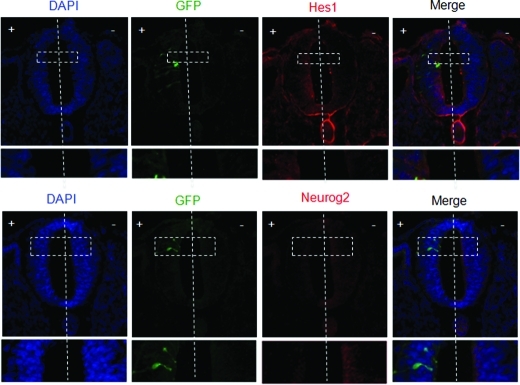
To further examine SIRT1 regulation of Hes1 and Neurog2 expression in vivo, SIRT1 was overexpressed in chick embryos. Chick embryos at HH10–11 were electroporated with SIRT1 cDNA on one side of the neural tube, with the opposite side serving as a control. Electroporated embryos were incubated at 38°C until HH15–16, at which time the neural tubes were harvested and stained for Hes1 and Neurog2. Analysis of sectioned embryos demonstrated that Hes1 expression increased, whereas Neurog2 expression decreased on the side of SIRT1 cDNA electroporation (Figure 8). These results suggest that, because Neurog2 is a proneural transcription factor, SIRT1 overexpression would affect neurogenesis adversely.
FIGURE 8:
Regulation of Hes1 and Neurog2 expression by SIRT1 overexpression in vivo. (A) Murine SIRT1 cDNA and GFP containing plasmids were coelectroporated into the + side of chick embryonic neural tubes, and the tubes stained with anti-Hes1 and anti-Neurog2 antibodies. GFP expression was used as a positive control for electroporation. The vertical line demarcates the two sides. SIRT1 cDNA electroporation increased Hes1 and decreased Neurog2 immunostaining as shown in the inset (n = 6).
DISCUSSION
Many transcription factors are controlled by posttranslational modifications. Understanding how posttranslational modifications affect transcription factor function will answer critical questions about the mechanisms through which these molecules regulate key developmental processes (Singh et al., 2010). Miller et al. (2008) showed that Pax3 is phosphorylated at serine 205 in proliferating cells but not in differentiating cells. In the present study, we suggest that Pax3 is a member of the group of acetylated transcription factors and that regulation of Pax3 acetylation plays a role in the switch-over from stem cell proliferation to differentiation in caudal neural tube development.
Our results showed the following: 1) Pax3 is acetylated at lysine residues K437 and K475, and these residues are substrates for SIRT1 deacetylase. 2) K437 is involved in Hes1 regulation; deletion or substitution of this residue to arginine up-regulates Hes1 promoter-luciferase activity. 3) K437 and K475 are involved in regulating Neurog2; deleting or substituting these residues to arginine down-regulates Neurog2 promoter-luciferase activity. 4) SIRT1 over-expression results in increased Hes1 expression and decreased Neurog2 in vivo (chick embryos) and also in nondifferentiating ND7 cells. Similarly, siRNA-mediated silencing of SIRT1 decreases Hes1 but increases Neurog2 and Brn3a expression in ND7 cells. 5) SIRT1 associates with Hes1 and Neurog2 in murine embryonic caudal neural tube at E9.5, suggesting that nonacetylated Pax3 may be responsible for regulating Hes1 during this stage. SIRT1 does not associate with Hes1 or Neurog2 promoters in mouse embryonic caudal neural tube at E12.5. This suggests that acetylated protein(s) which are substrates of SIRT1 may be contributing to decreased stem cell proliferative activities and increased neurogenic activity, in particular acetylated Pax3 may down-regulate Hes1 and therefore stem cell proliferation and up-regulate Neurog2 to promote neurogenesis. Lysine residues K437 and K475 may play a role in the switch from stem cell proliferation to differentiation.
In these studies SIRT1 overexpression decreased Neurog2 and Brn3a (sensory neurogenesis markers) and increased Hes1 expression in nondifferentiated ND7 cells. Hisahara et al. (2008) reported that SIRT1 enhanced neuronal differentiation and decreased Hes1 expression in neural precursor cells. These results were observed under differentiating conditions during which SIRT1 transiently translocated into the nucleus from the cytoplasm. Our experiments were not performed under differentiating conditions; therefore it is unlikely that all SIRT1 translocated to the nucleus. Non-nuclear or cytoplasmic SIRT1 would deacetylate only transcription factors that are en route to the nucleus, whereas nuclear SIRT1 may deacetylate histones in addition to transcription factors. Prozorovski et al. (2008) suggested that SIRT1 participated in oxidation-mediated suppression of neurogenesis. It is possible that SIRT might play a distinct role on differentiation under different conditions and potentially in different cells. Hisahara et al. (2008) and Prozorovski et al. (2008) were working with murine embryonic brain cells, whereas our lab is working with murine embryonic caudal neural cells. Further work is needed to dissect these differences.
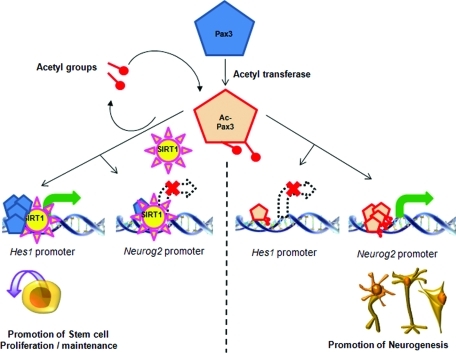
Based on our work, a hypothetical model for the interaction of Pax3 and SIRT1 is proposed in Figure 9. In this model Pax3, acetylated by acetyltransferase, interacts with Hes1 and Neurog2 promoters. In the presence of SIRT1, as seen on these promoters at E9.5 (see Figure 5), Pax3 is deacetylated. This results in up-regulation of Hes1 and promotes stem cell proliferation and maintenance. Neurog2 is not expressed in the presence of deacetylated Pax3. At E12.5, when SIRT1 does not associate with Hes1 and Neurog2 promoters, acetylated Pax3 down-regulates Hes1 and up-regulates Neurog2 promoter activities. This promotes neurogenesis and reduces stem cell proliferation/maintenance. Although this model is overly simplistic, it highlights the possible role of acetylated Pax3 as a switch involved in regulating stem cell proliferation/maintenance and sensory neuronal differentiation.
FIGURE 9:
Hypothetical model for the regulation of Hes1 and Neurog2 via Pax3 and SIRT1. Pax3 is acetylated by acetyltransferase; acetylated Pax3 binds to Hes1 and Neurog2 promoters. When SIRT1 is associated with Hes1 promoter, deacetylated Pax3 up-regulates Hes1 expression. When SIRT1 is associated with Neurog2 promoter, deacetylated Pax3 does not up-regulate Neurog2. The overall effect is stem cell proliferation and maintenance. When SIRT1 is not associated with these promoters, acetylated Pax3 binds the promoters resulting in down-regulation of Hes1 and up-regulation of Neurog2. The overall effect is promotion of neurogenesis.
In a recent study, we showed up-regulation of several miRNAs during early cell proliferation stages in Pax3 mutant “Splotch” homozygous embryos, which do not express functional Pax3 (Ichi et al., 2010). Some of these miRNAs target the H3K27 histone demethylase KDM6B, decreasing KDM6B expression and increasing H3K27 methylation. We also reported that association between methylated H3K27 and Hes1 promoter resulted in decreased Hes1 expression and lowered stem cell proliferation. At the same time, we observed low association between methylated H3K27 and Neurog2 promoter but high association between acetylated H3K9 and H3K18 and Neurog2 promoter. These association patterns of methylated/acetylated histones on Hes1 and Neurog2 promoters could possibly result in premature neurogenesis as observed by Nakazaki et al. (2008). Overall, a complex model of Pax3 regulation of Hes1 and Neurog2 is emerging, in which Pax3 acetylation status along with H3K27 methylation and H3K9 and H3K18 acetylation may all interact to regulate these genes.
Another point at which Pax3 and SIRT1 may interact is at p53. Apoptosis is a component of neurogenesis (Nijhawan et al., 2000). Pani et al. (2002) showed that p53 deficiency rescues not only apoptosis but also neural tube defects in Splotch (Pax3 mutant) embryos. They concluded that Pax3 deficiency increased p53 protein levels. These results suggest that Pax3 regulates neural tube closure by inhibiting p53-dependent apoptosis, rather than by inducing neural tube-specific gene expression. Similarly, Gnc5 acetyltransferase is required for cranial neural tube closure in mouse embryos. Loss of Gnc5 acetyltransferase activity has been linked to neural tube closure defects and exencephaly. Deletion of p53 allows Gnc5−/− embryos to survive longer (Bu et al., 2007). These results indicate that deletion of p53, which is a substrate of SIRT1, rescues neural tube defects in Splotch as well as in Gnc5−/− embryos. Our results showing Pax3 as a SIRT1 substrate suggest that Pax3 could potentially be a substrate for Gnc5 acetyltransferase. Thus a complex interaction may exist between p53, Pax3, SIRT1, and Gnc5 acetyltransferase. An anti-apoptotic role for SIRT1 has been demonstrated for apoptosis induced by p53 (Luo et al., 2001; Vaziri et al., 2001), and this may be another regulatory mechanism involved in neurogenesis. Overall, Pax3, SIRT1, p53, and Gnc5 acetyltransferase and the many transcription factors delineated as regulators of neurogenesis (LaBonne and Bronner-Fraser, 1999) will most likely function in complex regulatory loops that have yet to be elucidated.
MATERIALS AND METHODS
Antibodies and reagents
The following antibodies were used: anti-p300/CBP rabbit polyclonal antibody (Ab2832), anti–acetylated protein rabbit polyclonal antibody (AP) (Ab193), and anti-HDAC1 rabbit polyclonal antibody (Ab7028) from Abcam (Boston, MA); anti-SIRT1 rabbit polyclonal antibody (sc-15404); anti-Hes1 rabbit polyclonal (sc-25392), anti-Neurog2 goat polyclonal (sc-19233), and anti–Lamin B goat polyclonal (sc-6217) from Santa Cruz Biotechnologies (Santa Cruz, CA); anti-Pax3 rabbit polyclonal antibody (CA1010) from EMD Biosciences (Calbiochem; San Diego, CA); anti-Tuj1 mAb from Covance (Princeton, NJ); anti-Brn3a (Ab5945) rabbit polyclonal antibody from Chemicon (Billerica, MA); anti–acetyl-lysine (9441) rabbit polyclonal antibody from Cell Signaling Technology (Beverly, MA); anti-Pax3 mouse mAb from Developmental Studies Hybridoma Bank (Iowa City, IA). Other reagents used: IgG from rabbit serum (I-5006), curcumin (C1386–5G) and EGF (E9644) from Sigma (St. Louis, MO); sirtinol (566321) from EMD Biosciences; bFGF (233-FB) from R&D Systems (Minneapolis, MN); and growth factor reduced Matrigel (35–6230) from BD Biosciences (San Diego, CA).
Removal of embryonic neural tube
The day the vaginal plug was found was noted as E0.5. Embryos were collected at E9.5 and E12.5. Caudal neural tubes were isolated as described in Bennett et al. (1998).
Transfection, immunoprecipitation, immunoblotting, and immunofluorescence
Lipofectamin2000 (11668–027; Invitrogen, Carlsbad, CA) reagent was used for all cotransfection as per the manufacturer’s instructions. siPORT NeoFX Transfection Agent (AM4510; Ambion, Austin, TX) and MegaTran 1.0 (OriGene Technologies TT200002, Rockville, MD) were used for siRNA transfection and cDNA transfection respectively, as per the manufacturers’ instructions. To lyse cells, DAOY or ND7 cells were washed and collected in cold PBS and then lysed on ice for 30 min in cell lysis buffer containing 20 mM Tris-HCl (pH 7.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM sodium vanadate. Protease inhibitor (Sigma P2714) was added just before lysis. Immunoprecipitation was carried out with polyclonal anti–acetylated lysine antibody (9441), polyclonal anti-Pax3 antibody (CA1010), monoclonal anti-Pax3 antibody (Developmental Studies Hybridoma Bank), and polyclonal anti-SIRT1 antibody (sc-15404) as per the manufacturers’ instructions. The immunoprecipitates were collected with 50 μl of 50% slurry of protein A Agarose (Sigma P7786). The precipitates were washed three times in lysis buffer, boiled in 2× SDS sample buffer for 5 min, and centrifuged for 1 min. Proteins in the supernatant were resolved in 8% SDS–PAGE and immunoblotted with antibodies described below. For immunofluorescence, cultured ND7 cells, both nondifferentiated and differentiated, were fixed with 4% paraformaldehyde in PBS and blocked with 10% normal donkey serum/0.1% Triton X-100/0.01% NaAzide in PBS. Cells were then incubated overnight with appropriate antibodies. The next day, cells were incubated with secondary antibodies followed by nuclear staining with DAPI (D8417; Sigma).
Primary antibodies used for immunoblotting included anti-Pax3 rabbit polyclonal (1:1000), anti-SIRT1 rabbit polyclonal (1:1000), anti-Hes1 rabbit polyclonal (1:500), anti–Lamin B goat polyclonal (1:2000), anti–acetyl-lysine rabbit polyclonal (1:500), and anti-Pax3 mouse monoclonal (1:100). Secondary antibodies included anti–rabbit and anti–goat HRP-conjugated antibodies. Antibody binding was revealed by enhanced chemiluminescence detection (Amersham Pharmacia Biotech, Piscataway, NJ). Primary antibodies for immunofluorescence included anti–rabbit SIRT1 primary polyclonal, anti–mouse Brn3a primary monoclonal, and anti–rabbit Brn3a polyclonal. Secondary antibodies were donkey anti–rabbit AlexaFluor 488 (green fluorescence) and donkey anti–mouse Cy3 (red fluorescence).
Chromatin immunoprecipitation (ChIP)
ChIP assays on caudal neural tube from E9.5 and E12.5 embryos were performed as described in Mayanil et al. (2006). Mouse monoclonal Pax3 antibody, rabbit polyclonal p300, HDAC1, SIRT1, and acetylated lysine antibody were used for ChIP. PCR was performed with primers for murine Hes1 and Neurog2 promoter regions (Nakazaki et al., 2008). All ChIP samples were tested for false-positive PCR amplification by sequencing the 200-bp amplified product. False-positive PCR amplification was ruled out by amplifying a sequence from the murine β-actin promoter: forward 5′-gtgggccgccctaggcaccag-3′ and reverse 5′-ctctttgatgtcacgcacgatttc-3′. This sequence failed to give any PCR product in the immunoprecipitates. Primers for PCR amplification of murine Hes1 promoter were as follows: forward (bp98–118) 5′-ttggctgaaagttactgtgg- 3′ and reverse (bp279–299) 5′-tcttagggctacttagtgat-3′ and for murine Neurog2 promoter: forward (bp6881–6900) 5′-ggcagatctgattgttttct-3′ and reverse (bp7061–7080) 5′gcagctcgcccgagtctcgt-3′. A 200-bp fragment was amplified in each case (Nakazaki et al., 2008).
Electromobility shift assays
Probes were prepared for EMSA by annealing complementary oligonucleotides representing selected regions of murine Hes1 and Neurog2 promoters, followed by 5′-end labeling with [γ-32P]ATP by T4 polynucleotide kinase. For EMSA experiments, nondifferentiated ND7 cells were treated with 100 μM of curcumin or sirtinol for 4 h, 0.5 μg of nuclear extract protein was then purified and incubated in 20 μl of reaction mixture containing 15 mM Tris (pH 7.5), 6.5% glycerol, 90 mM KCl, 0.7 mM EDTA, 0.2 mM dithiothreitol, 1 mg/ml bovine serum albumin, 50 μM pyrophosphate, 300 ng of salmon sperm DNA as a competitor to reduce nonspecific DNA binding, and 30,000 dpm per nmol of 32P-labeled double-stranded oligonucleotide probe. EMSA was done as described (Mayanil et al., 2001, 2006; Nakazaki et al., 2008). Double-stranded 30-mer oligonucleotide probes were made for the following regions: Hes1 promoter Hes1oligo: 5′-cattggccgccagaccttgtgcctagcggc-3′ and Neurog2 promoter Neurog2oligo: 5′- gacaccgtgctcggttccgggctgcgggga-3′. For supershift assays 0.1 μg polyclonal Pax3 antibody was preincubated on ice for 30 min with nuclear extract prior to the addition of labeled oligonucleotides. After 20-min incubation, free DNA and DNA protein complexes were resolved in 4% polyacrylamide gels using 0.25× TBE as the running buffer. Electrophoresis was performed at 4°C at 300 mV and 30 mA for 3 h. Gels were dried and subjected to PhosphorImager (Amersham Biosciences) analysis to view shifted bands.
Quantitative real-time PCR
Each 20 μl of real-time (RT)-PCR reaction included 1.25 μl forward primer, 1.25 μl reverse primer, 2.5 μl water, 5 μl immunoprecipitated DNA (1 ng/μl), and 10 μl of the double-stranded DNA binding dye SYBR Green I to detect PCR product (PerfeCTa SYBR Green FastMix, Low ROX [95074–250]; Quanta Biosciences, Gaithersburg, MD). RT-PCR cycle parameters were 95°C for 10 min and 95°C for 10 s, followed by 58°C for 50 s and 72°C for 32 s (7500 Fast Real-Time Thermal Cycler; Applied Biosystems, Foster City, CA). The following murine primers were used: Hes1 forward 5′-ggtcctaacgcagtgtcacctt-3′ and reverse 5′-cagtggcctgaggctctca-3′; Neurog2 (Accession# AF303001) forward 5′-agaggtggcccttgcaatc-3′; and reverse 5′-cacacgccatagtcctctttga-3′; β-actin forward 5′-acggccaggtcatcactattg-3′ and reverse 5′-tggatgccacaggattcca-3′. Primers were designed using Primer Express software (PerkinElmer Life Sciences, Waltham, MA) and synthesized by Eurofins MWG Operon (Huntsville, AL).
Analysis of Hes1 and Neurog2 promoter activities
To investigate the effect of Pax3 C-terminal lysine residues, K437 and K475, on Hes1 and Neurog2 promoter activities, plasmids containing Hes1 promoter-luciferase (−467 to +46, 0.2 μg) or Neurog2-promoter luciferase (1.2-kb fragment; bp5498–7254, 0.2 μg) were transiently cotransfected with Pax3-pcDNA3, pcDNA3 vector control, or different Pax3 lysine mutants into DAOY cells. Luciferase assays were done as described in Mayanil et al. (2006), using the Dual Luciferase kit from Promega (Madison, WI). Renilla luciferase plasmid, pRL-null (5 ng/well), was simultaneously transfected as an insertional control to assess transfection efficiency. Hes1 and Neurog2 promoter activities were evaluated as described in Nakazaki et al. (2008). Hes1 promoter construct Hes1 (−467 to +46) luciferase was provided by R. Kageyama (Takebayashi et al., 1994), and Neurog2 promoter constructs were kindly provided by Jane Johnson (Simmons et al., 2001).
Pax3 mutants were made with the QuickChangeXL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Six mutants were made in a Pax3-pcDNA3 expression construct, three deletion mutants: Pax3ΔK437 (lysine 437 deleted), Pax3ΔK475 (lysine 475 deleted), and Pax3ΔK437ΔK475 (lysine 437 and 475 deleted), and three substitution mutants, with lysine substituted by arginine: Pax3K437→R, Pax3K475→R, and Pax3K437→R+K475→R mutants.
Nuclear extract preparation
Nuclear extracts from ND7 cells were prepared by using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (78833; Pierce Biotechnology, Rockford, IL) as per manufacturer’s instructions.
Chick embryo electroporation and immunohistochemistry
White Leghorn fertilized chicken eggs were purchased from Phil’s Fresh Egg (Forreston, IL) and incubated at 38°C for an appropriate period. Embryos were staged according to Hamburger and Hamilton (1951) and processed as described in Sauka-Spengler and Barembaum (2008). Embryos at HH10–11 were used for in ovo electroporation. Plasmid-encoding SIRT1 (pcDNA3-Sir2α, 2.0 μg/μl, a gift from Wei Gu) together with EGFP (pcDNA3-EGFP, 2.0 μg/μl, addgene 13031) was injected into nearly closed neural tubes, with a PICOSPRITZERIII microinjector (Parker, Cleveland, OH). After injection, a parallel fixed and angled tip electrode (Pomona Electronics, Everett, WA) was placed on both sides of the neural tube and five 30-ms, 18-V electric pulses were applied with an electroporator (TSS20 Ovodyne Electroporator and EP21 Current Amplifier; INTRACEL, Royston, UK). Embryos were then incubated at 38°C until HH15–16, and electroporated neural tubes expressing EGFP were dissected under a Leica MZ16F microscope. Dissected neural tubes were then fixed in 4% PFA/PBS with 0.01% Triton X-100 and embedded in gelatin. Frozen neural tube sections (10 μm) were immunostained with anti–Hes1 and Neurog2 antibodies followed by staining with Cy3-conjugated donkey antibodies (711–165-152 and 705–165-147, respectively; Jackson ImmunoResearch, West Grove, PA). DAPI (Sigma; D8417) was used for counterstaining nuclei. Immunostained sections were observed and photographed with a Leica DMIRB inverted microscope.
Statistical analysis
P values were determined by Student’s t test using the GraphPad Prism Version 5.0.
Supplementary Material
Acknowledgments
We thank Peter Gruss (Max Planck Institute, Gottingen, Germany) for the pBH3.2 plasmid containing Pax3; Wei Gu (Columbia University) for SIRT1 cDNA; M. Calissano (Institute of Child Health, London), for the generous gift of ND7 cells; and R. Kageyama and Jane Johnson for Hes1 and Neurog2 promoter constructs, respectively. This work was supported by the McLone Professorship Fund (CSKM), a State of Illinois Excellence in Academic Medicine Award (CSKM), grants from the Spastic Paralysis Research Foundation of Illinois-Eastern Iowa District of Kiwanis (CSKM and DGM) and the Spina Bifida Association (CSKM).
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- Hes1
hairy and enhancer of split 1
- Neurog2
Neurogenin2
- SIRT1
sirtuin (silent mating type information regulation 2 homologue) 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0541) on December 17, 2010.
REFERENCES
- Araki T, Sasaki Y, Milbrandt J. Nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/non-histone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Miska EA. Regulation of gene expression by transcription factor acetylation. Cell Mol Life Sci. 2000;57:1184–1192. doi: 10.1007/PL00000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Sardet C, Coux O. Lessons from interconnected ubiquitylation and acetylation of p53: think metastable networks. Biochem Soc Trans. 2010;38:98–103. doi: 10.1042/BST0380098. [DOI] [PubMed] [Google Scholar]
- Bennett GD, An J, Craig JC, Gefrides LA, Calvin JA, Finnell RH. Neurulation abnormalities secondary to altered gene expression in neural tube defect susceptible Splotch embryos. Teratology. 1998;57:17–29. doi: 10.1002/(SICI)1096-9926(199801)57:1<17::AID-TERA4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G, Gruss P. Identification of DNA recognition sequences for the Pax3 paired domain. Gene. 1995;162:267–270. doi: 10.1016/0378-1119(95)00345-7. [DOI] [PubMed] [Google Scholar]
- Das C, Kundu TK. Transcriptional regulation by the acetylation of nonhistone proteins in humans—a new target for therapeutics. IUBMB Life. 2005;57:137–148. doi: 10.1080/15216540500090629. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Vekemans M, Gros P. Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell. 1991;67:767–774. doi: 10.1016/0092-8674(91)90071-6. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Vogan KJ, Trasler DG, Gros P. A mutation within intron 3 of the Pax-3 gene produces aberrantly spliced mRNA transcripts in the splotch (Sp) mouse mutant. Proc Natl Acad Sci USA. 1993;90:532–536. doi: 10.1073/pnas.90.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esner M, Meilhac SM, Relaix F, Nicolas JF, Cossu G, Buckingham ME. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133:737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding MD, Sterrer S, Fleming J, Balling R, Nadeau J, Moore KJ, Brown S, Steel KP, Gruss P. Analysis of the Pax-3 gene in the mouse mutant splotch. Genomics. 1993;17:355–363. doi: 10.1006/geno.1993.1332. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morph. 1951;88:49–92. [PubMed] [Google Scholar]
- Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisahara S, Chiba S, Matsumoto H, Tanno M, Yagi H, Shimohama S, Sato M, Horio Y. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci USA. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichi S, et al. Folic acid remodels chromatin on Hes1 and Neurog2 promoters during caudal neural tube development. J Biol Chem. 2010;285:36922–36932. doi: 10.1074/jbc.M110.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Koblar SA, Murphy M, Barrett GL, Underhill A, Gros P, Bartlett PF. Pax-3 regulates neurogenesis in neural crest-derived precursor cells. J Neurosci Res. 1999;56:518–530. doi: 10.1002/(SICI)1097-4547(19990601)56:5<518::AID-JNR7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Kuang S, Chargé SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- Latchman DS, Polak JM. Analysis of neuronal gene regulation and differentiation using the ND (neuroblastoma-dorsal root ganglion) series of cell lines. Methods. 1995;7:277–284. [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Li J, Liu KC, Jin F, Lu MM, Epstein JA. Transgenic rescue of congenital heart disease and spina bifida in Splotch mice. Development. 1999;126:2495–503. doi: 10.1242/dev.126.11.2495. [DOI] [PubMed] [Google Scholar]
- Libert S, Cohen D, Guarente L. Neurogenesis directed by Sirt1. Nat Cell Biol. 2008;10:373–374. doi: 10.1038/ncb0408-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Dormand E, Greenwood A, Anderson DJ. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homolog in cultured neural progenitor cells. Development. 2002;29:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Mayanil CSK, George D, Mania-Farnell B, Bremer CL, McLone DG, Bremer EG. Overexpression of murine Pax3 increases NCAM polysialylation in a human medulloblastoma cell line. J Biol Chem. 2000;275:23259–23266. doi: 10.1074/jbc.M002975200. [DOI] [PubMed] [Google Scholar]
- Mayanil CSK, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG, Bremer EG. Microarray analysis detects novel Pax3 downstream target genes. J Biol Chem. 2001;276:49299–49309. doi: 10.1074/jbc.M107933200. [DOI] [PubMed] [Google Scholar]
- Mayanil CSK, Pool A, Nakazaki H, Reddy AC, Mania-Farnell B, Yun B, George D, McLone DG, Bremer EG. Regulation of murine TGFβ2 by Pax3 during early embryonic development. J Biol Chem. 2006;281:24544–24552. doi: 10.1074/jbc.M512449200. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Dietz KN, Hollenbach AD. Identification of serine 205 as a site of phosphorylation on Pax3 in proliferating but not differentiating primary myoblasts. Protein Sci. 2008;17:1979–1986. doi: 10.1110/ps.035956.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH gene Hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazaki H, Reddy A, Mania-Farnell B, Yueh-Wei S, McCabe C, Ichi S, George D, McLone DG, Tomita T, Mayanil CSK. Key basic helix-loop-helix transcription factor genes Hes1 and Ngn2 are regulated by Pax3 during mouse embryonic development. Dev Biol. 2008;316:510–523. doi: 10.1016/j.ydbio.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Honarpour N, Wang X. Apoptosis in neural development and disease. Annu Rev Neurosci. 2000;23:73–87. doi: 10.1146/annurev.neuro.23.1.73. [DOI] [PubMed] [Google Scholar]
- Pani L, Horal M, Loeken MR. Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3-dependent development and tumorigenesis. Genes Dev. 2002;16:676–680. doi: 10.1101/gad.969302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- Reeves FC, Burdge GC, Fredericks WJ, Rauscher FJ, Lillycrop KA. Induction of antisense Pax-3 expression leads to the rapid morphological differentiation of neuronal cells and an altered response to the mitogenic growth factor bFGF. J Cell Sci. 1999;112:253. doi: 10.1242/jcs.112.2.253. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Ribes V, Stutzmann F, Bianchetti L, Guillemot F, Dollé P, Le Roux I. Combinatorial signalling controls Neurogenin2 expression at the onset of spinal neurogenesis. Dev Biol. 2008;321:470–481. doi: 10.1016/j.ydbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004;556:281–286. doi: 10.1016/s0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–56. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Cross-regulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- Simmons AD, Horton S, Abney AL, Johnson JE. Neurogenin2 expression in ventral and dorsal spinal neural tube progenitor cells is regulated by distinct enhancers. Dev Biol. 2001;229:327–339. doi: 10.1006/dbio.2000.9984. [DOI] [PubMed] [Google Scholar]
- Sun YE, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Singh BN, Zhang G, Hwa YL, Li J, Dowdy SC, Jiang SW. Nonhistone protein acetylation as cancer therapy targets. Exp Rev Anticancer Ther. 2010;10:935–954. doi: 10.1586/era.10.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1: negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- Theriault FM, Nuthall HN, Dong Z, Lo R, Barnabe-Heider F, Miller FD, Stifani S. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng-Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Kumar S, Slevin M, Kumar P. Functional analysis of alternative isoforms of the transcription factor PAX3 in melanocytes in vitro. Cancer Res. 2006;66:8574–8580. doi: 10.1158/0008-5472.CAN-06-0947. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.