A combination of mathematical modeling and live-cell measurements was applied to determine the dynamics of small nuclear ribonucleoprotein (snRNP) formation in Cajal bodies of living cells. Our results indicate that a substantial fraction of tri-snRNPs is formed in Cajal bodies in cells with many Cajal bodies per nucleus.
Abstract
The U4/U6·U5 tri-small nuclear ribonucleoprotein particle (tri-snRNP) is an essential pre-mRNA splicing factor, which is assembled in a stepwise manner before each round of splicing. It was previously shown that the tri-snRNP is formed in Cajal bodies (CBs), but little is known about the dynamics of this process. Here we created a mathematical model of tri-snRNP assembly in CBs and used it to fit kinetics of individual snRNPs monitored by fluorescence recovery after photobleaching. A global fitting of all kinetic data determined key reaction constants of tri-snRNP assembly. Our model predicts that the rates of di-snRNP and tri-snRNP assemblies are similar and that ∼230 tri-snRNPs are assembled in one CB per minute. Our analysis further indicates that tri-snRNP assembly is approximately 10-fold faster in CBs than in the surrounding nucleoplasm, which is fully consistent with the importance of CBs for snRNP formation in rapidly developing biological systems. Finally, the model predicted binding between SART3 and a CB component. We tested this prediction by Förster resonance energy transfer and revealed an interaction between SART3 and coilin in CBs.
INTRODUCTION
Numerous distinct nonmembrane structures and bodies have been identified in the cell nucleus. Splicing factor compartments (SFCs; also called nuclear speckles) and Cajal bodies (CBs) among them represent the places of small nuclear ribonucleoprotein particle (snRNP) accumulation (Lamond and Spector, 2003; Stanek and Neugebauer, 2006). Whereas SFCs likely serve as storage places for inactive snRNPs (Jimenez-Garcia and Spector, 1993), increasing evidence is being gathered that CBs actively participate in snRNP biogenesis and recycling (Gall et al., 1999; Matera, 1999; Nesic et al., 2004; Stanek and Neugebauer, 2006; Morris, 2008).
The Cajal body was found in many organisms including plants, fruit flies, zebrafish, and humans (Gall, 2000; Matera and Shpargel, 2006; Stanek and Neugebauer, 2006). CBs are also commonly visible in many in vitro cultured cell lines. CB represents a highly dynamic compartment that moves throughout the nucleus and constantly exchanges its components with the surrounding nucleoplasm (Platani et al., 2000, 2002; Carmo-Fonseca, 2002; Handwerger et al., 2003; Dundr et al., 2004; Cioce and Lamond, 2005). Recent data show that CBs can be artificially formed de novo by tethering CB components to DNA (Kaiser et al., 2008). The RNA processing rate and snRNP levels probably determine the CB abundance in living cells (Sleeman et al., 2001; Lemm et al., 2006); the number and size of CBs quickly respond to changes in transcription rates (Lafarga et al., 1998), their integrity depends on transcription level and snRNP biogenesis, and inhibition of either of these processes results in disintegration of CBs (Carmo-Fonseca et al., 1992; Stanek et al., 2003). Distinct molecular processes that underlie CB formation and integrity are mostly unknown, however. In addition to components of the snRNP biogenesis pathway, CB contains many factors involved in the metabolism of other RNPs, including small nucleolar ribonucleoprotein particles (snoRNPs), telomerase, and U7 snRNP (Gall, 2000; Carmo-Fonseca, 2002; Ogg and Lamond, 2002; Schumperli and Pillai, 2004; Matera and Shpargel, 2006). Twenty years ago, a CB protein (coilin) was described that has been serving as a marker of the CB since then (Andrade et al., 1991; Raska et al., 1991).
Pre-mRNA splicing is catalyzed by a ribonucleoprotein complex called the spliceosome. The spliceosome is composed of five snRNPs and additional non-snRNP proteins (Wahl et al., 2009). Each snRNP consists of a small nuclear RNA (snRNA), a ring of seven Sm or Like-Sm (LSm) proteins, and a set of proteins specific for each snRNP (Will and Lührmann, 1997). There are five major snRNPs: U1, U2, U4, U5, and U6, named according to the snRNA they contain. U1, U2, U4, and U5 snRNAs are transcribed by RNA polymerase II. After synthesis they are exported to the cytoplasm where the Sm ring is assembled and the 5′ end hypermethylated to form the 2,2,7-trimethyl guanosine cap (Will and Luhrmann, 2001). The Sm ring and the trimethyl guanosine cap serve as signals for nuclear import (Matera and Shpargel, 2006). In the cell nucleus, snRNPs first appear in Cajal bodies (Sleeman and Lamond, 1999) where snRNAs are further modified by pseudouridylation and ribose methylation guided by small Cajal body–specific RNAs (Darzacq et al., 2002; Kiss et al., 2002; Jady et al., 2003). Furthermore, addition of specific proteins and the final snRNP maturation likely occur in the CB as well (Stanek and Neugebauer, 2006). The U6 snRNA is synthesized by RNA polymerase III, and its modifications are guided by snoRNPs in the nucleolus. The U6 snRNP contains a ring formed from LSm proteins and during biogenesis does not leave the cell nucleus (Mayes et al., 1999; Kiss, 2004; Listerman et al., 2007).
Three of the snRNPs, U4, U5, and U6, enter the splicing reaction as a preassembled complex called the tri-snRNP. The first step of tri-snRNP assembly is catalyzed by LSm proteins and the protein SART3 (also named hPrp24 or p110), and involves base pairing between U4 and U6 snRNAs and addition of U4/U6 di-snRNP specific proteins (Ghetti et al., 1995; Raghunathan and Guthrie, 1998; Achsel et al., 1999; Bell et al., 2002). The U5 snRNP is then attached via protein–protein interactions (Liu et al., 2006). In this step, binding between U4/U6-specific protein hPrp31 (also called 61K) and U5-specific hPrp6 (also named 102K) plays a crucial role, and depletion of either of these proteins results in inhibition of the tri-snRNP assembly (Makarova et al., 2002; Schaffert et al., 2004). SART3 is not a part of the mature tri-snRNP and leaves before or during the tri-snRNP formation (Bell et al., 2002; Schneider et al., 2002).
Several lines of evidence suggest that di- and tri-snRNP assembly occurs in the CB. Proteins promoting U4/U6 snRNP formation, SART3 and LSm proteins, are localized to CBs (Stanek et al., 2003). In addition, assembly intermediates that contain SART3 and U4/U6-specific proteins are highly concentrated in the CB (Stanek and Neugebauer, 2004). Finally, inhibition of tri-snRNP formation results in the accumulation of di-snRNP in CBs, further supporting the role of CB in di-snRNP metabolism and strongly suggesting that tri-snRNP is assembled in this nuclear compartment (Schaffert et al., 2004).
In silico modeling predicted that U4/U6 snRNP assembly rates in the cell nucleus containing CBs should increase 10-fold compared to the nucleus lacking this compartment (Klingauf et al., 2006). It suggests that accumulation of snRNP components in CB promotes snRNP assembly and recycling. Importantly, coilin knockout (KO) mice display significant fertility and fecundity defects (Walker et al., 2009), and coilin-depleted zebrafish embryos die during embryogenesis (Strzelecka et al., 2010b). The latter phenotype can be rescued by injection of mature snRNPs (Strzelecka et al., 2010b).
Direct measurements of di- and tri-snRNP formation in vivo have never been documented, however. In this study, we addressed in vivo kinetics of tri-snRNP assembly and, by using fluorescence recovery after photobleaching (FRAP), we monitored the kinetics of snRNP exchange between the CB and the nucleoplasm. We constructed a mathematical model of di- and tri-snRNP formation and we used the model to estimate (using only the FRAP data) kinetic parameters of the tri-snRNP assembly process.
RESULTS
GFP-tagged hPrp4, Brr2, and SART3 report the pools of snRNP complexes
To study dynamics of tri-snRNP formation in Cajal bodies, it is imperative to visualize trafficking of its basic building blocks within the nuclear volume. For this purpose, we fluorescently tagged protein markers specific for individual U4, U5, and U6 snRNP complexes. Specifically, we visualized the U4 snRNP pool with hPrp4-green fluorescence protein (hPrp4-GFP), U5 with Brr2-GFP, and U6 with SART3-GFP. To keep the expression of GFP-tagged proteins at endogenous levels, recombineering on bacterial artificial chromosomes (BACs) was used for construction of expression vectors. This approach preserves endogenous promoters as well as additional regulatory elements. BACs coding for GFP-tagged proteins were subsequently introduced into HeLa cells, and cells stably expressing these proteins were selected for further experiments (Poser et al., 2008).
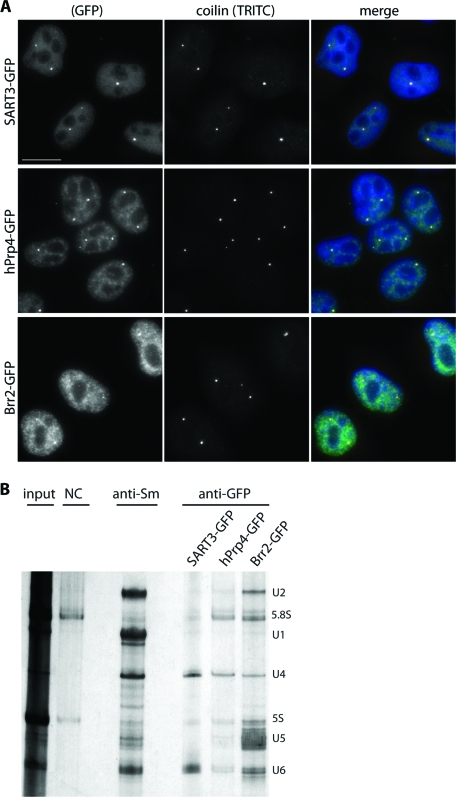
We verified that all constructed fluorescent snRNP markers were localized into the cell nucleus (Figure 1A): SART3-GFP was dispersed throughout the nucleoplasm and accumulated in CBs as its endogenous counterpart (Stanek et al., 2003), hPrp4-GFP was localized to CBs and SFCs as previously described (Stanek and Neugebauer, 2004; Stanek et al., 2008), and Brr2-GFP distribution corresponded with the localization of other U5 snRNP markers (e.g., hPrp8-GFP or hSnu114; Stanek et al., 2008) detected in the SFCs and in CBs. To further test whether the GFP-tagged proteins are properly incorporated into snRNP complexes, GFP-tagged proteins were immunoprecipitated using antibody against GFP, and their association with snRNAs was analyzed (Figure 1B). SART3-GFP precipitated down U4 and U6 snRNAs, which indicated that SART-GFP interacted with U4/U6 snRNPs, but was not present in the U4/U6·U5 tri-snRNP (Bell et al., 2002; Stanek et al., 2003). Consistent with hPrp4 association with U4 and U4/U6 snRNPs, hPrp4-GFP coprecipitated U4 and U6 snRNAs and smaller amounts of U5 snRNAs reflecting its integration into the U4/U6·U5 tri-snRNP. Brr2-GFP as a U5 snRNP core protein coprecipitated mainly the U5 snRNA. In addition, Brr2-GFP interacted with U6 and U2 snRNAs, which indicated its incorporation into the active spliceosome. A similar observation was previously reported for hPrp8-GFP–labeled U5 snRNP (Huranova et al., 2010). The presence of smaller amounts of U4 snRNA among Brr2-GFP precipitates suggests that this protein interacts with U4 snRNA only transiently within the tri-snRNP complex. Immunoprecipitation together with protein localization confirmed that the GFP-tagged proteins were integrated into snRNPs.
FIGURE 1:
Distribution of U4, U5, and U6 snRNPs as revealed by GFP-tagged markers. (A) HeLa cell lines stably expressing snRNP-specific proteins SART3-GFP, hPrp4-GFP, or Brr2-GFP. GFP-tagged proteins are properly localized to the cell nucleus visualized by DAPI staining and to CBs depicted by coilin immunostaining. Bar: 10 μm. (B) GFP-tagged snRNP specific proteins were immunoprecipitated from stable cell lines expressing individual snRNP-GFP proteins using anti-GFP antibody and coprecipitated snRNAs visualized by silver staining. Immunoprecipitation with the anti-Sm antibody served as a positive control, and nonspecific goat serum was used as a negative control. Positions of snRNAs and small rRNAs are depicted.
Model of tri-snRNP formation
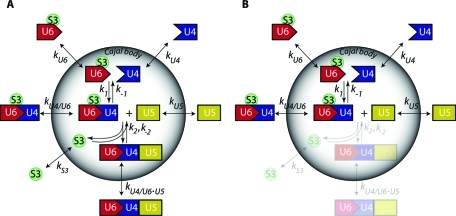
Previously, we proposed a model of stepwise U4/U6·U5 tri-snRNP assembly in CB (Stanek and Neugebauer, 2006). In this model, the maturation and export of tri-snRNPs involves a number of interactions and depends on the availability of basic building blocks in the CB. We described the dynamics of the model by a system of ordinary differential equations in terms of individual snRNP concentrations inside and outside the CB, transfer rates of snRNPs across the CB boundary, and proper formation and dissociation constants (Figure 2A). The complete set of 12 differential equations is presented in the Supplemental Material (Eqs. S1–S12). In general, it is difficult to measure concentrations of individual free snRNPs and their complexes in nuclear compartments. Fortunately, the total snRNP pools can be visualized directly using fluorescently tagged proteins specifically binding U4, U5, or U6. In this representation, the measured fluorescence signal is a sum of contributions from all marked snRNPs and their complexes. Assuming that the measured fluorescence intensities are proportional to the concentrations of the fluorescent proteins, we can write:
 |
where terms MARKERUi refer to the concentrations of the fluorescently tagged snRNP marker proteins, and [S3] denotes the concentration of free SART3 protein that is not incorporated in snRNPs. Considering the model of tri-snRNP formation (Figure 2), it seems advantageous to use SART3 as a U6 marker because SART3 not only interacts with U6 snRNP and U4/U6 di-snRNP, but, in addition, is capable to report U4/U6·U5 assembly as this step is accompanied by SART3 release from U6.
FIGURE 2:
Kinetic model of tri-snRNP assembly in the CB. (A) Full (WT) model of tri-snRNP formation in CBs. The CB is modeled as a spherical object with a permeable surface. Coefficients kU4, kU6, kU5, kU4/U6, and kU4/U6.U5 describe transfer rates of U4, U6, U5, U4/U6, and U4/U6.U5 complexes, respectively, between nucleoplasm and CB. kS3 denotes the transfer rate of SART3 protein (S3). Constant k1 characterizes formation rate of U4/U6 di-snRNP complex from U4 and U6 components. Constant k2 describes the conjugation rate of U4/U6 di-snRNP and U5 into U4/U6.U5 tri-snRNP. (B) Simplified (KO) model with blocked tri-snRNP formation. Depletion of hPrp6 inhibits tri-snRNP assembly and reduces the model to di-snRNP assembly.
The mass conservation for each of the tagged components should always hold true:
 |
in which Vin and Vout stand for volume of CB and nucleoplasm, respectively. Equations 1 and 2 together with Eqs. S1–S12 represent a complete kinetic description of the proposed model. Values of Vin and Vout as well as Cajal body surface S participating in Eqs. S1–S12 and modulating influx and efflux rates were taken from Klingauf and colleagues (2006). For all the model components, initial concentrations, rate constants kx (rates of components transfer between Vin and Vout), and reaction rates describing the formation/disassembly of U4/U6 di-snRNP and U4/U6·U5 tri-snRNP have to be optimized to fit the experimental data. Quantities [MARKERUi]in·Vin (Eqs. 1 and 2), however, are proportional to overall fluorescence intensities directly measurable in the region of interest corresponding to one CB. A combination of multiple FRAP experiments, in which the spatiotemporal redistribution of fluorescently labeled markers of U4, U5, and SART3 after the bleaching pulse are separately monitored (Supplemental Movie S1), thus contains information sufficient for the complete description of tri-snRNP formation kinetics.
hPrp6 knockdown simplifies the model of tri-snRNP formation
Complete analysis of the FRAP data, which would reveal all kinetic parameters of our model of tri-snRNP formation, requires fitting of the entire system of equations to the measured data. To do it at once is a rather difficult task mainly due to unknown initial values of the parameters. Without a reasonably accurate first guess, it is difficult to achieve convergence. In the first step, we therefore decided to simplify the model and freeze some of its degrees of freedom. The fitted subset of parameters could be subsequently used as a starting point for the complete calculation. It was previously shown that hPrp6 knockdown (KD) resulted in inhibition of tri-snRNP formation and accumulation of di-snRNP components in the CB (Schaffert et al., 2004). Therefore, hPrp6 depletion significantly simplifies the proposed model to di-snRNP assembly only (Figure 2B). Importantly, the simplified model is described by a smaller number of parameters.
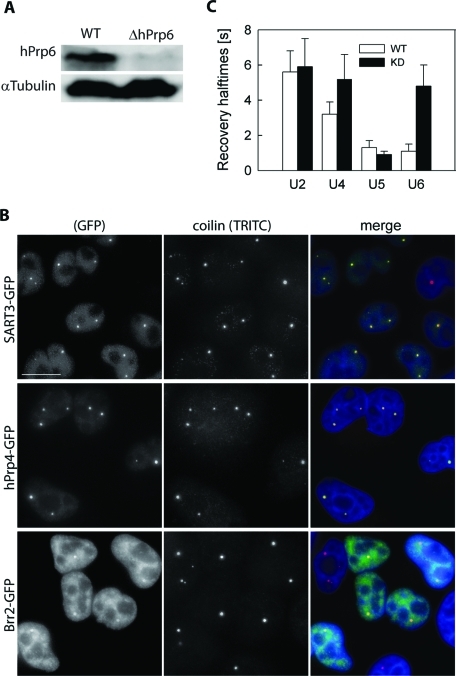
Specific siRNA was used to selectively inhibit hPrp6 expression (Schaffert et al., 2004). After the treatment, the amount of hPrp6 in the cell lysate decreased to 17 ± 2% of the original level (Figure 3A). KD of hPrp6 resulted in further accumulation of U4 and U6 but not U5 snRNPs in the CB (Figure 3B, compare with Figure 1A). In addition, we assessed residence times of individual snRNPs in CBs before and after hPrp6 KD by FRAP. For this purpose, we fitted fluorescence recovery data with a double-exponential model and determined recovery halftimes (t1/2). Consistently with U4 and U6 snRNP accumulation, fluorescence recoveries of hPrp4-GFP and SART3-GFP measured after hPrp6 KD exhibited prolonged t1/2 values compared to mock-treated cells. The decrease of Brr2-GFP (U5) retention in CBs was statistically insignificant (Figure 3C). It has to be noted that systems containing a network of multiple linked equilibria exhibit complex kinetic behavior and, as a consequence, heterogeneous multicomponential FRAP curves. Therefore, t1/2 is an intricate function of numerous kinetic constants and concentrations of all involved kinetic species, and it is better to use it only as an indicator of system perturbation rather than for drawing detailed conclusions. Valid conclusions can be made only after complete analysis of the system and characterization of all underlying kinetic processes. Taken together, these results confirmed that hPrp6 KD inhibited tri-snRNP formation as shown previously.
FIGURE 3:
hPrp6 KD. (A) HeLa cells were treated with siRNA specific for hPrp6 mRNA, and the level of hPrp6 protein was determined by Western blotting before and after siRNA KD. α-Tubulin served as a loading control. (B) Depletion of hPrp6 resulted in accumulation of SART3-GFP and hPrp4-GFP in CBs (depicted by coilin immunostaining). Localization of Brr2-GFP was not significantly altered. Compare with Figure 1A. Bar: 10 μm. (C) Mobility of GFP-tagged snRNP markers was monitored in CBs by FRAP, and mean fluorescence recovery halftimes (t1/2) were calculated in untreated cells (empty bars) and after hPrp6 KD (full bars). The halftime values were obtained from double-exponential fits of the measured fluorescence intensities. Mean values and standard deviations (error bars) were calculated from at least 10 FRAP curves for each cell line.
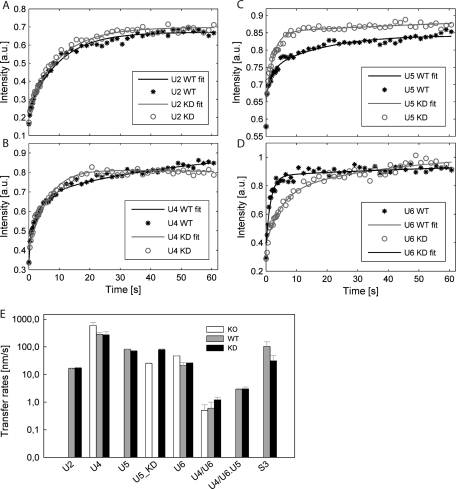
Subsequently, the FRAP curves measured in the KD cells were fitted by our model. For simplicity, we first neglected any amount of tri-snRNP formed under the KD conditions and treated the situation as a total hPrp6 KO. This approximation implies no formation of tri-snRNP (k2 = 0) as well as no exchange of released SART3 and tri-snRNP between the CB and the nucleoplasm (kS3 = kU4/U6·U5 = 0; Figure 2B). This simplifies Eqs. S6 and S8, and Eqs. S9–S12 are reduced to triviality. The normalized data sets from FRAP experiments with hPrp4-GFP, Brr2-GFP, and SART3-GFP proteins were globally fitted (Eisenfeld and Ford, 1979; Beechem et al., 1983; Knutson et al., 1983) by this simplified system of equations. Fit examples are shown in Figure 4, A–D. The fit provided us with the first guess of transfer rates that refer to the exchange of snRNP complexes between the CB and the nucleoplasm and implicitly depend on diffusion of individual snRNPs across the CB boundary. These transfer rates (kU4 = 590 ± 170 nm·s−1, kU6 = 47 ± 1 nm·s−1, kU5 = 25 ± 1 nm·s−1, and kU4/U6 = 0.5 ± 0.3 nm·s−1) decreased with increasing molecular weight of snRNP complexes (Figure 4E and Supplemental Table S1), in agreement with our assumption of freely diffusing snRNP particles.
FIGURE 4:
Global analysis of FRAP data. Examples of fitted FRAP data for the GPF-tagged U2 (A), U4 (B), U5 (C), and U6 complexes (D) before (asterisks) and after the hPrp6 KD (open circles). Lines represent the best fits. Whereas hPrp6 depletion changed recovery profiles of tri-snRNP specific snRNPs, the character of U2 curves remained unaltered. Compared to the initial KO fits with the simplified model, χ2 decreased by factors of 2.9, 2.5, and 8 for the WT fits, and of 1.1, 2.0, and 13 for KD fits of U4, U5, and U6 data sets, respectively. (E) Fluorescence recoveries of snRNP specific markers were analyzed in hPrp6 KD cells using a simplified model (empty bars; KO model on Figure 2B), and subsequently in untreated cells (gray, WT) and KD cells (black, KD) using the full model (Figure 2A). Coefficients kU2, kU5, kU5_h6, kU4, kU6_S3, kS3, kU4/U6, and kU4/U6·U5 represent apparent transfer rates for diffusion of snRNP particles between nucleoplasm and CB. Error bars were estimated as standard deviations from multiple experiments and fittings (n = 10–12).
To verify that the effect of hPrp6 depletion was specific for tri-snRNP components, we analyzed the dynamics of the U2 snRNP, which accumulates in CBs, but the biogenesis of which is independent of tri-snRNP assembly. U2 was visualized by GFP tagging of the U2-specific protein U2A′ on BAC as described earlier in the text for other snRNP proteins used in this study. Immunoprecipitation together with protein localization confirmed that U2A′-GFP was properly incorporated into snRNP complexes (Supplemental Figure S2). In contrast to U4 and U6 snRNP markers, no change in the halftime of the U2A′ fluorescence recovery was detected after siRNA treatment (Figure 3C). In addition, compartmental analysis of U2A′-GFP fluorescence recoveries before and after the siRNA treatment revealed that the U2 snRNP dynamics are independent of the hPrp6 level (Figure 4; for details of modeling, see Supplemental Material). We concluded that the hPrp6 KD had a specific effect on tri-snRNP formation.
Proposed model estimates the kinetics of tri-snRNP assembly
Parameters derived from the hPrp6 KO model were used as initial estimates for global fits of FRAP data measured in mock-treated cells. First, we determined parameters inaccessible from the simplified KO model: tri-snRNP transfer rate (kU4/U6.U5), tri-snRNP assembly/disassembly rates (k2/k-2), as well as the corrected value for the U5 transfer rate (kU5). These parameters were fixed in further global analysis of the U4 and U6 snRNP data sets. Finally, we obtained values of all 22 parameters of the tri-snRNP assembly process, including the corrected transfer rates of U4, U5, and U6 snRNPs and U4/U6 di-snRNP (Figure 4E and Supplemental Table S1). The adjustment of parameter values after transition from the simplified to the full model was only moderate (approximately a factor of two; Figure 4E). At the same time, the quality of the fit considerably increased: Reduced χ2 dropped 2.9-, 2.5-, and 8-fold for U4, U5, and U6 data sets, respectively (see also Figure 4, B–D for the fit quality). Altogether, these findings indicate that the initial guess for fitting of the full model was relatively close to the correct parameter values. Similarly to the KO model, transfer rates of larger complexes were in general lower, with the exception of U6 snRNP and U4/U6 di-snRNP, which moved more slowly than expected. The simple relation between the molecular weight and the diffusion rate predicted, for example, by the Stokes–Einstein equation can be substantially modulated by the shape and hydration of the diffusing species and their interactions with the surrounding environment (De La Torre and Bloomfield, 1977).
Global fitting of the full model also yielded formation rate constants of di-snRNP U4/U6 and tri-snRNP assemblies, k1 = 0.7 ± 0.1 mol−1s−1 and k2 = 0.10 ± 0.07 mol−1s−1, respectively. As expected, the corresponding dissociation rates (∼2.10−4 s−1 and ∼5.10−4 mol−1 s−1 per cell) were many orders of magnitude lower, indicating relatively high stability of the di- and tri-snRNP complexes. To estimate the number of complexes formed in one cell per second, we tried to assess the absolute amounts of snRNPs involved in the assembly reactions. Due to varying normalization factors applied to individual FRAP curves of different snRNP markers, relative concentrations of the individual components were used in the mathematical model. We scaled these concentrations by a factor yielding the reported amount of 1.3 × 105 free U6 snRNP complexes per cell (Klingauf et al., 2006). After this rescaling, the formation rates of di-snRNP U4/U6 and tri-snRNP assemblies correspond to 3.1 ± 0.8 and 3.6 ± 2.5 molecules s −1 per cell. To the best of our knowledge, these values represent the first estimate of the snRNP assembly rates in living cells.
The model predicts hPrp6 KD efficiency
Having in hand the complete description of snRNP dynamics, we attempted to calculate efficiency of the hPrp6 KD based solely on FRAP data measured in the siRNA-treated cells. Because the result critically depends on values of all internal model parameters, its agreement with directly measured value would strongly support the model validity. The KD was introduced to the model as follows: The U5 snRNP pool was subdivided into two fractions, each undergoing different dynamics. Only complete U5 snRNP complexes were allowed to enter the tri-snRNP assembly reaction, whereas those lacking hPrp6 protein could not interact with the U4/U6 di-snRNP and took part only in the U5 exchange between CB and nucleoplasm. Fitting of this heterogeneous model, i.e., combination of the complete (wild type [WT], Figure 2A), and the simplified (KO, Figure 2B) models, to FRAP data acquired under the KD conditions allowed us to estimate the fraction of the complete, unaffected U5 remaining in the KD cells.
Kinetic parameters obtained in the WT fit (Supplemental Table S1, middle column) were used as initial values for the KD fit. According to the description provided earlier in the text, three new parameters related to the U5 complex without hPrp6 (U5_KD), specifically [U5_KD]out, [U5_KD]in, and kU5_KD, were added to the WT model. Both U5 fractions were treated separately during optimization. The fit revealed transfer rates of kU5_KD = 79 ± 5 nm.s−1 and kU5_WT = 70 ± 5 nm.s−1 for both U5 species, the slightly higher rate corresponding to the smaller complex lacking the hPrp6, as expected. The ratio of the U5 fractions predicted a KD efficiency of 84 ± 6%. This result is in good agreement with the average KD efficiency measured by Western blot analysis (83 ± 2%, Figure 3A). We conclude that the quantitative agreement between the calculated and directly measured KD efficiency strongly supports the model validity and its internal consistency.
The model predicts levels of snRNP accumulation in CB
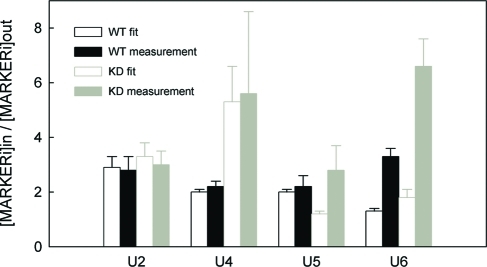
Based on the solutions describing the behavior of snRNP complexes within the CB compartment, the accuracy of the proposed model was further tested. From the computed concentrations of all studied snRNP complexes in the CB and the nucleoplasm, the expected accumulation levels of individual snRNPs in the CB before and after the hPrp6 KD were calculated. The same values were directly measured in raw images, as a ratio of the average fluorescence intensity inside and outside the CB (Figure 5). We found the experimental and computed data to be in good qualitative agreement except for the U5 snRNP. The model predicts release of the U5 snRNP from the CB after the hPrp6 KD, which was not observed experimentally. The unexpectedly long capture (compared to the predicted behavior) of the U5 snRNP in the CB cannot be explained within the frame of the proposed model and indicates some additional interactions of U5 inside the CB. The discernible immobile phase in the fluorescence recovery profiles of Brr2-GFP in untreated cells (Figure 4C) supports this hypothesis. In all other cases, however, the predicted values were qualitatively in good agreement with the measured data. For U2 and U4, the agreement is even quantitative. The levels of U2A′ (U2 snRNP) accumulation are predicted to remain unchanged, which is consistent with direct measurements (see also Supplemental Figure S2). Predicted accumulation of both hPrp4 (U4 snRNP) and SART3 (U6 snRNP) proteins in CBs after hPrp6 KD was also experimentally observed (compare Figures 1A and 3B). Higher accumulation of the U6 snRNP than predicted by the model could indicate an additional interaction of U6 (SART3) inside the CB that was not included in our model. We decided to further test this particular option (see next section).
FIGURE 5:
Accumulation of snRNP complexes in CB. The ratio of the mean equilibrium fluorescence intensity in the CB and the nucleoplasm (Iin/Iout) was calculated using the results of the global FRAP-based analyses (full model predictions; empty bars) and directly measured from the fluorescence micrographs (raw data; full bars). Predicted and raw-data values in mock-treated (black) and KD (gray) cells expressing the U2, U4, U5, and U6 markers are compared. Mean intensities were obtained from raw fluorescence data as integral intensities over the whole CB and the whole nucleoplasm normalized to the elapsed pixel count. In all cases, at least 20 CBs located in more than 10 cells were analyzed.
The model predicts the interaction of SART3 and coilin
As already mentioned, the model revealed unexpectedly slow U6 and U4/U6 snRNP transfer rates between the CB and the nucleoplasm (Figure 4E) and predicted lower U6 accumulation in CBs after the hPrp6 KD than we observed experimentally (Figure 5). One possible explanation for these discrepancies could be the existence of an interaction between SART3, which is a part of both these complexes, and a CB component(s). We tested the interaction of SART3 with coilin that is a stable CB component. Coilin forms homo-oligomers that could serve as a “docking” place for different snRNP complexes inside the CB (Stanek and Neugebauer, 2006), and its residence time in CBs is significantly higher than those of snRNPs (Sleeman et al., 2003; Dundr et al., 2004).
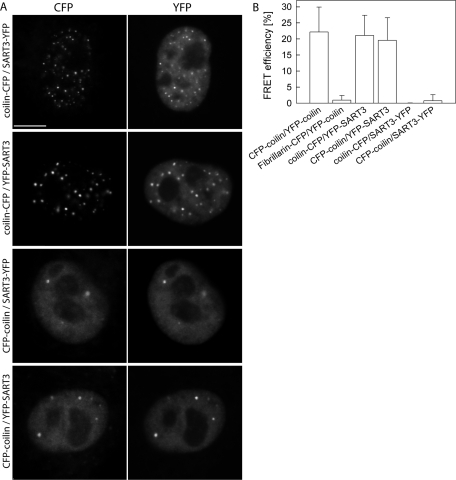
CFP-tagged coilin was coexpressed with YFP-tagged SART3 in HeLa cells, and their interaction in CBs was tested by the Förster resonance energy transfer (FRET). Expression of coilin-CFP induced formation of aberrant CBs as shown before (Hebert and Matera, 2000), and SART3 surprisingly localized to these structures (Figure 6A). A previously used acceptor photobleaching method (Stanek and Neugebauer, 2004) was used to determine FRET efficiency. A positive FRET signal was detected for CFP-coilin/YFP-SART3 (20 ± 7%) and coilin-CFP/YFP-SART3 (21 ± 6%) pairs (Figure 6B). The SART3-YFP fusion protein, however, did not exhibit any FRET signal with coilin tagged on either terminus despite the same localization of YFP-SART3 and SART-YFP (Figure 6A). These data provide additional control and show that SART3/coilin FRET is not simply due to accumulation of both proteins in CBs. Furthermore, FRET results also indicate that YFP at the C terminus does not support FRET either due to a structural hindrance or because the SART3 C terminus is too far from coilin in a complex containing SART3 and coilin.
FIGURE 6:
FRET analysis of SART–coilin interaction. (A) HeLa cells transiently expressing pairs of fluorescently tagged coilin and SART3 variants are shown. Note the presence of numerous aberrant CBs in cells expressing coilin-CFP construct. Bar: 7.5 μm. (B) FRET was measured in CBs by acceptor photobleaching, and FRET efficiency was determined as a relative increase of donor fluorescence. Positive FRET signal between coilin and SART3 was detected when SART3 was tagged at the N terminus but not at the C terminus despite the same localization of both constructs. CFP-coilin/YFP-coilin served as a positive control, and YFP-coilin/fibrillarin-CFP as a negative control.
DISCUSSION
The composition of snRNPs and the factors involved in their maturation and formation have been described in molecular detail. Little is known, however, about the dynamics of snRNP assembly especially in vivo. Here, we focused on the kinetics of U4/U6·U5 snRNP assembly in CBs of living cells. We took advantage of the fact that the first step (di-snRNP formation) and the second and final step (tri-snRNP assembly) are both occurring in CBs (Schaffert et al., 2004; Stanek and Neugebauer, 2004). In addition, inhibition of tri-snRNP assembly or recycling leads to accumulation of the di-snRNP in CBs, indicating that only the fully formed mature tri-snRNP is able to leave CBs (Schaffert et al., 2004; Stanek et al., 2008).
Live cell-imaging techniques followed by mathematical modeling have previously been used to investigate the dynamic properties of several macromolecular complexes, including splicing factors and RNA polymerases (Dundr et al., 2002; Darzacq et al., 2007; Rino et al., 2007; Gorski et al., 2008). In this study we proposed a full mathematical model describing tri-snRNP formation in CBs and used it to determine the dynamics of individual snRNP complexes. We are aware that the presented model is a simplification of the real situation in living cells. We modeled CB as a sphere with isotropic properties and a well-defined border between CB and the nucleoplasm instead of a more realistic membraneless body with a diffused boundary and spatiotemporal variation of the internal viscosity. Such a level of model complexity, however, would require extremely detailed knowledge of the CB structure that is not available at the moment. Therefore, our task was rather to build the simplest possible mathematical model that could outline the mechanisms of experimentally observed trafficking of snRNPs through CBs with reasonable simplifications and offer the first quantitative estimates of the snRNP dynamics. Even the simplest model requires six transfer and four rate constants to be estimated to mimic the dynamics of snRNP accumulation in CB, maturation of tri-snRNP, and their delivery to the nucleoplasm (Figure 1). This “minimal” reaction scheme was thoroughly tested and was found to be qualitatively (and, in most cases, quantitatively) consistent with experimental data. Some discrepancy between calculated and measured accumulation levels of Brr2 strongly indicates that U5 snRNP may interact not only with U4/U6 di-snRNP but also with other CB components (e.g., via Sm protein interaction with coilin; Xu et al., 2005).
Global analysis (Eisenfeld and Ford, 1979; Beechem et al., 1983; Knutson et al., 1983) is a powerful method for discerning between alternative models and for accurate recovery of model parameters. The method is based on an ordinary nonlinear least-squares minimization (Bevington and Robinson, 2002) and allows simultaneous analysis of multiple data with a single model that encompasses them all. Many parameters are therefore common for multiple curves, and the resulting set of parameters must be consistent with all data sets. Overdetermination of the model sharpens the minimum of the χ2 surface with concomitant decrease of parameter correlation and increase of parameter accuracy (Knutson et al., 1983; Herman and Lee, in press). As a consequence, models inconsistent with data can therefore be eliminated with higher confidence. Global analysis has been previously used for a large variety of experimental data and techniques (Ackers et al., 1975; Johnson et al., 1981; Consler et al., 1992; Ionescu and Eftink, 1997; Ucci and Cole, 2004; Boo and Kang, 2005; Bednarkiewicz and Whelan, 2008). Recently, a similar approach was successfully used for analysis of complex allosteric systems with multiple linked equilibria (Herman and Lee, 2009a, 2009b, 2009c). In all cases, the global approach provided performance far beyond resolution of the conventional analysis. In addition, several parameters of the CB-localized tri-snRNP formation (e.g., accumulation of snRNP proteins or interaction between SART3 and coilin in CBs) predicted by the model were subsequently confirmed by direct measurements, which further support the validity of the model.
We show that di-snRNP and tri-snRNP are formed with similar kinetics and that the rate of di- and tri-snRNP formation is estimated to 186 di-snRNP and 228 tri-snRNP molecules per minute and CB. The average CB number in our HeLa cell line is approximately three CBs per cell (Klingauf et al., 2006), which results in ∼680 tri-snRNP complexes assembled in CBs of one cell per minute. How is that relevant to the total snRNP turnover? Numerous in vitro studies as well as our in vivo data indicate that tri-snRNP is disassembled during spliceosome activation and must be recycled after each round of splicing (Wahl et al., 2009). If a steady state of the snRNP recycling pathway is assumed, then the number of recycled tri-snRNPs should equal the number of synthesized and removed introns. How many introns are synthesized per minute? The total RNA synthetic rate was estimated to be 2 × 108 nucleotides (nt)/min, with 58% of this amount corresponding to RNA polymerase II synthesis (Brandhorst and McConkey, 1974). The average length of human exon plus intron is ∼5,000 nt. Taken together, there are 23,200 introns (2 × 108 × 0.58/5 × 103 = 23,200) synthesized and likely spliced out every minute and a corresponding number of tri-snRNPs has to be recycled. According to the presented prediction, a ∼3% fraction is recycled in CBs, which would explain why HeLa cells without CBs are viable albeit with reduced proliferation (Lemm et al., 2006). In biological systems with larger CB numbers, the CB-associated tri-snRNP assembly might represent a significant portion of recycled complexes. For example, in zebrafish early embryos with an average number of 19 CBs per cell (Strzelecka et al., 2010a), the number of tri-snRNPs assembled in CBs would, according to our model, increase to almost 20%, which is consistent with the lethal embryonic phenotype observed in zebrafish lacking CBs. In addition, the importance of CBs in the snRNP assembly process is further emphasized by the observation that the supply of assembled snRNPs can rescue the coilin depletion phenotype (Strzelecka et al., 2010b). Moreover, the portion of snRNP assembled in CBs is likely an underestimation as it depends on the total rate of intron synthesis. Many active genes transcribed by Pol II, however, are intronless (e.g., histone or snRNAs), and some introns are not removed due to alternative splicing. Moreover, a significant portion of Pol II enzymes could be engaged in synthesis of short regulatory RNAs (e.g., miRNAs or RNAs at promoters and enhancers) or in synthesis of cryptic transcripts (Berretta and Morillon, 2009). Thus, the total number of recycled tri-snRNPs can be significantly lower than the earlier estimate, which would increase the percentage of snRNPs recycled in CBs.
A CB with a 0.5-μm radius occupies a volume of ∼0.50 fl, and the volume of the whole nucleus (without nucleoli) is ∼620 fl (Klingauf et al., 2006). The volume of three CBs thus represents 0.25% of the nuclear volume, which indicates that the efficiency of tri-snRNP formation is 11 times higher in CBs than in the surrounding nucleoplasm. Our measurements are fully consistent with the model of nuclear compartmentalization proposing that the concentration of different factors in a cellular structure (such as CBs and promyelotic leukemia (PML) bodies in the nucleus or P-bodies or stress granules in the cytoplasm) promotes their interaction (Klingauf et al., 2006; Strzelecka et al., 2010b). Quantitative data describing individual steps in gene expression are necessary for understanding the molecular processes that underlie formation and maintenance of nuclear structures and represent one of the future challenges of cell biology.
MATERIALS AND METHODS
Cell lines, cell treatment
We used HeLa cells stably expressing GFP-tagged snRNP specific proteins from BACs (Poser et al., 2008). SART3-GFP and Brr2-GFP cell lines were gifts from Karla Neugebauer (MPI-CBG, Dresden, Germany). hPrp4-GFP and U2A′-GFP were provided by Ina Poser and Tony Hymann (MPI-CBG) (Poser et al., 2008). Cells were cultured in DMEM supplemented with 10% fetal calf serum, penicillin, and streptomycin (Invitrogen, Carlsbad, CA).
Preannealed siRNA duplexes were obtained from Ambion (Austin, TX). The sequence of siRNA against hPrp6 mRNA was previously described (Schaffert et al., 2004). “Negative control 1” siRNA from Ambion (Austin, TX) was used as a negative control. siRNAs were transfected using Oligofectamine (Invitrogen) according to the manufacturer’s protocol, and cells were analyzed 48 h after transfection. Plasmid DNAs were introduced to cells using FuGene HD Transfection Reagent (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s protocol 24 h before the experiment and 24 h after siRNA transfection. Within the 48-h incubation period we did not observe any extensive cell death with respect to the treatment with the negative control siRNA.
Plasmids and antibodies
SART3-mRED was used for visualization of CBs in vivo. SART3 cDNA was cloned from SART3-GFP (Stanek and Neugebauer, 2004) into the mRed-C3 vector (Clontech, Mountain View, CA) using EcoRI/BglII restriction sites. Anti-coilin (5P10) antibody, provided by M. Carmo-Fonseca (Institute of Molecular Medicine, Lisbon, Portugal) (Almeida et al., 1998), was used as a marker of Cajal bodies. The efficiency of hPrp6 knockdown was tested by Western blotting and analyzed using ImageJ. Proteins were detected with rabbit antibody anti-PRP6 (H-300; Santa Cruz Biotechnology, Santa Cruz, CA) or mouse antibody anti–α-tubulin provided by P. Draber (IMG AS CR, Prague, Czech Republic) (Draber et al., 1989) and then with secondary antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA).
Immunofluorescence staining and image acquisition
Forty-eight hours after siRNA transfection, cells were fixed in 4% paraformaldehyde/piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) for 10 min, permeabilized for 5 min with 0.2% Triton X-100 (Sigma Aldrich, Steinheim, Germany), and incubated with primary antibody against coilin. Secondary anti–mouse antibodies conjugated with tetramethylrhodamine-5-(and 6)-isothiocyanate (TRITC) (Jackson ImmunoResearch Laboratories) were used. Images were acquired using the DeltaVision microscopic system (Applied Precision, Issaquah, WA) coupled to an Olympus IX70 microscope equipped with an oil immersion objective (60×/1.4NA). For all the samples, identical settings were used. Stacks of 20 xy-sections with 200-nm z step were collected per sample and subjected to image restoration using a measured point spread function (SoftWorx; Applied Precision).
FRAP
FRAP experiments were performed using the DeltaVision microscopic system equipped with an oil immersion objective (60×/1.4NA) and an environmental chamber controlling temperature (37°C) and CO2 concentration (5%). Photobleaching was achieved by a 100-ms laser pulse at 488 nm (50% of the laser power level, spot No. 1), and integral fluorescence intensity was monitored in a series of 50 images, in a circular area of 1 μm in diameter corresponding to one CB.
FRET
HeLa cells were transfected with vectors coding for CFP- and YFP-tagged proteins of interest using FuGENE HD, grown for 24–26 h, and fixed in 4% paraformaldehyde/PIPES (Sigma-Aldrich. St. Louis, MO) for 10 min at room temperature. After rinsing with phosphate-buffered saline (PBS) and water, cells were embedded in glycerol containing 1,4-diazabicyclo[2.2.2]octane (DABCO). Cell micrographs were acquired using the Leica SP5 confocal microscope. FRET was measured by the acceptor photobleaching method as described previously (Stanek and Neugebauer, 2004). Before the YFP (acceptor) photobleaching, intensities of CFP (donor, excited by 405-nm laser set to 10–15% of maximum power) and YFP fluorescence (excited by 514-nm laser line set to 3–6% of maximum power) were measured. Then, YFP was bleached in a region of interest containing one CB by three to five intensive (50% maximum power) pulses of a 514-nm laser line, and CFP and YFP fluorescence was measured again. FRET efficiency was calculated from CFP fluorescence intensities in bleached CBs as FRETefficiency[%] = (CFPafter – CFPbefore) × 100 / CFPafter. Unbleached CBs of the same cell were used as a negative control. Cells (10–20) were measured per each FRET pair, and the mean and standard deviation were calculated.
Compartmental analysis
The measured FRAP curves were analyzed by means of compartmental analysis (Jacques, 1996). CB was modeled as a uniform organelle occupying a volume of Vin = 0.5 fl surrounded by an isotropic homogeneous nucleoplasm of a volume Vout = 620 fl (Klingauf et al., 2006). Both Vin and Vout were kept constant. For each of the labeled snRNP complexes, we constructed a compartmental system where its components (i.e., different snRNP particles and their complexes) partitioned between Vin and Vout. A transfer rate of the species i across the CB boundary was described by a time-invariant transfer coefficient ki. Biochemical interactions taking place inside the CB and involving studied species were characterized by corresponding reaction rates kj, j = 1, 2, 3 (see Figure 2). Each compartmental system was described by a set of ordinary differential equations written in terms of component concentrations (see Supplemental Material). Initial conditions were selected to reflect the situation in FRAP experiments. In particular, during the experiment a strong light pulse at t = 0 depletes the fluorescent label in a small volume coinciding with the volume of the CB. Accordingly, we adjusted concentrations of all fluorescently tagged species inside the CB close to zero. Due to the incomplete and often variable depletion degree, concentrations immediately after the bleaching pulse at t = 0 had to be fitted and were kept specific for each experiment. The bleaching pulse created nonequilibrium conditions driving the system evolution when photodestructed labels inside the CB were exchanged with the fresh ones diffusing from outside the photobleached volume.
For estimation of the rate constants ki and kj, each modeled compartmental system was fitted to normalized FRAP data by a nonlinear least-squares method (Johnson, 1994; Bevington and Robinson, 2002) using the NLINFIT iterative optimization routine (Matlab, The MathWorks, Natick, MA). Goodness of fit was evaluated by standard statistical procedures, mainly by a value of reduced χ2 and distribution of residuals. Fit accuracy was quantified by confidence intervals determined for the 67% confidence level. To increase overdetermination of the model and the accuracy of recovered parameters, we used a simultaneous analysis of multiple FRAP curves measured with the same fluorescently labeled complexes in different CBs of different cells. During this global fitting (Eisenfeld and Ford, 1979; Beechem et al., 1983; Knutson et al., 1983), the transfer coefficients ki and rate constants kj were common for all analyzed curves. Initial concentrations of individual system components were kept specific for each experiment, and their values were adjusted during the fitting process.
During the data analysis, we were aware that the multidimensional χ2 surface could contain local minima. To avoid being trapped in such a minimum, the global fitting was done with numerous sets of the initial parameter guesses to make sure that the fitting always converged to the same global minimum. In addition, to verify consistency of the global data set and to identify curves potentially biased by systematic errors, the fitting was performed multiple times with different data subsets (Herman and Lee, in press).
Supplementary Material
Acknowledgments
We are grateful to Karla Neugebauer, Maria Carmo-Fonseca, Pavel Draber, Tony Hyman, and Ina Poser for the gift of reagents. This work was supported by the Grant Agency of the Czech Republic (CZ:GA CˇR:GA204/07/0133), the Grant Agency of Charles University (79508 to M.B.; 153310 to I.N.), grants from the Academy of Sciences of the Czech Republic (KAN200520801, AV0Z50520514, AV0Z50390703, AV0Z50390512), and the Ministry of Education, Youth and Sports of the Czech Republic (MSM 0021620835, to P.H.). D.S. was supported by a Max Planck Institute (MPI) partner group grant from the Max Planck Society.
Abbreviations used:
- BAC
bacterial artificial chromosome
- CB
Cajal body
- FRAP
fluorescence recovery after photobleaching
- FRET
Förster resonance energy transfer
- KD
knockdown
- LSm
Like-Sm
- SFC
splicing factor compartment
- snoRNP
small nucleolar ribonucleoprotein particle
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein particle
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0560) on December 22, 2010.
REFERENCES
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackers GK, Johnson ML, Mills FC, Halvorson HR, Shapiro S. The linkage between oxygenation and subunit dissociation in human hemoglobin. Consequences for the analysis of oxygenation curves. Biochemistry. 1975;14:5128–5134. doi: 10.1021/bi00694a017. [DOI] [PubMed] [Google Scholar]
- Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarkiewicz A, Whelan MP. Global analysis of microscopic fluorescence lifetime images using spectral segmentation and a digital micromirror spatial illuminator. J Biomed Opt. 2008;13:041316. doi: 10.1117/1.2950308. [DOI] [PubMed] [Google Scholar]
- Beechem JM, Knutson JR, Ross JBA, Turner BW, Brand L. Global resolution of heterogeneous decay by phase modulation fluorometry–mixtures and proteins. Biochemistry. 1983;22:6054–6058. [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences, New York: McGraw-Hill; 2002. [Google Scholar]
- Boo BH, Kang D. Global and target analysis of time-resolved fluorescence spectra of di-9H-fluoren-9-yldimethylsilane: dynamics and energetics for intramolecular excimer formation. J Phys Chem A. 2005;109:4280–4284. doi: 10.1021/jp0406621. [DOI] [PubMed] [Google Scholar]
- Brandhorst BP, McConkey EH. Stability of nuclear RNA in mammalian cells. J Mol Biol 85. 1974:451–463. doi: 10.1016/0022-2836(74)90444-6. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M. New clues to the function of the Cajal body. EMBO Rep. 2002;3:726–727. doi: 10.1093/embo-reports/kvf154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Consler TG, Jennewein MJ, Cai GZ, Lee JC. Energetics of allosteric regulation in muscle pyruvate kinase. Biochemistry. 1992;31:7870–7878. doi: 10.1021/bi00149a018. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre JG, Bloomfield VA. Hydrodynamic properties of macromolecular complexes. I. Translation. Biopolymers. 1977;16:1747–1763. doi: 10.1002/bip.1977.360160813. [DOI] [PubMed] [Google Scholar]
- Draber P, Draberova E, Linhartova I, Viklicky V. Differences in the exposure of C- and N-terminal tubulin domains in cytoplasmic microtubules detected with domain-specific monoclonal antibodies. J Cell Sci. 1989;92(Pt 3):519–528. doi: 10.1242/jcs.92.3.519. [DOI] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Eisenfeld J, Ford CC. A systems-theory approach to the analysis of multiexponential fluorescence decay. Biophys J. 1979;26:73–83. doi: 10.1016/S0006-3495(79)85236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Company M, Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol Cell. 2008;30:486–497. doi: 10.1016/j.molcel.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Murphy C, Gall JG. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J Cell Biol. 2003;160:495–504. doi: 10.1083/jcb.200212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P, Lee JC. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 1. calorimetric study. Biochemistry. 2009a;48:9448–9455. doi: 10.1021/bi900279x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P, Lee JC. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 2. Fluorescence study. Biochemistry. 2009b;48:9456–9465. doi: 10.1021/bi900280u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P, Lee JC. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 3. Mechanism. Biochemistry. 2009c;48:9466–9470. doi: 10.1021/bi900281s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P, Lee JC. The advantage of global fitting of data involving complex linked reactions. In: Allostery: Methods and Protocols. In: Fenton AW, editor. New York: Springer; in press. [DOI] [PubMed] [Google Scholar]
- Huranova M, Ivani I, Benda A, Poser I, Brody Y, Hof M, Shav-Tal Y, Neugebauer KM, Stanek D. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J Cell Biol. 2010;191:75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu RM, Eftink MR. Global analysis of the acid-induced and urea-induced unfolding of staphylococcal nuclease and two of its variants. Biochemistry. 1997;36:1129–1140. doi: 10.1021/bi9609681. [DOI] [PubMed] [Google Scholar]
- Jacques JA. Compartmental Analysis in Biology and Medicine, Ann Arbor: MI:BioMedware; 1996. [Google Scholar]
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Johnson ML. Use of least-squares techniques in biochemistry (reprinted from Analytical-Biochemistry, Vol 206, 1992). Numerical Comput Methods Pt B. 1994;240:1–22. [Google Scholar]
- Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Darzacq X, Verheggen C, Bertrand E, Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson JR, Beechem JM, Brand L. Simultaneous analysis of multiple fluorescence decay curves—a global approach. Chem Phys Lett. 1983;102:501–507. [Google Scholar]
- Lafarga M, Berciano MT, Garcia-Segura LM, Andres MA, Carmo-Fonseca M. Acute osmotic/stress stimuli induce a transient decrease of transcriptional activity in the neurosecretory neurons of supraoptic nuclei. J Neurocytol. 1998;27:205–217. doi: 10.1023/a:1006937032068. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Bledau AS, Grishina I, Neugebauer KM. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Rauhut R, Vornlocher HP, Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Liu S, Vornlocher HP, Luhrmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–1157. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matera AG, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- Ogg SC, Lamond AI. Cajal bodies and coilin—moving towards function. J Cell Biol. 2002;159:17–21. doi: 10.1083/jcb.200206111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Rino J, Carvalho T, Braga J, Desterro JM, Luhrmann R, Carmo-Fonseca M. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput Biol. 2007;3:2019–2031. doi: 10.1371/journal.pcbi.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Will CL, Makarova OV, Makarov EM, Luhrmann R. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol Cell Biol. 2002;22:3219–3229. doi: 10.1128/MCB.22.10.3219-3229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and coilin show differential dynamic behaviour in vivo. J Cell Sci. 2003;116:2039–2050. doi: 10.1242/jcs.00400. [DOI] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- Stanek D, Pridalova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal snRNPs repeatedly cycle through Cajal bodies. Mol Biol Cell. 2008;19:2534–2543. doi: 10.1091/mbc.E07-12-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Oates AC, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus. 2010a;1:13. doi: 10.4161/nucl.1.1.10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010b;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- Ucci JW, Cole JL. Global analysis of non-specific protein-nucleic interactions by sedimentation equilibrium. Biophys Chem. 2004;108:127–140. doi: 10.1016/j.bpc.2003.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the Cajal body marker protein, coilin. PLoS One. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. snRNP structure and function. In: Krainer AR, editor. Eukaryotic mRNA Processing. Oxford: IRL Press; 1997. pp. 130–173. [Google Scholar]
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schumperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005;114:155–166. doi: 10.1007/s00412-005-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.