Summary
Bacterial populations frequently act as a collective by secreting a wide range of compounds necessary for cell-cell communication, host colonization and virulence. However, how such behaviors avoid exploitation by spontaneous ‘cheater’ mutants that use but do not contribute to secretions remains unclear. We investigate this question using Pseudomonas aeruginosa swarming, a collective surface motility requiring massive secretions of rhamnolipid biosurfactants. We first show that swarming is immune to the evolution of rhlA− ‘cheaters’. We then demonstrate that P. aeruginosa resists cheating through metabolic prudence: wild-type cells secrete biosurfactants only when the cost of their production and impact on individual fitness is low, therefore preventing non-secreting strains from gaining an evolutionary advantage. Metabolic prudence works because the carbon-rich biosurfactants are only produced when growth is limited by another growth limiting nutrient, the nitrogen source. By genetically manipulating a strain to produce the biosurfactants constitutively we show that swarming becomes cheatable: a non-producing strain rapidly outcompetes and replaces this obligate cooperator. We argue that metabolic prudence, which may first evolve as a direct response to cheating or simply to optimize growth, can explain the maintenance of massive secretions in many bacteria. More generally, prudent regulation is a mechanism to stabilize cooperation.
Introduction
Rather than being isolated entities, bacteria communicate with each other (Bassler & Losick, 2006), form biofilms (Costerton et al., 1999)and secrete many molecules during the course of an infection (Arvidson, 2000, Visca et al., 2007, Griffin et al., 2004). The realization that bacteria interact in many ways is challenging our traditional view of microbes, and may affect the way we treat infections caused by pathogenic bacteria (Foster, 2005). Specifically, bacteria secrete numerous substances necessary for nutrient scavenging, host colonization, and pathogenesis (Arvidson, 2000, Visca et al., 2007, Cascales et al., 2007). Understanding the widespread evolution of secretions in microbes is a challenge. These substances can be costly to synthesize but, once released, will also benefit other cells within range (Crespi, 2001). Microbial secretions are therefore expected to be vulnerable to exploitation by ‘cheater’ mutants that do not contribute but still benefit from the secretions of others (West et al., 2006). Empirical studies have demonstrated that ‘cheating’ can indeed occur for a number of microbial products including iron scavenging molecules (Griffin et al., 2004), digestive enzymes and quorum-sensing molecules (Diggle et al., 2007), and antibiotic resistance factors (Dugatkin et al., 2005, Chuang et al., 2009).
Despite such predictions of the rise of non-secretors and the collapse of microbial secretions, bacteria do make use of a wide range of secretions both in nature and pathogenesis. How then are these systems maintained? Recent years have witnessed a surge in the application of social evolution theory to answer this question (Crespi, 2001, Foster et al., 2006, Nadell et al., 2009, West et al., 2007a, West et al., 2006). One explanation is simply that secreting cells are unlikely to benefit non-secreting cells (Griffin et al., 2004). For example, the spatial structure that is expected to naturally emerge within cell groups can keep secretors together and away from non-secretors that would exploit them (Nadell et al., 2010). Pleiotropic constraints provide another candidate mechanism to stabilize cooperation in microbes (Foster et al., 2004), and there is recent evidence that such constraints can limit the rise of ‘cheaters’ for the case of the secretion of iron scavenging molecules (Harrison & Buckling, 2009). However, strong spatial structure and pleiotropy are unlikely to be ubiquitous raising the question of whether other factors are needed to explain microbial cooperation, particularly for high abundance secretions for which potential costs are very high.
An important feature of bacterial lifestyles is that environments change constantly. It is therefore very likely that bacteria evolved mechanisms to regulate cooperative secretions, which may, in turn, affect their evolutionary costs and benefits (Perkins & Swain, 2009). Here we indentify a regulatory mechanism that stabilizes cooperative secretions against cheating competitors within microbial groups. We use swarming, a collective form of surface motility in the opportunistic pathogen Pseudomonas aeruginosa (Rashid & Kornberg, 2000, Kohler et al., 2000, Kearns, 2010). In order to swarm, individual bacteria must secrete rhamnolipid biosurfactants (Caiazza et al., 2005, Deziel et al., 2003) which are synthesized through a well characterized pathway involving the rhl gene family (Zhu & Rock, 2008). Gene rhlA encodes for the enzyme RhlA which uses metabolic intermediates from fatty acid biosynthesis to produce 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the lipid precursors of rhamnolipids (Zhu & Rock, 2008). Two enzymes, RhlB and RhlC, are metabolically downstream of RhlA and each adds a single rhamnose to produce mono-rhamnolipids, and di-rhamnolipids respectively (Zhu & Rock, 2008). The secreted biosurfactants are thus a mixture of HAAs, mono- and di-rhamnolipids. RhlA, RhlB and RhlC have no other known functions, and loss-of-function mutants in the gene rhlA are incapable of any biosurfactant production and, consequently, swarming (Deziel et al., 2003, Caiazza et al., 2005). RhlA expression is the only requirement for the initiation for rhamnolipid synthesis in P. aeruginosa (Zhu & Rock, 2008), which makes the genetic regulation of the rhlAB operon key to the use of rhamnolipids biosurfactants as cooperative secretions.
We show that P. aeruginosa lowers the cost of biosurfactant secretion by regulating rhlAB expression to ensure that biosurfactants are produced only when carbon source is in excess of that needed for growth. This mechanism, which we call ‘metabolic prudence’, makes swarming colonies less susceptible to exploitation by rhlA− cheaters, which lack biosurfactant secretion but can swarm using the secretions of others. This mechanism works by ensuring that cells only invest carbon into rhamnolipid synthesis when growth is limited by another nutrient, the nitrogen source. By constructing a strain where the rhlAB operon is regulated by an inducible promoter, we show that swarming can become susceptible to cheating. We end by discussing how metabolic prudence explains why certain pathogenic secretions are evolutionary stable, as well as the broader implications of prudence for the evolution of cooperative behaviors.
Results
Swarming motility in P. aeruginosa is a cooperative trait that resists ‘cheating’
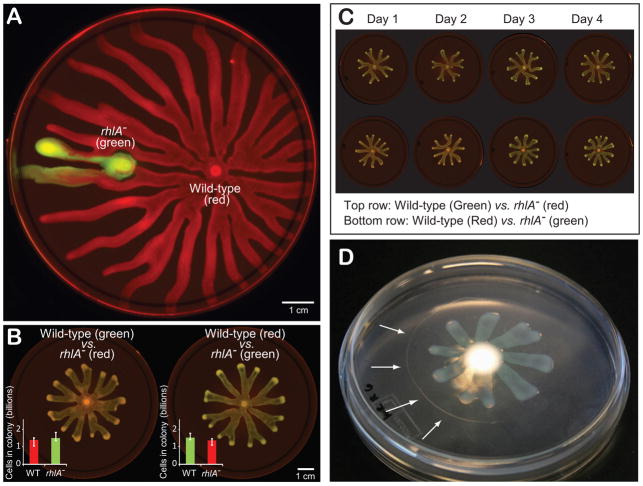
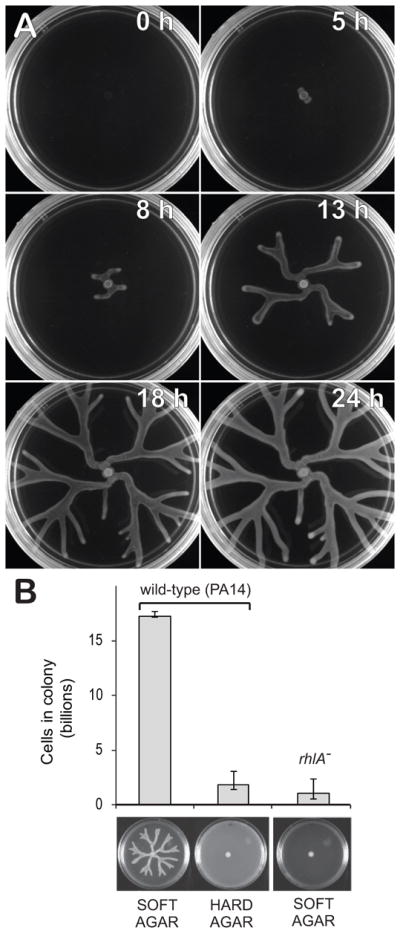
Swarming assays were initiated by inoculating P. aeruginosa at the center of an agar Petri dish. A growing colony swarms towards the edges of the dish, covering the entire distance in less than 20 h to produce spectacular star-shaped patterns (fig 1A, video SV1). Swarming colonies consistently produced seven times more cells compared to those that were prevented from swarming (P<2×10−9). We tested this both by placing cells on hard agar, which physically prevents swarming, and by removing the rhlA gene, which is essential for biosurfactant synthesis (Ochsner et al., 1994, fig 1B). Although the secreted biosurfactants are visible to the naked eye, the precise quantification of biosurfactants on swarming agar is difficult. However, we measured the biosurfactants produced in a shaking planktonic shaking culture with same medium composition and it amounted to 0.14–0.16 g/L, which corresponds to 19.8–22.5 % of the total cell dry mass (0.717 g/L, see methods). We then tested whether the rhlA− mutant was capable of swarming using the secretions provided by wild-type (WT) cells. For this, we inoculated the rhlA− mutant and WT in different locations of the same Petri dish. The experiment showed that the rhlA−, while unable to swarm alone, can indeed swarm in the presence of a biosurfactant-producing strain (Fig 2A, video SV2) and thus that rhlA− cells can indeed use exogenous biosurfactants for swarming.
Figure 1.
Swarming is a collective form of surface motility in Pseudomonas aeruginosa that benefits the colony but requires individual cells to synthesize and secrete rhamnolipid biosurfactants. (A) Frames from time-lapse imaging of swarming. See video SV1. (B) Swarming colonies achieve much higher cell numbers than colonies grown on hard agar (1.5 % agar, which prevents wild-type swarming, P<2×10−9) or colonies of mutants lacking the rhlA gene necessary for biosurfactant synthesis (P<2×10−9), showing that swarming benefits the colony.
Figure 2.
The mutant rhlA−, lacking biosurfactant secretion, can use the secretions of others to swarm yet has no measurable competitive advantage. (A) Wild-type bacteria and rhlA− were placed on the same plate in colonies initially separated by 2 cm. rhlA− bacteria (labeled in green), incapable of swarming on their own, became motile once they became mixed with biosurfactant-secreting wild-type cells (labeled in red, see video SV2). (B) rhlA− swarms equally as well as the wild-type when mixed in the same colony at 1:1. The two genotypes show no fitness difference within the mixat 24 h, suggesting a low cost of biosurfactant secretion. (C) The wild-type remains robustly at 1:1 frequency (24 h of growth) when mixed with the rhlA− even overfour days of consecutive passages (P>0.6) with no visible loss in swarming over time. (D) The massive ring of biosurfactants secreted (indicated with white arrows) is visible by naked eye.
Following these initial results, our initial expectation was that rhlA− cells would behave as ‘cheaters’ when mixed with WT in the same swarming colony. Specifically, we expected the rhlA− to outcompete the WT since they can use the biosurfactants produced by others without contributing to their production. When we carried out competitions between rhlA− and WT cell types in 1:1 mixtures, saw that the presence of the rhlA− mutant decreased mean fitness as the colonies at 24 h were smaller than those of the wild-type (fig 2B). However, we were surprised to not find any evidence of fitness differences between the two strains (fig 2B inset plots, P>0.3). We carried out experiments at ratios of 100:1, 10:1, 1:10 and 1:100, which also showed no fitness difference (Supporting material fig. SF1). To test this observation with greater accuracy, we extended the experiment over four cycles of swarming (fig 2C). At day four, the two strains still had indistinguishable cell numbers (P>0.8). Accordingly, the calculated ratio of the fitness of WT and rhlA− and was statistically indistinguishable from 1 (0.99±0.05, where interval represents 95 % confidence level). This lack of a measurable fitness difference was at odds with the copious amounts of biosurfactants secretions both measured in liquid (~20 % of the biomass produced) and visible in swarming assays (fig 2D), which we expected should translate into a major cost for the secreting cells.
Biosurfactant secretion occurs when cells have excess carbon
We sought to understand biosurfactant secretion further by turning to a liquid culture assay, where the medium is well mixed by constant shaking and cells are grown in their planktonic state. We used the same nutrient composition as in swarming assays except for the agar, which is the solidifying agent and therefore was omitted from liquid media. This method allowed the monitoring of growth and gene expression with high time resolution, together with the ability to do end-point measurements of rhamnolipids secreted. As mentioned above, the rhamnolipids secreted by WT amounted to 20 % of their biomass, whereas as the rhlA− secreted no rhamnolipids as expected. Consistent with the lack of fitness difference on agar, experiments in liquid showed no evidence of a difference in growth rate or final density between the WT and rhlA− when both strains were grown separately (fig 3A). We also grew both strain in competition by mixing WT and rhlA− in the same medium. We still found no detectable change in the ratio of the two cell types, even after four daily passages (P>0.6, fig 3A insert). We then studied the timing of biosurfactant synthesis expression by integrating a reporter fusion of the rhlAB promoter (PrhlAB) and green florescent protein (GFP) (Boles et al., 2005) into the WT genome. Time series of OD600 (cell density in liquid) and GFP (expression of rhlAB) measurements revealed that gene expression initiates only at high cell density, coinciding with the time that growth slows down and the bacteria enter stationary phase (fig 3B, see also Lequette & Greenberg, 2005). We then tested whether such a delay in the expression of rhlAB still occurs in swarming plates. We did this by comparing fluorescence by colonies of the strain carrying the PrhlABGFP fusion with that of a strain where GFP was under the regulation of a constitutive promoter (fig 3C). The time series of rhlAB expression confirmed that the delay also occurs in swarming assays (fig 3D). The observation that rhlAB expression is delayed provided a potential explanation for our findings: Cell-density (quorum) sensing ensures that biosurfactant synthesis is delayed until cultures reach a high cell density, thus limiting its impact doubling timing (fitness).
Figure 3.

(A) Growth curves in shaken liquid cultures shows no difference between wild-type (black) and rhlA− (red), in spite of the large amounts of biosurfactants secreted by WT. Inset: The ratio of WT to rhlA− CFUs in a mixed culture grown in liquid (starting ratio of 1:1) is maintained even after four daily passages to fresh medium, again showing a lack of a fitness difference. (B) A GFP fusion to the rhlAB promoter in the WT shows that rhlAB expression is delayed until stationary phase. In A and B-thick lines are the median among 6 replicates with thinner lines showing the limits of 66 percentile among replicates. The same criterion was used in all following plots of growth curves. (C) Time course of rhlAB expression was measured in swarming assays using the PrhlABGFP reporter fusion and compared with constitutive GFP expression. The pictures shown use pseudocolor to highlight the fluorescence level. (D) The PrhlABGFP expression was quantified by image analysis and normalized relatively to constitutive expression to confirm that the delay in rhlAB expression observed in liquid culture (panel B) occurs also in the swarming assay.
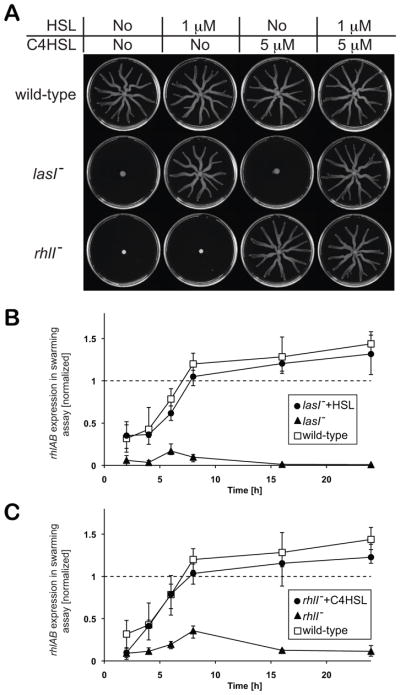
We tested this idea by investigating the role of quorum sensing. The rhlAB operon is regulated by two hierarchical quorum sensing systems: lasI/lasR and rhlI/rhlR (Latifi et al., 1996; fig 4A). Each system has its own quorum sensing signal. The signal HSL is produced by LasI and the signal C4HSL is produced by RhlI. We reasoned that if these systems were central to the delay in rhlAB expression, then exogenous supplementation of the two respective quorum-sensing signals should trigger early rhlAB expression. We first measured the concentrations of signal that restore WT levels of biosurfactant secretion in the signal negative mutants lasI− and rhlI− (fig 4B). Then, we tested the effect of adding signals at saturating levels to WT cultures at the beginning of growth. Surprisingly, we found that adding either or both signals did not detectably affect WT growth (fig 4C–D) nor did it induce over-secretion (fig 4F). We also confirmed that the signal negative mutants, lasI− and rhlI−, are incapable of swarming but that swarming is recovered by complementing the medium with the appropriate autoinducer (fig 5A). Using the PrhlABGFP reporter fusion, we further confirmed that while autoinducers are necessary for rhlAB expression in swarming assays, autoinducer presence by itself is not enough to remove the observed delay (fig 5B,C). This is consistent with previous reports suggesting that additional regulatory elements can modulate rhlAB expression in addition to the hierarchical lasI/lasR - rhlI/rhlR quorum sensing (Yarwood et al., 2005, Medina et al., 2003, Heurlier et al., 2004).
Figure 4.
The HSL and C4HSL Quorum sensing signals are required for biosurfactant secretion, yet their addition to the medium addition does not cause over secretion. (A) Diagram of the hierarchical quorum sensing system lasR/lasI - rhlR/rhlI regulating expression of the rhlAB operon. (B) Mutants that are unable to synthesize either of the two quorum sensing signals (lasI− lacks synthesis of HSL and rhlI− lacks synthesis of C4HSL) do not secrete biosurfactants in the absence of exogenous signal. These signal dose response curves allowed measuring the autoinducer concentrations needed to restitute WT level of rhamnolipid secretion: 1 μM for HSL and 5 μM for C4HSL. (C–E) Adding saturating amounts of either signal (C, D) or both together (E) to growth media did not visibly affect growth of the wild-type. (F) The rhamnolipids measured at the end of growth also showed no significant difference (P > 0.03, P > 0.3 and P > 0.1 respectively for HSL, C4HSL and both signals when compared to WT grown without signal addition).
Figure 5.
The genes for synthesis of quorum sensing autoinducers (lasI and rhlI) are necessary for rhlAB expression and swarming but adding the autoinducers does not anticipate rhlA expression. (A) lasI− and rhlI− lack swarming motility and swarming is rescued by adding the corresponding autoinducer, HSL and C4HSL respectively, to the swarming medium. (B) The lasI− mutant does not express rhlAB unless its product, HSL, is added to the medium. Nevertheless, HSL addition does not anticipate rhlAB expression relative to the wild-type. (C) The rhlI− mutant and its product, the autoinducer C4HSL, show the same effect – complementation by signal addition does not remove the delay in rhlAB expression. In both B and C, the PrhlABGFP fluorescence was normalized by GFP expression regulated by a constitutive promoter.
We next investigated the effect of nutrient on the timing of the expression of biosurfactant synthesis gene. The common minimal medium used for swarming uses casamino acids (5 g/L) as the sole source of carbon and nitrogen. The addition of more casamino acids, which increases both carbon and nitrogen source levels in the media, allowed growth to higher optical density (OD600) levels (fig 6A). This showed, as expected, that entry into the stationary phase was regulated by nutrients. Importantly, we found that when plotting rhlAB expression against growth rate (fig 6B) across the entire range of casamino acids used all the data collapsed to a single curve. On the other hand, the same expression data did not collapse to a single curve when plotted against OD (fig 6B inset). This suggests that rhlAB expression is tightly coupled to the growth rate, but not cell density per se, and therefore that cells initiate biosurfactant synthesis when growth decreases.
Figure 6.
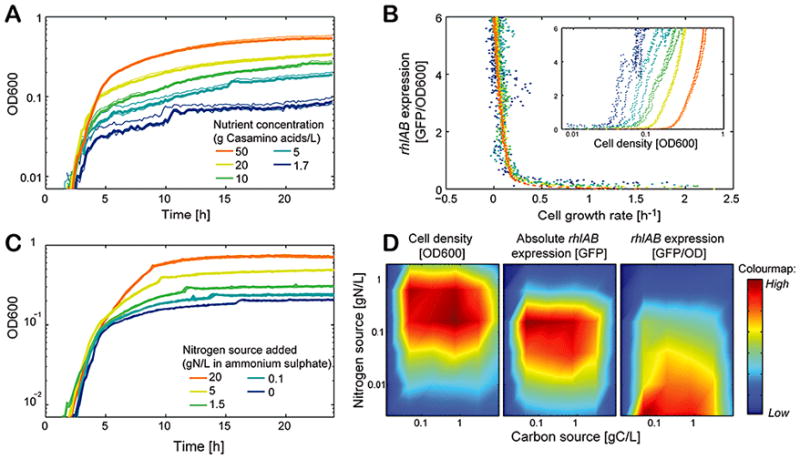
The expression of genes for biosurfactant synthesis is growth-rate dependent and is triggered by nitrogen limitation. (A) Increasing nutrients (casamino acids, the sole carbon and nitrogen source in the medium) increases growth, showing that entry into the stationary phase is due to nutrient depletion. (B) rhlAB expression data measured at all nutrient levels collapse to a single curve when plotted against growth rate - but not OD600 (inset panel) - revealing that expression is a function of growth rate, not cell density per se. (C) Complementing media with a nitrogen source (ammonium sulfate) has the same effect as increasing casamino acids, showing that growth was originally limited by nitrogen in the standard medium. (D) 2-D matrices of OD, GFP expression and GFP/OD measured at 24 h for a range of carbon (glycerol) and nitrogen (ammonium sulfate) source levels. These data show that rhlAB expression is favored at lower nitrogen/carbon ratios, when compared to the conditions producing optimal cell growth (highest OD).
Earlier studies aimed at optimizing industrial rhamnolipid production (Guerrasantos et al., 1984) found that rhamnolipid secretion depends strongly on the ratio of carbon to other essential nutrients. Consistent with this, we supplemented our growth medium with a nitrogen source (ammonium sulfate) and found the same effect of increased OD (fig 6C) as obtained by increasing casamino-acids. This result suggested that indeed growth in the casamino acids medium is limited by nitrogen source and suggested that nitrogen depletion is what triggers rhlAB expression in our assay. We then carried out additional growth experiments in ranges of carbon and nitrogen levels, which we could manipulate independently by using glycerol as the sole carbon source and ammonium sulfate as sole the nitrogen source. The results confirmed that nitrogen limitation induces rhlAB expression, but not carbon limitation (Fig 6D).
Taken together, our data suggest that the cells delay expression of biosurfactant genes to only secrete when excess carbon is present, which can occur when growth becomes limited by other nutrients such as nitrogen. This model explains why there is little or no growth cost to biosurfactant production. Biosurfactants are only produced when the cells are not dividing, due to nitrogen limitation, and use carbon source that under these circumstances cannot be used for growth. Notably, secretion also depends on the hierarchical quorum sensing systems regulating rhlAB (Latifi et al., 1996, figs 4,5). Therefore, P. aeruginosa appears to combine environmental information on the availability of excess carbon and quorum sensing to trigger the synthesis of biosurfactants.
Removing the native regulation of biosurfactants allows cheaters to invade
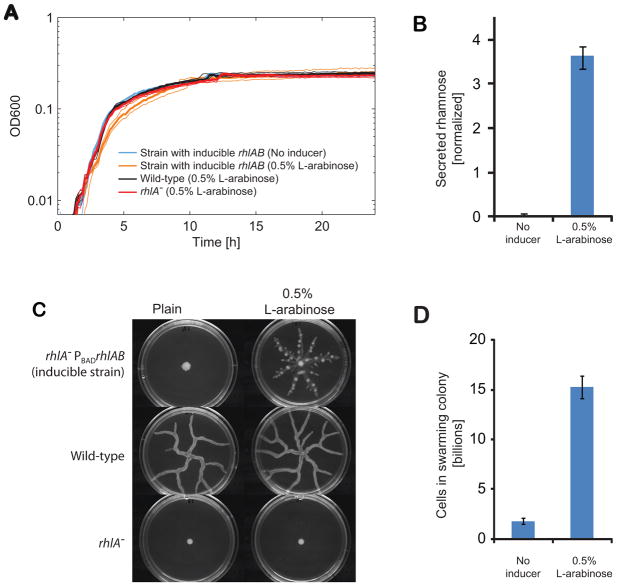
A key prediction of our model is that it would be costly for bacteria to produce biosurfactants during exponential growth. We, therefore, engineered a strain that produces biosurfactants even before reaching stationary phase by inserting rhlAB into the rhlA− background under the regulation of the inducible PBAD promoter (Boles et al., 2005). This inducible strain (PA14 rhlA− PBADrhlAB) expresses biosurfactant synthesis genes in the presence of an inducer (0.5 % L-arabinose), thus bypassing their native transcriptional regulation. We measure growth curves in liquid and saw that induction caused a significant impact for this engineered strain in terms of its exponential growth rate measured in liquid (fig 7A) when compared to the same strain without the inducer, the WT or the rhlA− strains (P<0.002). The induced strain secreted 3.6 fold the WT secretion levels by the end of the experiment (fig 7B, P<10−6), and therefore our manipulation is affecting both the timing of gene expression and the overall amount of secretions. However, the cost of this relative to the WT is only seen during exponential growth – when the WT does not express – showing the importance of timing (Fig 7A). Returning to the swarming assay, we confirmed that the inducible strain could swarm when L-arabinose was added to the medium (fig 7C). Despite differences to the WT swarm morphology, likely to be derived from differences in biosurfactant secretion (Caiazza et al., 2005), swarming motility still provided a enormous net benefit as shown by the final number of cells in swarming colonies (fig 7D). Next, we competed the inducible strain against the rhlA− mutant. For this we prepared mixed swarming assays where the inoculum was a 1:1 mix of the two strains. As expected, the inducible strain was both exploited and outcompeted in direct competition against rhlA− (fig 8A). Unlike the wild-type (fig 2C), the frequency of the inducible strain decreased in favor of rhlA− cells over daily passages (visible in color shift in fig 8A and plotted in fig 8B). By the end of the fourth daily passage, the colonies showed no swarming motility (fig 8A) and the final number of cells in colonies decreased to the numbers of cells in a colony of pure rhlA− (fig 8C, P>0.6).
Figure 7.
Unregulated biosurfactant secretion has a detectable impact to growth in liquid culture but still provides a benefit in swarming assays. (A) The addition of 0.5% L-arabinose to the inducible strain (rhlA− PBADrhlAB) results in constitutive biosurfanctant secretion. Induction affects its growth in the exponential phase in contrast to the same strain in the absence of the inducer as well as the WT and rhlA− strains in the presence of the inducer. (B) Rhamnolipids measured at 24 h of growth in liquid show that secretion by the inducible strain increases from virtually nil (without inducer) to 3.6X the wild-type secretion in medium with 0.5 % L-arabinose (values normalized by WT secretion level). (C) The inducible strain swarms in medium complemented with 0.5 % L-arabinose. The inducer does not affect the phenotypes of wild-type and rhlA−. (D) Cell counts from colonies of the inducible strain with and without the inducer show that induced swarming is very beneficial to the population.
Figure 8.
Biosurfactant secretion becomes exploitable in a biosurfactant secreting strain that lacks the native rhlAB regulation (the inducible strain rhlA− PBADrhlAB). (A) Unlike the wild-type (fig 2C), rhlA− PBADrhlAB is rapidly outcompeted by rhlA− and colonies lose swarming over four days of consecutive passages. The experiment was also conducted by swapping fluorescent labels to show the outcome is independent of the fluorescent protein used. (B) Cell count ratios measured from the competition experiments confirm that strain rhlA− PBADrhlAB has a strong competitive disadvantage against rhlA− (blue), while WT cells do not (red). (C) The productivity of swarming colonies (number of cells in colony) comprising a mixture of rhlA−PBADrhlAB and rhlA− is comparable to that of the wild-type at day 1 but decreases down to a level comparable to that of rhlA− at day 4.
We have shown that prudent regulation makes rhamnolipid secretion immune to rhlA- mutants, something that is not true of obligate secretion of rhamnolipids (Fig. 8). This provides a candidate evolutionary explanation for why P. aeruginosa regulates rhamnolipid secretion as it does, i.e. the frequent evolution of rhlA mutants favoured a molecular mechanism that prevented rhlA mutants from invading. However, an alternative explanation for the origin of prudent regulation is that it is simply more efficient, i.e. the WT is more efficient in its use of rhamnolipids than the obligate producer. There is no clear fitness difference between the obligate producer and the WT when alone, with both reaching comparable cell densities (Figs 1B and 7D). In order to test their performance in direct competition, we mixed the WT with the inducible strain in 1:1 mixes in media containing 0.5% L-arabinose. Our data show that the WT initially increases in frequency above the initial 1:1 at 2 h (P < 0.0016) but then shows no further advantage (Fig. S2). This is consistent with there being an efficiency cost to obligate secretion, but only when cells are growing most rapidly. Overall, our data suggest that prudent secretion can evolve either due to efficiency or due to costs associated with cheater mutants. But either way, the outcome is evolutionarily stable cooperation.
Discussion
The evolution of cooperation is a fundamental problem in biology because selection for selfishness and cheating should undermine group behaviors (Pennisi, 2005). Secretions by bacteria are often viewed as an example of this problem (West et al., 2007a, West et al., 2006, Diggle et al., 2007, Sandoz et al., 2007) since the secreted products of one cell that can be used by another would seem an exploitable trait. Here we propose that one way bacteria can solve this problem is by prudent regulation of secretion genes (fig 9A). We showed that P. aeruginosa regulates rhlAB expression to ensure massive secretions of biosurfactants occur only when it will not severely impact its growth. The direct consequence of this is a lower cost of biosurfactant synthesis to individual cells. This explains why wild-type swarming motility is stable in 1:1 competition with non-producers, and even over several passages (fig 2C). By engineering a strain that secretes but lacks the native regulation, we confirmed that the reverse is also possible. Unregulated biosurfactant secretion becomes costly (fig 7A), making swarming exploitable and ultimately leading to a loss of swarming motility at a cost to the entire colony (fig 8B).
Figure 9.

Our data suggest that Pseudomonas aeruginosa regulates gene expression to secrete rhamnolipid biosurfactants only when carbon source is in excess – a mechanism we call ‘metabolic prudence’. (A) Bacteria can grow and divide if both carbon-and nitrogen-source are available. In casamino acids media, carbon is initially in excess relative to nitrogen, and therefore bacteria grow exponentially until the nitrogen becomes limiting. Subsequently, cells express the biosurfactant synthesis genes and use the excess carbon at a low cost to their fitness (unlike cell biomass, rhamnolipid bisurfactants contain no nitrogen in their composition). (B) The enzyme RhlA plays a central role in biosurfactant synthesis. This enzyme is under the genetic regulation of rhlAB, the expression of which requires both quorum sensing and adequate nutrient conditions.
The regulation of biosurfactant synthesis in P. aeruginosa has proved puzzling (Medina et al., 2003, Lequette & Greenberg, 2005) because it deviates from the established paradigm of quorum sensing where gene expression is simply a function of cell density. Our study helps to resolve this puzzle by showing that rhlAB expression in the presence of excess carbon can be tightly coupled to the growth rate, and not just cell density (fig 6B). Accordingly, the wild-type does not secrete biosurfactants, or swarm, unless both quorum sensing and nutrient conditions are suitable. This suggests that P. aeruginosa has evolved to ensure biosurfactants are only secreted when both cell density is high enough for these secretions to be useful and there is an excess of carbon to minimize the impact of their synthesis (fig 9B).
The role of swarming and rhamnolipid secretion in nature
What is the role of swarming in natural systems? While often interpreted as an adaptation (Verstraeten et al., 2008, Venturi et al., 2010), the functional significance of swarming in nature has yet to be established. The inverse regulation of biofilm formation and swarming motility suggests a close link between these two key surface behaviors (Caiazza et al., 2007) and supports that swarming is a genuine adaptation. Nevertheless, it remains possible that swarming as studied in the laboratory does not occur in exactly the same way in nature. Our conclusions, however, do not rest on the relevance of swarming under natural conditions because there is considerable evidence that rhamnolipid biosurfactants have additional functions such as the emulsification of water-insoluble substrates (Ochsner et al., 1994), promoting biofilm detachment (Boles et al., 2005), antimicrobial activity (Haba et al., 2003), virulence (Zulianello et al., 2006) and the disruption of host defenses during infection (Read et al., 1992, Alhede et al., 2009).
An alternative explanation for maintenance of swarming might have been the discovery that rhlA− mutants are prevented from cheating due to a major pleiotropic cost to the loss of rhlA expression (Foster et al., 2004). However, there is no evidence in the literature of any pleiotropic effects in rhamnolipid synthesis genes (Zhu & Rock, 2008), and to be consistent with our data such a major pleiotropic cost would have to exactly match the cost of secretion to allow for growth curves of WT and rhlA− to match so closely as observed (fig 3A). Combined with our remaining data, therefore, we conclude that such a hidden cost is highly unlikely. It is still possible that small unmeasurable cost exist, for example due to transcription and translation of the gene rather than its actual function, but is not critical to our arguments that biosurfactant secretion is absolutely cost free. What we can conclude, supported by competitions both in swarming (figs 2C) and liquid (fig 3A inset), is that if any cost exists it is much smaller than simply expected from the large amounts of rhamnolipids secreted (~20 % of biomass) and it is not enough to allow cheaters to increase in the population (fig 2C). We showed that this is possible because gene regulation ensures biosurfactant synthesis genes expression does not coincide with fast growth (fig 3B, 6B).
Broader evolutionary implications
We propose that metabolic prudence is a molecular mechanism that renders bacterial secretions stable against ‘cheaters’, or at least greatly slows their evolution. This mechanism may play a key role in other microbial species as well. It is well accepted in microbial physiology that, when limited by a nutrient other than carbon and energy sources, cells will tend to redirect the non-limiting carbon flux to other functions, including secretions (Harder & Dijkhuizen, 1983). Recent mathematical models suggest that bacteria sense intracellular metabolites to perform systems-wide adjustments of metabolic fluxes (Kotte et al., 2010). Although the growth-limiting factor inducing rhamnolipid secretion was the nitrogen source in our experiments, a recent study supports our model by showing that iron limitation can also induces rhlAB expression in P. aeruginosa (Glick et al., 2010). A concrete example in another system is biofilm formation Salmonella typhimurium. There, the synthesis of the cellulose, carbon rich, matrix is positively regulated by csgD, a transcription regulator that is induced by depletion of phosphate, nitrogen and iron but not by depletion of carbon source (Gerstel & Romling, 2001).
It is important to distinguish between mechanistic (proximate) and evolutionary (ultimate) explanations for phenotypes (West et al., 2007b, Tinbergen, 1963). Most obviously, prudent regulation is a mechanistic phenomenon that involves responding to nutrient conditions in order to limit secretion to times where it has little or no effect on growth rate. However, this observation naturally feeds into ultimate considerations of why cooperation evolves. Namely, the existence of prudence can stabilize cooperation by altering its costs and benefits so as to reduce the fitness of cheating strategies. But why would prudence itself be favored by natural selection? Our data show that regulated biosurfactant secretion has the potential for large fitness returns (fig 1B) at effectively no cost (fig 2B,C). Therefore, so long as groups exist the tendency for a secreting cell to help itself and its genotype more than others in the population will allow prudent cooperation to evolve. It is important to emphasize that prudent cooperation may evolve without there ever being a cheating genotype, simply because prudence will tend to be an efficient strategy that allows minimum energy to be expended. It may also arise as a secondary modification of existing cooperation that is subject to cheating, such as unregulated secretion (fig 8A). Either way, the outcome is stable cooperation that is resistant to any subsequent cheating genotypes.
There is a growing body of evidence to show that social organisms modulate social behaviours in response to changing costs and benefits (Korb & Heinze, 2008). Work on bacteria shows that reducing costs can increase cooperation over evolutionary timescales (Brockhurst et al., 2008) and that some systems have phenotypic responses that only function to make cooperation more costly in the face of cheating (Brockhurst et al., 2008, Kümmerli et al., 2009). Closest to our findings is work showing that helping in social vertebrates can be increased by supplementary feeding (Clutton-Brock et al., 1998, Clutton-Brock et al., 1999). It is unclear in such cases whether the modulation of helping is sufficient to make it cost free and prevent the evolution of cheating, as demonstrated here. Nevertheless, the discovery of such responses in vertebrates suggests that prudence may play a role in diverse social species and provide a general explanation for the evolution of cooperation.
Experimental Procedures
Strains
Pseudomonas aeruginosa PA14 (also called wild-type or WT in the text) was donated by R. Kolter, Harvard Medical School. The rhlA− mutant strain was constructed from PA14 by deleting rhlA gene in its entirety using a suicide plasmid constructed by the gene splicing by overlap extension technique (Horton et al., 1990). rhlAB exists as a monocistronic operon in this parent strain, and therefore the rhlA− mutant was constructed in a fashion that rhlB may remain functional and under the control of its native promoter. GFP and DsRedExpress constitutively labeled varieties of WT and rhlA− were prepared using the miniTn7 transposon delivery plasmid, resulting in expression by the constitutive promoter PA1/04/03GFP (Lambertsen et al., 2004). Plasmids pYL122 (Lequette & Greenberg, 2005) (containing the PrhlABGFP fusion provided by E.P. Greenberg, U. Washington) and pEC16 (Boles et al., 2005)(containing PBADrhlAB, provided by P.K. Singh, U. of Washington) were used to integrate the constructs into the P. aeruginosa chromosome as a single copy. In the construction of the inducible strain PA14 rhlA− PBADrhlAB, although the rhlA− background strain keeps its functional rhlB under the control of its native promoter, the inclusion of rhlAB ensures that mono-rhamnolipid synthesis is completed in the inducible strain in the event that the native rhlB is inactive or its product stoichiometrically limiting thus allowing the measurement of secreted rhamnolipids.
Media and assays
The minimal media for plate and liquid assays was prepared using the following recipe: 800 mL of Milipore water (with no agar for liquid assays, with 0.625 % agar for swarming assays, with 1.857 % agar for hard agar assays), 200 mL of 5X stock phosphate buffer, 1mL of 1 M magnesium sulfate, 0.1 mL of magnesium sulfate, 25 mL of 200 g/L solution of casamino acids (Bacto™ from BD, Sparks, MD). 1 L of 5X stock phosphate buffer was prepared by dissolving 12 g of Na2HPO4 (anhydrous), 15 of KH2PO4 (anhydrows) and 2.5 g of NaCl into 1 L of Milipore water. The final pH of medium was 6.7. When necessary, media composition was altered as described in the text. Autoinducers N-(3-Oxododecanoyl)-L-homoserine lactone (called HSL in the text) and N-Butyryl-DL-homoserine lactone (called C4HSL in the text) were acquired from Sigma-Aldrich (St. Louis, US). Each swarming plate was prepared by pouring exactly 20 mL of medium onto a Petri dish and allowed to cool upright for 30 min. The plates were then turned upside down and left at room temperature to dry for 15 h. Inocula were prepared from 1 mL of overnight cultures washed twice with PBS. Plates inoculation was carried out by spotting a 2 μL drop of pre washed culture at the center of the swarming plate and allowed to dry. Plates were then placed upside down at 37C for 24 h. Each swarming experiment was repeated nine times (three different days with three experimental replicates each). All liquid assays were carried out at 37C with shaking, in 96-well microtiter plates using the Safire 2 (Tecan US, Inc) with OD600 and green fluorescence measured at 10 minute intervals.
Imaging and quantification
Still pictures were taken with a gel doc imager (AlphaInnotech ChemiImager). Time lapse videos were acquired using a Marshall electronics v-1070 surveillance camera, set up in a room acclimatized to 37C. Swarming plates with fluorescently labeled strains were imaged using the Amersham Typhoon 9400 (GE Healthcare). Colony forming units (CFU) were estimated by plating serial dilutions with different strains distinguished by fluorescent color. Data points for number of cells in colony and rhamnose secreted measurements shown in plots represent the median value among all experimental replicates, with error bars representing the 95- and 5-percentile. Cell number ratios were determined by dividing the CFU number of one color by the CFU number of the other color. Error bars for such ratio measurements were estimated from binomial distribution fitting (Johnson et al., 1993). rhlA expression in swarming assays was assessed by quantifying the total GFP expression in swarming colonies of a strain containing the PrhlABGFP construct. The total colony fluorescence was measured in Photoshop and normalized by the fluorescence of a colony expressing GFP constitutively (promoter PA1/04/03GFP). Secreted lipids were extracted from growth supernatants using a chloroform/methanol extraction protocol adapted from (Caiazza et al., 2007). The rhamnolipids in the extract were measured using the anthrone colorimetric assay (Zhu & Rock, 2008). The amount of rhamnose in culture supernatant (47.4 mg/L for the WT grown for 24 h in the standard minimal medium) was calibrated using a rhamnose calibration curve. This was converted into rhamnolipid concentration applying a conversion factor of 3.0 to 3.2 (Camilios Neto et al., 2008), leading to the concentration of biosurfactants of 0.14–0.16 g/L. The concentration of dry mass of cells (0.717 g/L) was measured by gravimetry. Nalgene sterile analytical filter units (Thermo Fisher Scientific, Rochester, NY) with 0.2 μm pore size where pre-dried for 24 h at 65 C and used to filter 120 mL of culture. The filters where then dry for 48 h until mass became stable over time.
Supplementary Material
Acknowledgments
We thank Mike Laub, Bodo Stern, Mike Cant, Joan Strassman and Karina Xavier for comments on the manuscript. We thank Bonnie Bassler for comments and for suggesting the experiments in figs 5B and 5C, and Justina Sanny for help in constructing reporter fusion strains and quantification of fluorescence in swarming assays. This work was supported by a National Institute of General Medical Sciences Center of Excellence grant (5P50 GM 068763-01) to K.R.F.
References
- Alhede M, Bjarnsholt T, Jensen PO, Phipps RK, Moser C, Christophersen L, Christensen LD, Gennip M van, Parsek M, Hoiby N, Rasmussen TB, Givskov M. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. 2009. pp. 3500–3508. [DOI] [PubMed] [Google Scholar]
- Arvidson S. Extracellular enzymes. In: Fischetti RPNVA, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens. Washington DC: ASM Press; 2000. pp. 379–385. [Google Scholar]
- Bassler B, Losick R. Bacterially Speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- Brockhurst M, Buckling A, Racey D, Gardner A. Resource supply and the evolution of public-goods cooperation in bacteria. BMC Biology. 2008 doi: 10.1186/1741-7007-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Merritt JH, Brothers KM, O’Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. Journal of Bacteriology. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Shanks RMQ, O’Toole GA. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. Journal of Bacteriology. 2005;187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilios Neto D, Meira J, de Araújo J, Mitchell D, Krieger N. Optimization of the production of rhamnolipids by Pseudomonas aeruginosa UFPEDA 614 in solid-state culture. Appl Microbiol Biotechnol. 2008;81:441–448. doi: 10.1007/s00253-008-1663-3. [DOI] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiology and Molecular Biology Reviews. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Rivoire O, Leibler S. Simpson’s paradox in a synthetic microbial system. Science. 2009;323:272–275. doi: 10.1126/science.1166739. [DOI] [PubMed] [Google Scholar]
- Clutton-Brock TH, Gaynor D, Kansky R, MacColl ADC, McIlrath G, Chadwick P, Brotherton PNM, O’Riain JM, Manser M, Skinner JD. Costs of cooperative behaviour in suricates (Suricata suricatta) 1998. pp. 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock TH, O’Riain MJ, Brotherton PN, Gaynor MD, Kansky R, Griffin AS, Manser M. Selfish Sentinels in Cooperative Mammals. 1999. pp. 1640–1644. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Crespi BJ. The evolution of social behavior in microorganisms. Trends Ecol Evol. 2001;16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- Deziel E, Lepine F, Milot S, Villemur R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the precursors of rhamnolipids. 2003. pp. 2005–2013. [DOI] [PubMed] [Google Scholar]
- Diggle S, Griffin A, Campbell G, West S. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- Dugatkin LA, Perlin M, Lucas JS, Atlas R. Group-Beneficial Traits, Frequency-Dependent Selection and Genotypic Diversity: An Antibiotic Resistance Paradigm. Proceedings: Biological Sciences. 2005;272:79–83. doi: 10.1098/rspb.2004.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K, Parkinson K, Thompson C. What can microbial genetics teach sociobiology? Trends in Genetics. 2006;23:74–80. doi: 10.1016/j.tig.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K, Shaulsky G, Strassmann J, Queller D, Thompson C. Pleiotropy as a mechanism to stabilize cooperation. Nature. 2004;431:693–696. doi: 10.1038/nature02894. [DOI] [PubMed] [Google Scholar]
- Foster KR. Biomedicine. Hamiltonian medicine: why the social lives of pathogens matter. Science. 2005;308:1269–1270. doi: 10.1126/science.1108158. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Romling U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ Microbiol. 2001;3:638–648. doi: 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- Glick R, Gilmour C, Tremblay J, Satanower S, Avidan O, Deziel E, Greenberg EP, Poole K, Banin E. Increase in Rhamnolipid Synthesis under Iron-Limiting Conditions Influences Surface Motility and Biofilm Formation in Pseudomonas aeruginosa. J Bacteriol. 2010;192:2973–2980. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- Guerrasantos L, Kappeli O, Fiechter A. Pseudomonas-Aeruginosa Biosurfactant Production in Continuous Culture with Glucose as Carbon Source. Applied and Environmental Microbiology. 1984;48:301–305. doi: 10.1128/aem.48.2.301-305.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haba E, Pinazo A, Jauregui O, Espuny MJ, Infante MR, Manresa A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by <I>Pseudomonas aeruginosa</I> 47T2 NCBIM 40044. 2003. pp. 316–322. [DOI] [PubMed] [Google Scholar]
- Harder W, Dijkhuizen L. Physiological-Responses to Nutrient Limitation. Annual Review of Microbiology. 1983;37:1–23. doi: 10.1146/annurev.mi.37.100183.000245. [DOI] [PubMed] [Google Scholar]
- Harrison F, Buckling A. Siderophore production and biofilm formation as linked social traits. ISME J. 2009;3:632–634. doi: 10.1038/ismej.2009.9. [DOI] [PubMed] [Google Scholar]
- Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Camara M, Williams P, Haas D. Positive Control of Swarming, Rhamnolipid Synthesis, and Lipase Production by the Posttranscriptional RsmA/RsmZ System in Pseudomonas aeruginosa PAO1. 2004. pp. 2936–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Johnson NL, Kotz S, Kemp AW. Univariate Discrete Distributions. Wiley-Interscience; Hoboken, NJ: 1993. [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T, Curty LK, Barja F, Delden C van, Pechere J-C. Swarming of Pseudomonas aeruginosa Is Dependent on Cell-to-Cell Signaling and Requires Flagella and Pili. 2000. pp. 5990–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb J, Heinze J. Ecology of Social Evolution. Heidelberg: Springer; 2008. p. 266. [Google Scholar]
- Kotte O, Zaugg JB, Heinemann M. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol Syst Biol. 2010;6:355. doi: 10.1038/msb.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerli R, Jiricny N, Clarke LS, West SA, Griffin AS. Phenotypic plasticity of a cooperative behaviour in bacteria. 2009;22:589–598. doi: 10.1111/j.1420-9101.2008.01666.x. [DOI] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Molecular microbiology. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. Journal of Bacteriology. 2005;187:37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina G, Juarez K, Soberon-Chavez G. The Pseudomonas aeruginosa rhlAB Operon Is Not Expressed during the Logarithmic Phase of Growth Even in the Presence of Its Activator RhlR and the Autoinducer N-Butyryl-Homoserine Lactone. 2003. pp. 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Foster KR, Xavier JB. Emergence of Spatial Structure in Cell Groups and the Evolution of Cooperation. PLoS Comput Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Koch AK, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. 1994. pp. 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. How did cooperative behavior evolve. Science. 2005;309:93–93. doi: 10.1126/science.309.5731.93. [DOI] [PubMed] [Google Scholar]
- Perkins TJ, Swain PS. Strategies for cellular decision-making. Mol Syst Biol. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. 2000. pp. 4885–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RC, Roberts P, Munro N, Rutman A, Hastie A, Shryock T, Hall R, McDonald-Gibson W, Lund V, Taylor G, et al. Effect of Pseudomonas aeruginosa rhamnolipids on mucociliary transport and ciliary beating. 1992. pp. 2271–2277. [DOI] [PubMed] [Google Scholar]
- Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinbergen N. On Aims and Methods of Ethology. Zeitschrift für Tierpsychologie. 1963;20:410–433. [Google Scholar]
- Venturi V, Bertani I, Kerényi Ádám, Netotea S, Pongor Sándor. Co-Swarming and Local Collapse: Quorum Sensing Conveys Resilience to Bacterial Communities by Localizing Cheater Mutants in <italic>Pseudomonas aeruginosa</italic>. PLoS ONE. 2010;5:e9998. doi: 10.1371/journal.pone.0009998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. Living on a surface: swarming and biofilm formation. Trends in Microbiology. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Visca P, Imperi F, Lamont I. Pyoverdine siderophores: from biogenesis to biosignificance. Trends in Microbiology. 2007;15:22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffins AS. The social lives of microbes. Annual Review of Ecology Evolution and Systematics. 2007a;38:53–77. [Google Scholar]
- West SA, Griffin AS, Gardner A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. Journal of Evolutionary Biology. 2007b;20:415–432. doi: 10.1111/j.1420-9101.2006.01258.x. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Yarwood JM, Volper EM, Greenberg EP. Delays in Pseudomonas aeruginosa quorum-controlled gene expression are conditional. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9008–9013. doi: 10.1073/pnas.0503728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Rock CO. RhlA Converts {beta}-Hydroxyacyl-Acyl Carrier Protein Intermediates in Fatty Acid Synthesis to the {beta}-Hydroxydecanoyl-{beta}-Hydroxydecanoate Component of Rhamnolipids in Pseudomonas aeruginosa. 2008;190:3147–3154. doi: 10.1128/JB.00080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulianello L, Canard C, Kohler T, Caille D, Lacroix J-S, Meda P. Rhamnolipids Are Virulence Factors That Promote Early Infiltration of Primary Human Airway Epithelia by Pseudomonas aeruginosa. 2006. pp. 3134–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.