Abstract
Purpose
Development of a radiosensitivity predictive assay is a central goal of radiation oncology. We reasoned a gene expression model could be developed to predict intrinsic radiosensitivity and treatment response in patients.
Methods and Materials
Radiosensitivity (determined by survival fraction at 2 Gy) was modeled as a function of gene expression, tissue of origin, ras status (mut/wt), and p53 status (mut/wt) in 48 human cancer cell lines. Ten genes were identified and used to build a rank-based linear regression algorithm to predict an intrinsic radiosensitivity index (RSI, high index = radioresistance). This model was applied to three independent cohorts treated with concurrent chemoradiation: head-and-neck cancer (HNC, n = 92); rectal cancer (n = 14); and esophageal cancer (n = 12).
Results
Predicted RSI was significantly different in responders (R) vs. nonresponders (NR) in the rectal (RSI R vs. NR 0.32 vs. 0.46, p = 0.03), esophageal (RSI R vs. NR 0.37 vs. 0.50, p = 0.05) and combined rectal/esophageal (RSI R vs. NR 0.34 vs. 0.48, p = 0.001511) cohorts. Using a threshold RSI of 0.46, the model has a sensitivity of 80%, specificity of 82%, and positive predictive value of 86%. Finally, we evaluated the model as a prognostic marker in HNC. There was an improved 2-year locoregional control (LRC) in the predicted radiosensitive group (2-year LRC 86% vs. 61%, p = 0.05).
Conclusions
We validate a robust multigene expression model of intrinsic tumor radiosensitivity in three independent cohorts totaling 118 patients. To our knowledge, this is the first time that a systems biology-based radiosensitivity model is validated in multiple independent clinical datasets.
Keywords: Intrinsic radiosensitivity, Gene expression, Predictive assay, Chemoradiation, Systems biology
INTRODUCTION
Personalized medicine holds the promise that the diagnosis, prevention, and treatment of cancer will be based on individual assessment of risk (1). Significant advances toward personalized radiation therapy (RT) have been largely achieved by physical advances in radiotherapy treatment planning and delivery (2). In contrast, the efforts in understanding the biological parameters that define intrinsic radiosensitivity have not met the same success. Thus, RT is prescribed without considering the potential individual differences in tumor and patient radiosensitivity. However, there is evidence to suggest that differences in intrinsic radiosensitivity exist (3) and understanding their biological basis could significantly impact clinical practice. Thus, a successful radiosensitivity predictive assay would be central to the development of biologically guided personalized treatment strategies in radiation oncology. A number of promising approaches have been developed in the past: (1) determination of ex vivo tumor SF2 (4–6); (2) electrodes to measure tumor hypoxia (7); and (3) determination of tumor proliferative potential (8, 9). However, none has become routine in the clinic.
The advent of high-dimensional and high-throughput technologies have provided an opportunity to address the development of biomarkers from a different perspective. For example, gene expression signatures have been shown to be prognostic in breast, lung, and head-and-neck (HNC) cancer (10–12). Further, recent studies have identified biomarkers predictive of patient response to drug treatment (13). Moreover, RT may represent a common denominator in cancer therapeutics, because approximately 60% of cancer patients are treated with RT (14). We have previously shown that gene expression can predict cellular intrinsic radiosensitivity (15). In addition, we developed a systems biology model of radiation sensitivity that identified 10 hub genes (see page 497 of this issue). We reasoned a gene expression model could be developed to predict radiosensitivity in patients from these hub genes.
In this article, we apply a novel multigene expression model of intrinsic tumor radiosensitivity. The model is based on the expression of 10 hub genes identified by the systems biology model of radiosensitivity. This model predicts a radiosensitivity index (RSI) that is directly proportional to tumor radioresistance. We clinically validate the model as a predictive factor of pathological response in two independent cohorts of esophageal (n = 12) and rectal (n = 14) cancer patients treated with preoperative concurrent chemoradiation in prospective clinical trials at Moffitt Cancer Center. In addition, we find RSI is of prognostic value in a third external dataset of HNC cancer patients (HNC, n = 92) treated with definitive concurrent chemoradiation within Phase 2 and 3 clinical trials at the Netherlands Cancer Institute. In conclusion, we think this model may play a central role in individualizing therapy in radiation oncology.
METHODS AND MATERIALS
Rectal cancer cohort
Fourteen patients were enrolled in an institutional review board–approved prospective Phase 1 trial evaluating escalating doses of oral topotecan as a radiosensitizing agent. Informed consent was obtained before enrollment. Eligibility criteria included patients with histologically confirmed rectal cancer, a primary tumor ≥3 cm, clinical stage ≥T2, and Eastern Cooperative Oncology Group performance status <2. All subjects were treated at Moffitt Cancer Center and were clinically staged by endoscopic ultrasound (EUS). Tumor biopsies (five core biopsies) were obtained before initiation of therapy and snap frozen in liquid nitrogen. No tumor macro- or microdissection was performed.
All subjects were treated with preoperative concurrent radiochemotherapy and underwent surgical resection (abdominoperineal resection or low anterior resection in 13/14) within 8 weeks of completion of preoperative treatment. The starting dose of oral topotecan was 0.25 mg/m2 and it was administered at least 3 h before RT on a daily basis. Patients were treated to 45 Gy to a standard pelvic field (three- or four-field three-dimensional conformal technique). Table 1 shows the clinical characteristics of this cohort.
Table 1.
Clinical characteristics: Rectal Cancer Trial
| Sex | ||
| Male | 10 | |
| Female | 4 | |
| Age (y) | ||
| Mean | 69.4 | |
| Median (range) | 72 | (50–90) |
| Chemotherapy dose | ||
| 0.25 mg/m2/day | 3 | (21) |
| 0.4 mg/m2/day | 5 | (36) |
| 0.55 mg/m2/day | 6 | (43) |
| Ultrasound tumor stage | ||
| T3 | 14 | (100) |
| Pathologic tumor stage | ||
| T0 | 2 | (14.3) |
| Tis | 1 | (7) |
| T1 | 2 | (14.3) |
| T2 | 3 | (21.4) |
| T3 | 5 | (36) |
| T4 | 1 | (7) |
| Downstaging | ||
| Yes | 8 | (57) |
| No | 6 | (43) |
Values are number (percentage) unless otherwise noted.
Response definition (rectal cancer)
Pathological response was defined by at least a decrease of one T stage in the primary tumor between the pretreatment EUS and the pathological evaluation of the specimen (16, 17). Pathological complete response was defined as no evidence of tumor in the surgical specimen (primary and nodes). Based on this definition, 57% (8/14) of the patients were considered responders.
Esophageal cancer cohort
Twelve patients were enrolled in an institutional review board–approved prospective tissue collection trial aimed at defining prognostic molecular signatures in esophageal cancer. Clinical management was not dictated by the protocol and left to the clinical judgment of the treating physicians. Treatment details are presented in Table 2. Eligibility criteria included a histological diagnosis of esophageal cancer, deemed a reasonable candidate for preoperative radiochemotherapy or esophagectomy, an Eastern Cooperative Oncology Group performance <2, and chemotherapy-naive. Subjects were clinically staged by EUS. All tumor biopsies were obtained before treatment and snap frozen in liquid nitrogen. No macro- or microdissection was performed.
Table 2.
Clinical characteristics: Esophageal Trial
| Sex | ||
| Male | 7 | (58.3) |
| Female | 5 | (41.7) |
| Age (y) | ||
| Mean | 67.08 | |
| Median (range) | 66 | (51–80) |
| Chemotherapy regimen | ||
| CDDP + 5-FU | 4 | (33) |
| 5-FU | 2 | (16.7) |
| Carbo/Tax + 5-FU | 1 | (8.3) |
| NA | 5 | (42) |
| Radiation dose | ||
| 45 | 1 | (8.3) |
| 50.4 | 5 | (42) |
| 54 | 2 | (16.7) |
| 55.8 | 1 | (8.3) |
| 61.2 | 1 | (8.3) |
| NA | 2 | (16.7) |
| Clinical tumor stage | ||
| T2N1 | 1 | (8.3) |
| T3N0 | 1 | (8.3) |
| T3N1 | 7 | (58.4) |
| T4N1 | 3 | (25) |
| Pathologic tumor stage | ||
| T0N0 | 4 | (33.3) |
| T0N1 | 1 | (8.3) |
| T1aN0 | 1 | (8.3) |
| T1N1 | 2 | (16.7) |
| T2bN1 | 1 | (8.3) |
| T2N1 | 1 | (8.3) |
| Progressive diagnosis | 2 | (16.7) |
| Downstaging | ||
| Yes | 7 | (58.3) |
| No | 5 | (41.7) |
Values are number (percentage) unless otherwise noted.
Abbreviations: CDDP = cisplatin; 5-FU = 5-flourouracil; carbo/tax = carboplatin + taxol.
Subjects were treated with concurrent radiochemotherapy to be followed by planned esophagectomy. Nine of 12 underwent planned esophagectomy. Three patients completed concurrent radiochemotherapy but were not operated on because of patient or physician preference (2 patients) or progressive disease (1 patient). The clinical characteristics of this cohort are summarized in Table 2.
Response definition (esophageal cancer)
This was defined as a decrease of at least two T stages between the pretreatment EUS evaluation and the pathological evaluation of the specimen (18). Three patients did not undergo esophagectomy. One had progressive disease during preoperative therapy, and 2 experienced clinical complete responses (documented by positron emission tomography and/or EUS and biopsy) and had no evidence of disease at least 1 year after completion of treatment. Because pathologic information was not available, response for these 3 patients was defined on clinical criteria. The patient with progressive disease was deemed a nonresponder, whereas the 2 patients with documented clinical complete responses and at least 1 year of follow-up were deemed responders. Based on this definition, 50% (6/12) of the patients were considered responders.
HNC cohort
Ninety-two patients were treated within prospective randomized Phase II–III trials at The Netherlands Cancer Institute (19). Tumors were mostly locally advanced (94% T3 and above, 74% N1 and above). The full clinical details of this cohort were previously published (19). All patients were treated with concurrent radiochemotherapy with cisplatin-based chemotherapy. Radiation dose was 70 Gy in 2-Gy fractions. Three schedules of cisplatin were given: 100 mg/m2 intravenously three times during radiotherapy; 150 mg/m2 given intra-arterially four times during radiotherapy; and 20 × 6 mg/m2 daily. No disease outcome differences were found between chemotherapy schedules.
Microarrays (rectal and esophageal cohorts)
Total RNA was isolated using the TRIzol Reagent (Invitrogen, Carlsbad, CA) and the manufacturer’s protocol and purified using the RNeasy cleanup procedure (Qiagen Inc., Valencia, CA). RNA quality was assessed by gel electrophoresis, A260/280 ratio, or the Agilent 2100 Bioanalyzer. Five micrograms of total RNA was processed. The poly(A) RNA was converted to cDNA, amplified, and labeled with biotin (20). Hybridization with the biotin-labeled RNA, staining, and scanning was performed as described (21). Affymetrix U133Plus2.0 chips were normalized using the robust multiarray analysis method (22).
Microarrays (HNC cohort)
These methods were previously published (19). Briefly, pretreatment biopsies were taken during examination under general anesthesia and snap frozen in liquid nitrogen. Biopsies containing >50% tumor cells were used. Extracted RNA was quality controlled by Bioanalyzer, amplified, labeled, and hybridized to 70-mer oligo microarrays containing 34,580 probes representing 24,650 genes (Operon v.3.0).
Ten gene systems model
The model was developed in 48 cancer cell lines from the National Cancer Institute panel of 60. Radiosensitivity measurements (SF2) were either determined in our lab (25 cell lines) or obtained from the literature (23 cell lines). Gene expression was from Affymetrix HU6800 Genechips from a previous study (23) and were preprocessed using MAS 5.0.
We developed a systems model of radiosensitivity by expanding a previously validated model (15) to include biological variables known to influence radiophenotype: tissue of origin, ras status (mut vs. wild-type), and p53 status (mut vs. wild-type). This analysis is given by the following equation (Tables E1–4):
The model consisted of all nonsingular terms (28 terms) including gene expression (yx), p53 mutation status, ras mutation status, tissue of origin, and all possible interactions among terms (Table E5). TO, p53 mutation and ras mutation status are categorical variables and were coded as dummy variables. This analysis is performed on a gene-by-gene basis, totaling 7,168 probe sets. The 500 gene-based models with the smallest sum of squared residuals (best linear fit) were selected for further analysis (Table E6).
The 500 selected genes were uploaded into the GeneGO Meta-Core software (GeneGO, Encinitas, CA). The primary edges (interconnections) were plotted using literature-based annotations and the model was reduced by identifying all genes (network hubs) with more than five edges and less than 50% of edges hidden within the network (Table E7). This resulted in the 10 genes described in this article. The Gather program was used to identify significant relationships of terms from the 10 genes (threshold p < 0.005).
Predictive model development
Gene expressions for the 10 genes were rank-ordered, with lowest expression ranked 1. Radiosensitivity was modeled using a linear regression model of the 10 genes in the 48 cell lines using the R software (see Results for equation). This model was applied to similarly rank-ordered patient data to generate the RSI.
Statistical analyses
A one-sided Mann-Whitney test was used to determine if the predicted RSI was significantly higher for nonresponders. Bar charts of patient response were graphed using mean and standard error values for each response group. Locoregional recurrence was defined previously (19). Locoregional control differences between low and high RSI values were calculated using the log–rank test. Because the model does not account for the radiosensitizing effect of chemotherapy, we expected the model would be most accurate in the most radiosensitive quartile. Thus, the RSI cut point for HNC patients was predefined at the 25th percentile.
Microarray platform translation—gene mapping
Probes were mapped from the HU6800 platform to the HG-U133 Plus 2.0 platform and NKI array format by mapping the probe sequences onto a corresponding NCBI refseq ID or genomic region, then identifying the closest probe match on the new microarray platform (Table 3).
Table 3.
Radiation network hub genes
| Gene name | HU6800 Probeset | U133Plus Probeset | NKI reporter |
|---|---|---|---|
| Androgen receptor | M23263_at | 211110_s_at | 324293 |
| c-Jun | J04111_at | 201466_s_at | 329987 |
| STAT1 | AFFX-HUMISGF3A/M97935_MA_at | AFFX-HUMISGF3A/M97935_MA_at | 308421 |
| PKC | X06318_at | 207957_s_at | 322907 |
| RelA (p65) | U33838_at | 201783_s_at | 326475 |
| c-Abl | X16416_at | 202123_s_at | 304192 |
| SUMO-1 | U83117_at | 208762_at | 308596 |
| CDK1 (p34) | U24153_at | 205962_at | 332859 |
| HDAC1 | D50405_at | 201209_at | 308690 |
| IRF1 | L05072_s_at | 202531_at | 310653 |
The probes used on each platform (Affymetrix HU6800, HGU133Plus2.0, and NKI cDNA arrays) are listed. Matches were identified via sequence similarity to the original HU6800 platform.
RESULTS
A radiosensitivity systems model captures central regulatory pathways in radiation response
Table 3 shows the 10 “hub” genes on which expression the radiosensitivity model is built. The selected genes are biologically important and are involved in regulating radiation signaling (24–32). In addition, 7/10 (HDAC1, PKC-beta, RelA, c-Abl, STAT1, AR, CDK1) have been studied as targets for radiosensitizer development (32–37). Furthermore, the Gene Ontology terms captured by the 10 gene systems model include DNA damage response, histone deacetylation, cell-cycle regulation, apoptosis, and proliferation, all of which play an important role in radiation response (34, 38, 39). In summary, the systems model captures central pathways and genes involved in regulating radiosensitivity.
Development of a radiosensitivity predictive model based on the systems model
We developed and optimized a linear regression algorithm to predict radiosensitivity, using gene expression of the 10 hub genes in the systems model. Translation of the model to other datasets was an important requirement; therefore, the hubs were assigned ranks by expression and the linear regression model was built from ranks (instead of absolute expression)(40). The model predicts a continuous RSI that is based on the survival fraction at 2 Gy (SF2), measured for the cell lines in the database. Thus, RSI is directly proportional to radioresistance (high index = radioresistance). Because the 10 hubs were selected from the cell line data, an estimate of accuracy generated by cross-validation of the model in the same cell lines would yield optimistically biased estimates of accuracy. Therefore, we used additional datasets for validation (clinical datasets). The rank-based linear regression equation is the following:
The radiosensitivity model predicts pathological response to chemoradiation in rectal and esophageal cancer
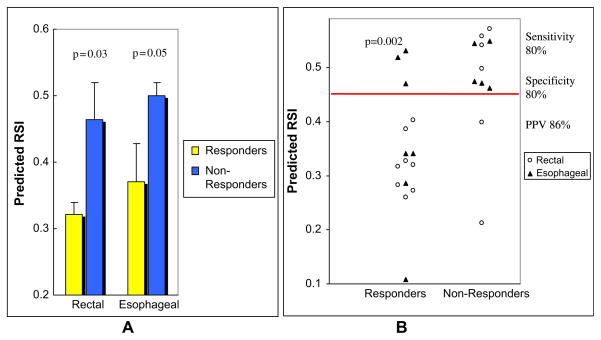
We applied the predefined model to the prediction of clinical response to concurrent radiochemotherapy in two independent prospectively collected pilot cohorts of patients with rectal (n = 14) and esophageal cancer (n = 12). Pathological response was defined by T stage criteria (see Methods). It should be emphasized that all features in the model were pre-defined, including the 10 genes, the rank-based linear regression approach, and the coefficients. The model significantly separated responders (R) from nonresponders (NR) in the pilot clinical cohort (Fig. 1) (all patients, mean predicted RSI, R vs. NR 0.34 vs. 0.48, p = 0.002). Importantly, the model was accurate in both disease cohorts despite the small number of patients (rectal cancer patients, mean predicted RSI, R vs. NR 0.32 vs. 0.46, p = 0.03) (esophageal cancer patients, mean predicted RSI, R vs. NR 0.37 vs. 0.50, p = 0.05).
Fig. 1.
Radiosensitivity index (RSI) is correlated with clinical response to concurrent radiochemotherapy in rectal and esophageal cancer patients. (A) The mean predicted RSI of responders is significantly lower than in nonresponders in both clinical cohorts (esophageal: p = 0.05, rectal: p = 0.03). (B) Predicted RSI of each individual patient in the cohorts (combined: p = 0.001511).
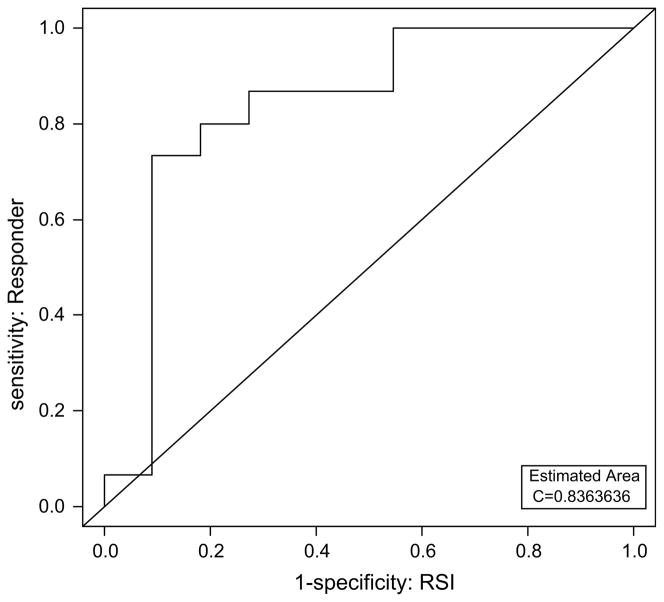
We generated a receiver-operating characteristic curve (Fig. 2) using the predicted RSI to determine the sensitivity and specificity of the predictor. Using a threshold RSI of 0.46, the model has a sensitivity and specificity of 80 and 82%, respectively, with a positive predictive value of 86%. Although preliminary, these numbers are encouraging because the predictor is not developed to account for the radiosensitizing effect of chemotherapy and we expected the inclusion of chemotherapy to account for prediction inaccuracies.
Fig. 2.
Receiver-operating characteristic curve using predicted radiosensitivity index (RSI) for radiosensitivity predictions. Using a threshold RSI of 0.4619592, the predictor has an 80% sensitivity and 82% specificity, with positive predictive value (PPV) of 86%. The estimated area under the curve (AUC) is 0.84.
The radiosensitivity predictive model is of prognostic value in HNC cancer
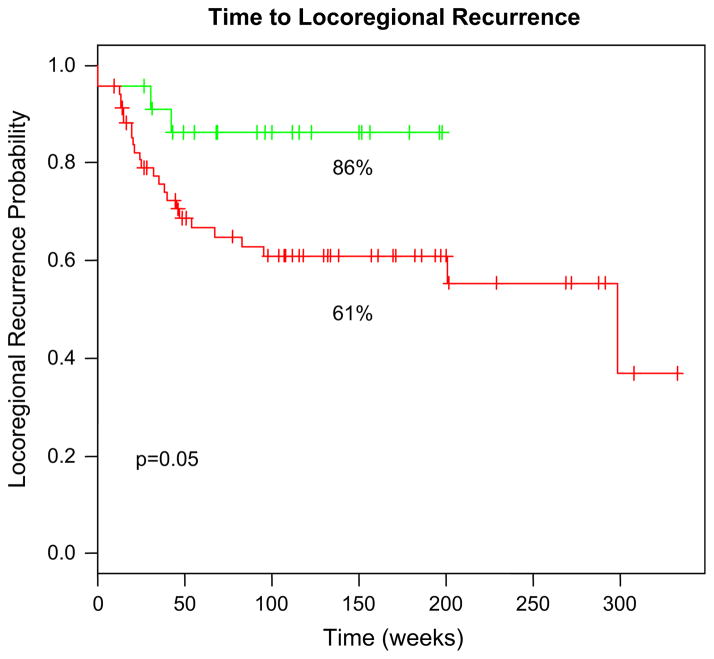
We further tested the model as a prognostic marker in locally advanced HNC patients treated with definitive concurrent radiochemotherapy. The clinical details of this cohort have been previously published (19). Briefly, the cohort included patients treated within Phase II and randomized Phase III trials at the Netherlands Cancer Institute (NKI). A total of 94% of patients presented with T3 or T4 disease and all patients were treated with concurrent radiochemotherapy (cisplatin-based). Gene expression profiles for all patients were generated using the NKI array. Using the same algorithm developed in cell lines and tested in the rectal and esophageal cohorts, we generated radiosensitivity predictions for this dataset. Interestingly, the average RSI prediction was lower in this disease site when compared with rectal and esophagus (predicted RSI, HNC vs. esophagus vs. rectal 0.06 vs. 0.43 vs. 0.39). Although this could be partly a function of radiosensitivity differences between these diseases, it could also be due to platform differences (Affymetrix U133 Plus vs. NKI array). Interestingly, in spite of these differences, the RSI was still of prognostic value within the HNC dataset. The predicted radiosensitive group had an improved 2-year locoregional control (2-year locoregional control 86% vs. 61%, p = 0.05), thus arguing that the model is capturing biological commonalities that determine tumor radiosensitivity across disease sites (Fig. 3).
Fig. 3.
Radiosensitivity index (RSI) distinguishes clinical populations with different disease-related outcomes in a head-and-neck cancer (HNC) cohort of 92 patients treated with definitive concurrent radiochemotherapy. Using the 25th percentile (RSI < 0.023), there is a superior 2-year locoregional control (LRC) in the predicted radiosensitive group (86% vs. 61%, p = 0.05).
DISCUSSION
The development of in vitro diagnostics to predict response to therapeutic agents is a central goal of molecular medicine (1). In this study, we validate a robust systems biology-based multigene expression model of intrinsic tumor radiosensitivity in three independent datasets totaling 118 patients. Although previous studies have shown that radiosensitivity signatures were possible (15, 41, 42), this is the first time to our knowledge that a systems biology-based radiosensitivity model is validated in multiple independent clinical datasets. We show that RSI when analyzed as a continuous variable is correlated with pathological response in rectal and esophageal cancer patients treated with preoperative concurrent chemoradiation. Furthermore, the receiver-operating characteristic analysis proposes a cut point (RSI = 0.46) where the test predictive accuracy is encouraging. The sensitivity, specificity, and positive predictive value of the assay were 80%, 82%, and 86%, respectively. Importantly, we also show that RSI is of prognostic significance in a cohort of 92 patients with locally advanced HNC. The applicability of the model in three different disease sites strongly suggests that the model captures commonalities that define radiosensitivity across disease sites. Therefore, it is possible that the model might be generally applicable to other disease sites (e.g., lung, prostate, cervix cancer). However, it should be emphasized that no disease-specific conclusions should be made at this juncture given the small nature of two of the validating clinical cohorts (rectal, esophageal).
In the molecular medicine era, high-throughput technologies (e.g., microarrays, proteomics) have led to the identification of numerous molecular signatures of prognostic or predictive significance (10–12, 43). However the initial enthusiasm that these signatures would lead to personalized medicine has been dampened by lack of robustness (44). The robustness of the radiosensitivity model is supported by several lines of evidence. First, the algorithm was validated in three independent prospectively collected datasets in three different diseases. Second, the model was valid across different gene expression platforms. The model is originally developed on an Affymetrix HU-6800 platform, but clinically validated in two different gene expression platforms (i.e., Affymetrix U133-Plus for esophageal and rectal cohorts, NKI cDNA array for HNC). This observation suggests the model can be transferred to a more practical clinical platform (i.e., RT-PCR/formalin-fixed tissue). Third, all patients in the validating clinical cohorts were treated with concurrent chemoradiation, because we were unable to obtain a dataset of patients treated with radiation alone. However, the algorithm was based on cellular radiosensitivity. Thus, in spite of this potential source of inaccuracy, the model was still validated. Finally, the model showed both predictive and prognostic value.
False negatives (predicted radioresistant that responded) were the main inaccuracy when the model was dichotomized in the esophageal and rectal datasets. This population represented 60% of the misclassified cases in these cohorts. This inaccuracy may be due to the radiosensitization effect of chemotherapy. The proportion of individuals that are classified in this group (11.5%) is consistent with the observed improvement in clinical responses with concurrent chemotherapy over radiotherapy alone (45–47). Therefore, this effect may be addressed by analyzing gene expression differences between R and NR that share a predicted radioresistant phenotype.
The model in this study is designed to predict tumor radiosensitivity. Interestingly RSI was prognostic in the HNC dataset, suggesting that the biologic factors that determine radiosensitivity are related to disease prognosis after treatment. This is not surprising because complete pathological response has been shown to have strong prognostic significance in several studies (17, 18, 48, 49).
This model may play a central role in the individualization of therapy in radiation oncology. For example, the model may provide an opportunity to individualize radiation dose parameters based on intrinsic radiosensitivity. Because higher doses of RT are associated with higher toxicity rate (50), dose personalization would result in a therapeutic ratio benefit. There is also a role for identifying patients that are likely to be downstaged, particularly in rectal cancer. For example, this knowledge might lead to better counseling of patients with low-lying rectal tumors where sphincter-sparing surgery is being considered. In addition, the model may provide a unique framework to understand the differences between R and NR that share a predicted radioresistant phenotype. This may allow the accurate identification of patients that benefit from the addition of concurrent chemotherapy.
In conclusion, we present evidence to support the clinical validity of a multigene expression model of intrinsic tumor radiosensitivity. To our knowledge, this is the first systems biology-based radiosensitivity model to have validation in multiple independent datasets. The model is versatile and robust as demonstrated by both its predictive and prognostic ability in three different disease sites using two different gene expression microarray platforms. The data presented justify further development and optimization of this technology in larger clinical populations.
Acknowledgments
Supported by NCI K08 CA 108926-05, NCI Grant R21CA101355, NFGC-DAMD 170220051.
Footnotes
Conflict of interest: S.E. and J.T.R. are named as inventors in a patent application of the technology described.
References
- 1.Dalton WS, Friend SH. Cancer biomarkers—an invitation to the table. Science. 2006;312:1165–1168. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 2.Bucci MK, Bevan A, Roach M., III Advances in radiation therapy: Conventional to 3D, to IMRT, to 4D, and beyond. CA Cancer J Clin. 2005;55:117–134. doi: 10.3322/canjclin.55.2.117. [DOI] [PubMed] [Google Scholar]
- 3.Zelefsky MJ, Fuks ZVI, Hunt M, et al. High dose radiation delivered by intensity modulated conformal radiotherapy improves the outcome of localized prostate cancer. J Urol. 2001;166:876–881. [PubMed] [Google Scholar]
- 4.Bjork-Eriksson T, West C, Karlsson E, et al. Tumor radiosensitivity (SF2) is a prognostic factor for local control in head and neck cancers. Int J Radiat Oncol Biol Phys. 2000;46:13–19. doi: 10.1016/s0360-3016(99)00373-9. [DOI] [PubMed] [Google Scholar]
- 5.Buffa FM, Davidson SE, Hunter RD, et al. Incorporating biologic measurements (SF(2), CFE) into a tumor control probability model increases their prognostic significance: A study in cervical carcinoma treated with radiation therapy. Int J Radiat Oncol Biol Phys. 2001;50:1113–1122. doi: 10.1016/s0360-3016(01)01584-x. [DOI] [PubMed] [Google Scholar]
- 6.West CM, Davidson SE, Roberts SA, et al. The independence of intrinsic radiosensitivity as a prognostic factor for patient response to radiotherapy of carcinoma of the cervix. Br J Cancer. 1997;76:1184–1190. doi: 10.1038/bjc.1997.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fyles A, Milosevic M, Hedley D, et al. Tumor hypoxia has independent predictor impact only in patients with node-negative cervix cancer. J Clin Oncol. 2002;20:680–687. doi: 10.1200/JCO.2002.20.3.680. [DOI] [PubMed] [Google Scholar]
- 8.Begg AC, Haustermans K, Hart AA, et al. The value of pretreatment cell kinetic parameters as predictors for radiotherapy outcome in head and neck cancer: A multicenter analysis. Radiother Oncol. 1999;50:13–23. doi: 10.1016/s0167-8140(98)00147-9. [DOI] [PubMed] [Google Scholar]
- 9.Corvo R, Giaretti W, Sanguineti G, et al. In vivo cell kinetics in head and neck squamous cell carcinomas predicts local control and helps guide radiotherapy regimen. J Clin Oncol. 1995;13:1843–1850. doi: 10.1200/JCO.1995.13.8.1843. [DOI] [PubMed] [Google Scholar]
- 10.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 11.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 13.Giles FJ, DeAngelo DJ, Baccarani M, et al. Optimizing outcomes for patients with advanced disease in chronic myelogenous leukemia. Semin Oncol. 2008;35:S1–S17. doi: 10.1053/j.seminoncol.2007.12.002. quiz S18–S20. [DOI] [PubMed] [Google Scholar]
- 14.Perez C. Principles and management of radiation therapy. Philadelphia: Lippincott-Raven; 1998. [Google Scholar]
- 15.Torres-Roca JF, Eschrich S, Zhao H, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65:7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 16.Janjan NA, Khoo VS, Abbruzzese J, et al. Tumor downstaging and sphincter preservation with preoperative chemoradiation in locally advanced rectal cancer: The M.D. Anderson Cancer Center experience. Int J Radiat Oncol Biol Phys. 1999;44:1027–1038. doi: 10.1016/s0360-3016(99)00099-1. [DOI] [PubMed] [Google Scholar]
- 17.Janjan NA, Crane C, Feig BW, et al. Improved overall survival among responders to preoperative chemoradiation for locally advanced rectal cancer. Am J Clin Oncol. 2001;24:107–112. doi: 10.1097/00000421-200104000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Chirieac LR, Swisher SG, Ajani JA, et al. Posttherapy pathologic stage predicts survival in patients with esophageal carcinoma receiving preoperative chemoradiation. Cancer. 2005;103:1347–1355. doi: 10.1002/cncr.20916. [DOI] [PubMed] [Google Scholar]
- 19.Pramana J, Van den Brekel MW, van Velthuysen ML, et al. Gene expression profiling to predict outcome after chemoradiation in head and neck cancer. Int J Radiat Oncol Biol Phys. 2007;69:1544–1552. doi: 10.1016/j.ijrobp.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 20.Van Gelder RN, von Zastrow ME, Yool A, et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobbin KK, Beer DG, Meyerson M, et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005;11:565–572. [PubMed] [Google Scholar]
- 22.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng X, Hofmann ER, Villanueva A, et al. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat Genet. 2004;36:906–912. doi: 10.1038/ng1396. [DOI] [PubMed] [Google Scholar]
- 25.Hallahan DE, Dunphy E, Kuchibhotla J, et al. Prolonged c-jun expression in irradiated ataxia telangiectasia fibroblasts. Int J Radiat Oncol Biol Phys. 1996;36:355–360. doi: 10.1016/s0360-3016(96)00327-6. [DOI] [PubMed] [Google Scholar]
- 26.Kao GD, McKenna WG, Muschel RJ. p34(Cdc2) kinase activity is excluded from the nucleus during the radiation-induced G(2) arrest in HeLa cells. J Biol Chem. 1999;274:34779–34784. doi: 10.1074/jbc.274.49.34779. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappa B through two distinct mechanisms. PNAS. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fryknäs M, Dhar S, Oberg F, et al. STAT1 signaling is associated with acquired cross-resistance to doxorubicin and radiation in myeloma cell lines. Int J Cancer. 2007;120:189–195. doi: 10.1002/ijc.22291. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T, Yukawa O, Azuma C, et al. Involvement of protein kinase C-related anti-apoptosis signaling in radiation-induced apoptosis in murine thymic lymphoma(3SBH5) cells. Radiat Res. 2004;161:528–534. doi: 10.1667/rr3176. [DOI] [PubMed] [Google Scholar]
- 30.Pamment J, Ramsay E, Kelleher M, et al. Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene. 2002;21:7776–7785. doi: 10.1038/sj.onc.1205981. [DOI] [PubMed] [Google Scholar]
- 31.Terzoudi GI, Jung T, Hain J, et al. Increased G2 chromosomal radiosensitivity in cancer patients: the role of cdk1/cyclin-B activity level in the mechanisms involved. Int J Radiat Biol. 2000;76:607–615. doi: 10.1080/095530000138268. [DOI] [PubMed] [Google Scholar]
- 32.Wang QE, Zhu Q, Wani G, et al. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell JS, Brady K, Burgan WE, et al. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 34.Ma BBY, Bristow RG, Kim J, et al. Combined-modality treatment of solid tumors using radiotherapy and molecular targeted agents. J Clin Oncol. 2003;21:2760–2776. doi: 10.1200/JCO.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 35.Cerna D, Camphausen K, Tofilon PJ. Histone deacetylation as a target for radiosensitization. Curr Top Dev Biol. 2006;73:173–204. doi: 10.1016/S0070-2153(05)73006-4. [DOI] [PubMed] [Google Scholar]
- 36.Milas L, Mason KA, Liao Z, et al. Chemoradiotherapy: Emerging treatment improvement strategies. Head Neck. 2003;25:152–167. doi: 10.1002/hed.10232. [DOI] [PubMed] [Google Scholar]
- 37.Kaminski JM, Hanlon AL, Joon DL, et al. Effect of sequencing of androgen deprivation and radiotherapy on prostate cancer growth. Int J Radiat Oncol Biol Phys. 2003;57:24–28. doi: 10.1016/s0360-3016(03)00539-x. [DOI] [PubMed] [Google Scholar]
- 38.Marples B, Collis SJ. Low-dose hyper-radiosensitivity: past, present, and future. Int J Radiat Oncol Biol Phys. 2008;70:1310–1318. doi: 10.1016/j.ijrobp.2007.11.071. [DOI] [PubMed] [Google Scholar]
- 39.Lindsay KJ, Coates PJ, Lorimore SA, et al. The genetic basis of tissue responses to ionizing radiation. Br J Radiol. 2007;80(Spec1):S2–S6. doi: 10.1259/bjr/60507340. [DOI] [PubMed] [Google Scholar]
- 40.Xu L, Tan AC, Winslow RL, et al. Merging microarray data from separate breast cancer studies provides a robust prognostic test. BMC Bioinformatics. 2008;9:125. doi: 10.1186/1471-2105-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe T, Komuro Y, Kiyomatsu T, et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 2006;66:3370–3374. doi: 10.1158/0008-5472.CAN-05-3834. [DOI] [PubMed] [Google Scholar]
- 42.Cao C, Subhawong T, Albert JM, et al. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res. 2006;66:10040–10047. doi: 10.1158/0008-5472.CAN-06-0802. [DOI] [PubMed] [Google Scholar]
- 43.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 44.Simon R, Radmacher MD, Dobbin K, et al. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 45.Herskovic A, Martz K, al-Sarraf M, et al. Combined chemotherapy and radiotherapy compared with radiotherapy alone in patients with cancer of the esophagus. N Engl J Med. 1992;326:1593–1598. doi: 10.1056/NEJM199206113262403. [DOI] [PubMed] [Google Scholar]
- 46.Al-Sarraf M, Martz K, Herskovic A, et al. Progress report of combined chemoradiotherapy versus radiotherapy alone in patients with esophageal cancer: An intergroup study [published erratum appears in J Clin Oncol 1997 Feb;15(2):866] J Clin Oncol. 1997;15:277–284. doi: 10.1200/JCO.1997.15.1.277. [DOI] [PubMed] [Google Scholar]
- 47.Bosset J-F, Collette L, Calais G, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114–1123. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- 48.Gavioli M, Luppi G, Losi L, et al. Incidence and clinical impact of sterilized disease and minimal residual disease after preoperative radiochemotherapy for rectal cancer. Dis Colon Rectum. 2005;48:1851–1857. doi: 10.1007/s10350-005-0133-6. [DOI] [PubMed] [Google Scholar]
- 49.Capirci C, Valentini V, Cionini L, et al. Prognostic value of pathologic complete response after neoadjuvant therapy in locally advanced rectal cancer: Long-term analysis of 566 ypCR patients. Int J Radiat Oncol Biol Phys. 2008;72:99–107. doi: 10.1016/j.ijrobp.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 50.Peeters ST, Heemsbergen WD, van Putten WL, et al. Acute and late complications after radiotherapy for prostate cancer: Results of a multicenter randomized trial comparing 68 Gy to 78 Gy. Int J Radiat Oncol Biol Phys. 2005;61:1019–1034. doi: 10.1016/j.ijrobp.2004.07.715. [DOI] [PubMed] [Google Scholar]