Abstract
Intraspecific hybrids between the Arabidopsis thaliana accessions C24 and Landsberg erecta have strong heterosis. The reciprocal hybrids show a decreased level of 24-nt small RNA (sRNA) relative to the parents with the decrease greatest for those loci where the parents had markedly different 24-nt sRNA levels. The genomic regions with reduced 24-nt sRNA levels were largely associated with genes and their flanking regions indicating a potential effect on gene expression. We identified several examples of genes with altered 24-nt sRNA levels that showed correlated changes in DNA methylation and expression levels. We suggest that such epigenetically generated differences in gene activity may contribute to hybrid vigor and that the epigenetic diversity between ecotypes provides increased allelic (epi-allelic) variability that could contribute to heterosis.
Keywords: yield increase, epigenome, transposons, transacting siRNA, transmethylation
Hybrid vigor, or heterosis, is characterized by the superior performance of a hybrid over its parents in traits such as growth rate, biomass, and stress tolerance, leading to yield increases (1). Hybrids are important in many agricultural and horticultural crops including maize, rice, sunflower, and canola (reviewed in refs. 2–4).
Genetic QTL analyses attempting to pinpoint the genes responsible for heterosis have pointed to large numbers of genes contributing to the final assayed phenotype (5–9). There is the possibility that the mechanism of hybrid vigor is one of successive cascades of gene activity responding to underlying regulatory controls. As well as the multigene contributions toward increased plant performance, there are examples of single gene mutations resulting in large increases in biomass and yield in plants (10 and reviewed in ref. 6).
In general, the degree and frequency of heterosis correlates with the genetic distance between the parents, as evidenced by the significant heterosis generated from interspecies crosses (reviewed in refs. 2, 6, 11). Relative to interspecies hybrids, intraspecies hybrids generally have fewer genetic differences between parents. However, many intraspecies crosses do exhibit substantial heterosis, including those between rice subspecies, closely related tobacco varieties and Arabidopsis accessions (12–14). Allelic variation may be provided by the epigenome, where epigenetic modifications create epi-alleles with altered states of gene expression (15–19). These epi-alleles can contribute differences in gene activity between genetically close parents (20, 21) and may provide the variability needed to generate heterosis in hybrid combinations.
We examined small RNA (sRNA) populations in the Arabidopsis accessions C24, Landsberg erecta (Ler), and their reciprocal hybrids, which display strong intraspecies heterosis in a number of vegetative traits such as rosette diameter and biomass. The parental accessions had significant differences in their sRNA populations and associated methylation profiles. The hybrids differed greatly from the parents in their sRNA populations including a marked reduction in 24-nt sRNA associated with loci that differed in the frequency of 24-nt sRNAs between the parents. At these loci, the hybrids showed alterations in RNA-directed DNA methylation (RdDM), resulting in a decrease in CHH methylation. We give examples of representative genes that in the hybrids have changed levels of short interfering RNA (siRNA) and DNA methylation correlated with altered gene expression.
We suggest that the altered hybrid epigenome provides allelic variants that can influence the activity of a large number of metabolic and regulatory genes that could contribute to the heterotic phenotype.
Results
C24 and Ler Hybrids Show Significant Heterosis.
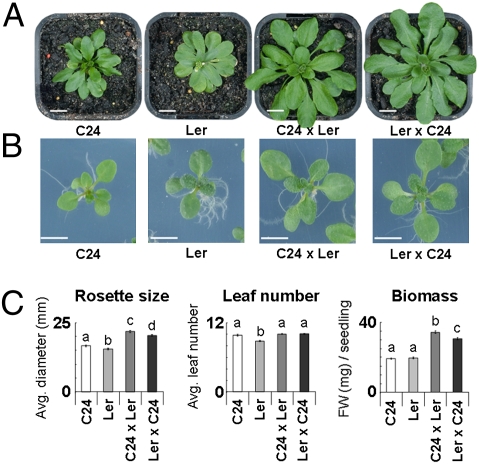
The F1 hybrids from crosses between the C24 and Ler accessions have significant vegetative heterosis (Fig. 1). The final hybrid rosette diameter is 60% greater than the midparent value and has a biomass approximating 250% of that of the parents (Fig. 1A and Fig. S1 A and B). The increased biomass of the hybrids is not a consequence of an extended period of growth due to a later flowering time, because the hybrids initiate flowering at times intermediate between the two parents (Fig. S1A). The hybrid phenotype is evident as early as 14–15 d after sowing, at which time biomass is already double that of the parents (Fig. 1 B and C). This early vigor is not correlated with differences in seed size or germination rates (Fig. S1 C and D). We have chosen this early developmental time point for molecular analysis.
Fig. 1.
(A) Vegetative heterosis of 4-wk-old F1 hybrids. (Scale bar, 10 mm.) (B and C) Hybrids of C24 and Ler show clear heterosis as early as 14–15 d after sowing. (Scale bar, 5 mm.) Error bars indicate SEM. FW, fresh weight. Lettering above each bar indicates statistical differences (t test, P ≤ 0.05). No. of seedlings: C24, 74; Ler, 71; C24 × Ler 71; and Ler × C24 72.
Abundance and Genomic Distribution of 24-nt and 21-nt sRNAs Differ in C24 and Ler.
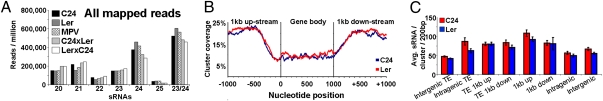
Deep sequencing of parental and hybrid sRNA populations (Dataset S1, Table S1) showed that the 24-nt sRNAs accounted for 56.5% (24-nt, 41.5%; 23-nt, 15%; two-parent average) of the sRNA population with 39.8% in the 21-nt class (21-nt, 18.5%; 20-nt, 14.9%; 22-nt, 6.4%; two-parent average) and 3.7% within “other” size classes (Fig. 2A). As expected, microRNA (miRNA) (>95%) and transacting short interfering RNA (tasiRNA) (>95%) species were in the 21-nt category. The frequency of the 24-nt sRNAs relative to the 21-nt class was greater in the Ler parent than in the C24 parent (Fig. 2A). The parental lines were similar between size classes of sRNAs in unique and repeat mapped reads and also in the unique nonredundant sequences (Fig. S2A).
Fig. 2.
(A) sRNAs (20- to 25-nt) from total mapped population. An additional category (23/24-nt) was added, as these two classes likely represent a single class, generated at similar loci through identical biogenesis pathway, exerting similar downstream effects (16). (B and C) siRNA frequency and density over genes associated with siRNA clusters. (B) Probability of a given nucleotide being covered by a 24-nt siRNA cluster (gene body incorporates 5′ and 3′ UTR; position 0 indicates transcriptional start site). Only genes ≥1 kb were considered. All genes and respective coverage is scaled to 1 kb for parity. (C) Average siRNAs per cluster for a genomic feature (standardized to 200 bp, i.e., average cluster length). “TE 1kb up- or down-stream” is a TE situated within ±1 kb of a gene. Intergenic TEs are transposable elements residing at least ±1 kb from a gene. Error bars represent SEM.
Genes involved in the 24-nt sRNA biogenesis pathway, DICER LIKE 3 (DCL3), ARGONAUTE 4 (AGO4), and AGO5, were all more highly expressed in Ler than in C24 (Fig. S2B). There was no consistent pattern of difference in the expression levels of DCL1, DCL4, AGO1, and AGO2, involved in the production of the 21-nt sRNA class, between the two parents (Fig. S2B).
We used a dynamic two-tier clustering approach to identify regions represented by substantial numbers of sRNAs. sRNA islands were recognized when there was a minimum of three overlapping uniquely mapped reads. These islands were then clustered if one island resided within 100-nt of another island and were labeled according to the majority sRNA species (SI Material and Methods, Dataset S1, Tables S2 and S3, and Fig. S2C). sRNA reads within clusters were standardized to reads per million (uniquely mapped), for each of the parental and the reciprocal hybrid samples. Genomic regions associated with sRNA clusters will be referred to as sRNA loci.
We obtained 9,591 (C24) and 11,533 (Ler) sRNA clusters, incorporating 81–83% of all uniquely mapped reads and covering 1.60–2.08% of the genome (Dataset S1, Table S2). Clusters were profiled against four genomic features: (i) transposons (TEs), (ii) gene body, (iii) ±1-kb flanking regions of a gene, and (iv) intergenic regions. Both parents showed a similar nonrandom (χ2 test, P < 2.20 × 10−16; Dataset S1, Table S4) distribution of 21-nt and 24-nt clusters across the Columbia reference genome, although there were localized differences (Figs. S3 and S4A). The 21-nt clusters were distributed across the genome in regions of high gene density (Fig. S3). The bulk of the 21-nt clusters were localized to known miRNA/tasiRNA generating loci and their targets (Fig. S4B and Dataset S1, Table S5). In both parents, almost two-thirds of the 24-nt clusters (24-nt siRNA clusters) were associated with TEs and pericentric heterochromatic regions (Figs. S3 and S4B). In all, 21% of the 24-nt siRNA clusters were within genic regions, the majority within the flanking ±1-kb sequences (Fig. S4B). The 24-nt siRNA clusters were two to three times more frequently associated with the distal end of the flanking regions, and there was a low likelihood of clusters overlapping the point just before the transcriptional start site of genes (Fig. 2B). Clusters not only mapped more frequently to flanking regions of genes, but also had greater densities of siRNAs compared with intergenic regions beyond 1 kb from a gene (Fig. 2C). Higher siRNA densities held true for TEs located within 1 kb of a gene (Fig. 2C). In C24 and Ler, respectively, 3,145 and 3,731 genes had siRNA associated within flanking regions and could potentially be epigenetically regulated (Dataset S1, Table S6).
Of the 15,336 clusters identified in the parents, only 5,884 occurred in both C24 and Ler, with only ∼10% showing significant differences in sRNA levels (Fisher exact test, P ≤ 0.01; Dataset S1, Table S3). A further 6,843 clusters had reads primarily in one parent, with the reads in the other parent being insufficient to form a recognizable cluster (Dataset S1, Table S3). The remaining 2,609 clusters had reads in only one parent. Of these clusters, 2,169 (C24, 709; Ler, 1,460) were the solitary clusters associated with a particular genomic feature (Dataset S1, Table S7). Included in this set were 257 (C24) and 540 (Ler) protein coding genes that had within their genic and flanking regions a siRNA cluster in only one parent (Dataset S1, Table S8).
F1 Hybrids Have Reduced Levels of 24-nt siRNAs Relative to Parents.
The null expectation for the hybrids is that their sRNA populations would correspond to the additive level of the haploid frequencies in each parent, i.e., the midparent value (MPV). This was not true for 24-nt sRNAs, for which both reciprocal hybrids had an average decrease in the range of 17% (23/24-nt) to 27% (24-nt) from the MPV, even falling below the value of the low expressing parent (Fig. 2A). In contrast, in both reciprocal hybrids, the shorter sRNA classes had values above the MP expectation and in most cases above high expressing parent values (Fig. 2A). In both hybrids, expression levels of DCL1/3/4 and AGO1/2/4 were not significantly different from MP and therefore probably not a primary factor in altering the sRNA levels (Fig. S5A).
The reduction in the levels of 24-nt sRNAs held for uniquely mapped reads, repeat mapped reads, and for unique sequences (Fig. S5B). There was a slight increase in 20- to 22-nt unique sequences (Fig. S5B); however, there was no evidence of 24-nt sRNAs being processed into shorter-length molecules. This suggests that the increased 20- to 22-nt levels in hybrids were probably a consequence of deeper sequencing of these size classes due to the decreased levels of 24-nt sRNAs. Consistent with this possibility, the 20- to 22-nt sRNAs showed similar proportional increases over MPV (Fig. 2A).
Northern and qRT-PCR analysis did not show significant deviation from MPV for six miRNAs, which according to deep sequencing reads were increased in the hybrids (Dataset S1, Table S9A and Fig. S5 C and D). Furthermore, expression levels of all genes targeted by miRNAs did not differ from MPV (Dataset S1, Table S9B and SI Materials and Methods, mRNA sequence). We conclude that the apparent up-regulation of the 20- to 22-nt sRNAs was a result of sequencing bias caused by the decreased 24-nt levels.
Apart from the reduction in 24-nt siRNAs, the hybrids were similar to the parents in the general distribution of sRNA populations over the genome (Fig. S6A). The hybrids also showed a similar coverage frequency and density of 24-nt siRNAs over genic regions (Fig. S6 B and C). We did not define any newly generated clusters in the hybrids.
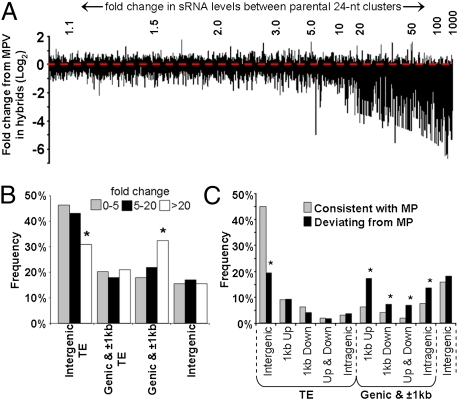
When all of the 14,175 parentally derived 24-nt siRNA clusters were ranked by their fold differences between the two parents, the frequency of siRNA clusters associated with genes and the ±1-kb flanking regions increased as the fold differences increased (Fig. 3 A and B); the frequency of the intergenic TE-associated clusters decreased (Fig. 3B). The 2,226 clusters that had a ≥15-fold difference between the parents contained 466 clusters which deviated significantly from MPV, with ∼98% below the MPV (Fig. 3A and Dataset S1, Table S10). Of the 11,949 clusters with a <15-fold difference between parents, only 162 deviated significantly from the MPV, with 80% being below MPV (Dataset S1, Table S10).
Fig. 3.
(A) Deviation of C24 × Ler hybrid siRNA values around the expected MPV. All 14,175 parentally derived 24-nt siRNA clusters are ranked left to right along the x axis, based on fold differences in siRNA levels between parents. The x-axis line itself represents MPV (red dotted line) with the vertical bars depicting log2 fold-change deviation from MPV in the hybrid (y axis). Ler × C24 hybrid has an almost identical pattern. (B) Profile of parental 24-nt siRNA clusters grouped based on fold differences between parental siRNA levels. (C) Genomic profile of clusters consistent with MPV (13,547) and those significantly deviating from MPV (628). *Significantly different (Fisher exact test, P < 0.01); Up, 1 kb upstream; Down, 1 kb downstream.
These data show that the greater the difference between parental 24-nt siRNA levels, the more likely the hybrid value is to deviate from MP and almost always toward the low expressing parent (Fig. 3A and Dataset S1, Table S10). This pattern applied only for the 24-nt siRNA clusters, as 20- to 22-nt clusters displayed a similar variation around the MPV across the entire ranked distribution of parental values (Fig. S7A). The 628 24-nt siRNA clusters that deviated significantly from MPV in both hybrids (Dataset S1, Table S10) were more frequently associated with genes and flanking regions than with TEs and intergenic regions (Fig. 3C and Fig. S7B). The underrepresentation of TEs occurred predominantly in intergenic TEs, whereas the overrepresentation in genic regions was present in both gene bodies and flanking regions, particularly in the 1-kb upstream flanking region (Fig. 3C).
The deviating clusters are associated with 438 protein-coding genes, which are categorized into a range of gene ontologies, including higher-order molecular functions affecting transcription factors, protein binding, signal transduction, and enzyme activity (Dataset S1, Table S11).
SNP Analysis Provides Insight into the Inheritance of siRNA Loci in Hybrids.
We searched all 14,175 24-nt siRNA clusters for SNPs to track the parental allelic contribution in the hybrids. This could be done for 2,341 24-nt siRNA clusters. In clusters with a number of siRNAs containing SNPs, any changes were similar across all of these siRNAs within the cluster. When only siRNAs containing SNPs were analyzed, the frequency of reads paralleled the changes in siRNA levels in the cluster as a whole. The expectation was that parental siRNA alleles retain their comparative contribution ratio in the hybrids. In the SNP-containing loci, the expected parental contributions were found in 78% of the loci indicating cis-regulation in production of the siRNA from the two alleles (Fig. S8A). The remaining 22% deviated significantly from the expected parental contributions (Fisher exact test, P ≤ 0.05) indicating a transeffect on siRNA expression. These transregulated loci were equally frequent in the reciprocal hybrids (Fig. S8A).
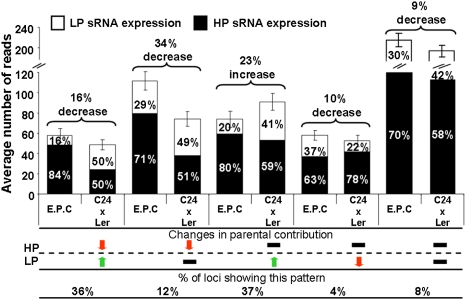
We categorized 97% of the transaffected loci into five major classes based on how the comparative parental siRNA contributions were altered (Fig. 4). Of these loci, 93% showed an equalizing of parental contributions indicating a change in the expression levels of the siRNA parental alleles in the hybrids (Fig. 4). Decreases in the high expressing parent values were greater than the increases in low expressing parent values, resulting in a net reduction in total siRNAs at these transregulated loci (Fig. 4).
Fig. 4.
A total of 97% of the transaffected loci categorized into five classes based on changed parental contributions in the hybrid. A siRNA locus can be up-regulated (1.5-fold), down-regulated (1.5-fold), or unaltered (within 1.5-fold of expected value) within the hybrid. The “expected parental contribution” (E.P.C) bars equal total siRNA levels that should be produced (bars represent SEM), with the relative contributions from high parent (HP; black) and low parent (LP; white). Ler × C24 is similar (Fig. S8B). Relative difference of average values located above parentheses.
Limitations of the SNP analysis prevented examination of loci that had a large difference in siRNA levels between parents (i.e., ≥10-fold difference; SI Materials and Methods). However, given the observed correlation between fold difference in parents and reduction in 24-nt siRNA levels in the hybrids, we predict that the frequency of transaffected loci in the hybrids would be greater in the ≥10-fold subset, as parental differences are greater. These loci contain the majority of the clusters that have less than the expected production of siRNAs.
Localized Differences in siRNA Levels Correlate with Changes in DNA Methylation Levels at the Same Locus Within the Hybrids.
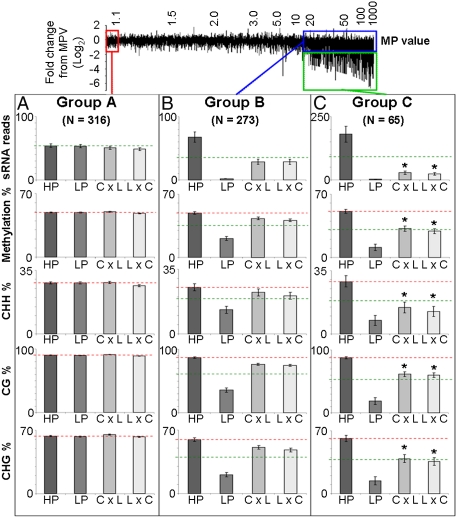
The 24-nt siRNAs direct de novo DNA methylation (15) and are required for the maintenance of CHH methylation. From a floral bud methylome dataset from C24, Ler, and the two reciprocal hybrids, we analyzed DNA methylation levels associated with three groups (A–C) of 24-nt siRNA clusters, each representing a distinctive profile of siRNA levels between parents and hybrids from the distribution in Fig. 3A (Fig. 5 and SI Materials and Methods). A total of 654 clusters in all three groups were identified with sufficient methylome coverage across all four samples (Fig. 5 and Dataset S1, Table S12). Because siRNAs have the ability to act in trans to initiate RdDM, a parental locus that is methylated and producing siRNAs could generate methylation of the homologous locus from the other parent, resulting in methylation of a previously unmethylated sequence or increasing the methylation level of a low methylated region. Therefore, methylation levels in the hybrids may be above MPV toward high methylation parent value (HPV).
Fig. 5.
DNA methylation levels associated with 24-nt siRNA clusters. Three distinct groups of clusters were examined. (A) Red box indicates no difference between parental or hybrid siRNA levels. (B) Blue box indicates large difference between parents but hybrids are near MPV. (C) Green box indicates large differences between parents and hybrids are significantly below MPV. C, C24; HP, high parent; L, Ler; LP, low parent. Red dotted line represents HP average; green dotted line represents MPV. Error bars represent SEM. The y axes are linear scale. *Significantly below values in group B (P ≤ 0.05).
In group A, in which parents and hybrids showed similar levels of siRNAs, there were equal levels of methylation in the hybrids (Fig. 5A). In groups B and C, in which parental siRNA levels differed by ≥15-fold, the levels of siRNA and methylation were correlated between parental lines (Fig. 5 B and C). In group B loci, in which parental siRNA levels differed greatly and hybrid values were near MPV, the degree of methylation in the hybrids was above the MPV in all contexts indicating transchromosomal methylation of the low parent loci (Fig. 5B). Group C loci also had 24-nt siRNA levels that differed greatly between parents and hybrid, but siRNA levels were significantly below MPV (Fig. 5C). The decreased siRNA levels in group C correlated with a reduction in total methylation compared with the increase in group B (Fig. 5C); CG methylation in group C was lower than in groups A and B (Fig. 5 A and B) but was still above MPV, indicating cis retention of methylation also with transchromosomal methylation of the low methylated or nonmethylated parental allele (Fig. 5C). CHG levels were reduced in group C compared with group B, indicating an absence of transchromosomal methylation (Fig. 5C). The most striking change was the decrease in CHH methylation, which was below MPV (Fig. 5C). This decrease suggests a reduction of cis-directed CHH methylation in addition to the expected absence of transchromosomal methylation as a consequence of the reduced 24-nt siRNAs in group C loci.
Loci with Altered Epigenetic States in the Hybrids Correlate with Changes in mRNA Levels.
We identified several genes in the hybrids that had altered epigenetic states in the flanking regions correlated with changes in gene expression (Fig. 6, Fig. S8C, and selection criteria in SI Materials and Methods). Parental epi-alleles were matched against those from the Columbia accession to draw conclusions between published expression profiles of wild type and various mutants affecting the RdDM pathway in Columbia. This search was not exhaustive and only intended to show that the altered hybrid sRNA levels can be associated with changes in gene activity.
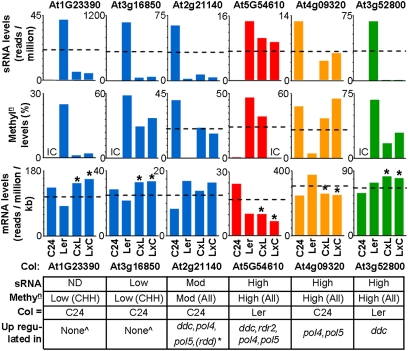
Fig. 6.
siRNA and methylation levels in hybrids associated with changes in gene expression. siRNA clusters are located within 1 kb upstream (At16850, At5g54610, At4g09320, and At3g52800) or downstream (At1g23390 and At2g21140). Table details sRNA and Methyl-C levels at the homologous gene in the Columbia accession (Col) matching the Col epi-allele to the C24 or Ler epi-allele. Bottom row indicates whether gene expression is altered in Col RdDM or DNA demethylation mutants. Examples cover four classes (color coded) based on changes in siRNAs and resulting gene activity in hybrids. ^Not expected to be altered in epigenetic mutants, as both C24 and Col alleles are not associated with substantial levels of siRNA or methylation. *At2g21140 down-regulated in hypermethylated Col ros1 demeter-like 2 demeter-like 3 (rdd) triple mutant. IC, insufficient coverage to determine methylation level; ND, not detected. *mRNA levels significantly different from MPV (P ≤ 0.05). References to Col data provided in SI Materials and Methods. qRT-PCR performed on three of six genes (Fig. S8C); consistent with mRNA-seq dataset used in this analysis.
Several different patterns of inheritance of sRNA, methylation and mRNA levels were identified. Genes such as At1g23390 and At3g16850 constitute the first correlative pattern where in the hybrids a decrease in siRNAs correlates with an increase in gene mRNA levels (Fig. 6, blue histograms). In each case, the parent with the higher level of siRNAs and methylation had lower gene activity. The lower level of 24-nt siRNAs in both hybrids correlated with an increase in mRNA levels. and both hybrids showed expression levels equal to or greater than that of the high parent. For both genes, transcriptome SNP analysis revealed that in the hybrids, the parental alleles contribute equally to total gene activity, deviating from the expected retention of comparative parental ratios (Dataset S1, Table S13 A and B). This nonadditive parental contribution to gene expression is consistent with increased mRNA levels of the low expressing parent allele through the reduction of 24-nt siRNA and methylation causing the observed high expressing parent mRNA levels (Fig. 6).
Genes such as At2g21140 are a subset of the first correlative class (Fig. 6, blue histograms), because although mRNA levels for At2g21140 are only slightly greater than and not significantly different from MPV (Fig. 6), the reduction in siRNA levels did correlate with nonadditive parental mRNA contributions (Dataset S1, Table S13A). The equal parental mRNA contributions are consistent with increased expression levels of the low parent allele through a reduction in 24-nt siRNA (Fig. 6). Near MPV methylation levels alone do not exclude At2g21140 as a potential candidate gene, as MPV may not always indicate additive inheritance. Transchromosomal methylation may have redistributed methylation between the two loci, causing an increase of the low parent expression and a decrease in high parent expression. This is consistent with equal parental mRNA contributions, whereas total expression levels in the hybrids remain between MPV and HPV. Such genes would be sensitive to subtle changes in siRNA and methylation, which At2g21140 appears to be, as indicated by up-regulation in mutants affecting RdDM and down-regulation in hypermethylated Columbia mutants (Fig. 6). This apparent sensitivity is consistent with the slight differences between the two hybrids in siRNA, methylation, and mRNA levels (Fig. 6).
A different correlative pattern was found with genes such as At5g54610, where the hybrids had an increased level of 24-nt siRNAs and methylation associated with reduced gene activity (Fig. 6). Increased gene activity is seen in RdDM mutants, suggesting that siRNA and methylation are capable of repressing the activity of these genes (Fig. 6). Parental contributions in the hybrids were again equal (Dataset S1, Table S13A), consistent with decreased high parent gene expression through transchromosomal methylation resulting in hybrids having low parent mRNA levels. Above-midparent levels of siRNA were not a prerequisite for transmethylation, because genes such as At4g09320 had siRNA levels no higher than midparent yet still produced above-expected methylation levels correlated with low parent mRNA levels (Fig. 6 and Dataset S1, Table S13A).
Unlike the previous examples, genes such as At3g52800 had the repressive epigenetic states associated with the high expressing parent, whereas the low expressing parent was devoid of siRNAs (Fig. 6). The elevated gene activity in the hybrids was due to an increased bias in the contribution from the high expressing parent allele (Dataset S1, Table S13A), which correlated with the reduction in siRNA and methylation levels in the hybrid (Fig. 6).
Discussion
The F1 hybrids between C24 and Ler showed a remarkable increase in vegetative growth throughout development, resulting in a large increase in biomass of the mature plant relative to the parents. This was not due to late flowering, as both reciprocal hybrids initiated flowering at a time intermediate to that of the parents. We chose 2-wk-old seedlings for molecular analysis, as this was an early time point showing evidence of significant heterosis in the hybrids.
The C24 and Ler accessions showed large differences between their sRNA profiles. The intraspecific hybrids generated by crossing the C24 and Ler accessions exhibited a unique composition of their sRNAomes and methylomes. The hybrids not only had siRNA epialleles inherited from each parent, but also hybrid-specific epialleles with altered frequencies of the parental siRNAs spawned through transeffects at a proportion of loci. We showed that changes in siRNA production were associated with changed methylation levels either through a reduction in methylation or through transchromosomal methylation, creating a hybrid epigenome different to that of either parent and to that of the expected midparent.
Hybrids Show a Significant Reduction of 24-nt siRNA Levels.
Unlike the interspecies hybrids between Arabidopsis thaliana and Arabidopsis arenosa, no large-scale upheaval of the total sRNA populations was observed (22). The differences were limited to 24-nt siRNAs, with a decrease in production of ∼27% occurring in each of the reciprocal hybrids. Similarly, a reduction in 24-nt siRNA has been reported in interspecies Arabidopsis and rice subspecies hybrids (13, 22). These analyses did not report whether the reduction was distributed among all loci in the genome or whether there was a particular subset of loci accounting for the reduced 24-nt levels. We show that a large proportion of the 24-nt siRNA decrease in the hybrids is associated with loci having highly different siRNA levels between parents. Regions of the genome primarily associated with genes and their upstream regions, showed marked differences between parental siRNA levels and were frequently represented among those loci showing decreased 24-nt siRNA levels in the hybrids.
The majority of siRNA loci in the hybrids were found to be additive in their inheritance, retaining their relative parental contribution ratios. Approximately one-fifth of all 24-nt siRNA loci, which could be tracked through SNPs, were influenced by transeffects between contributing parental siRNA loci, resulting in a net reduction in 24-nt siRNA levels at these loci. If it were possible to trace parental contributions at each 24-nt siRNA locus, it might show that such transinteractions could be one reason for the reduced 24-nt siRNA levels observed in the hybrids. Variations in the chromatin states, sequence composition, abundance of siRNA, and/or the feedback between methylation and siRNA production (23), may all account for why certain loci retain parental contributions, whereas others exhibit transeffects.
The loss of heterosis in generations beyond the F1 is generally assumed to be caused by segregation breaking up heterosis-promoting allelic combinations. If siRNA epigenetic mechanisms are involved, the breakup of advantageous allelic combinations beyond the F1 would be even greater. The dynamic nature of the siRNA system means that advantageous epialleles would not only segregate from each other during meiosis, but the epialleles themselves would alter because of changes in the controls of production of siRNAs. For instance, loci with lower than expected siRNA production in the F1 may show increased levels in subsequent generations as reported in interspecies Arabidopsis hybrids (22). These altered siRNA levels could then allow re-establishment of methylation levels at loci altered in the F1 hybrids in a manner similar to that seen in hypo-methylated recombinant lines (23). Furthermore, in generations beyond the F1, an increasing number of loci may show “balancing” interactions between parental epialleles, leading to loss of epiallelic diversity.
Altered siRNA Levels Lead to Changed Levels of Methylation in Hybrids.
In the parents, siRNA production and levels of methylation in all contexts were correlated, indicating that at these loci a significant proportion of methylation sites are affected by the RdDM pathway.
In the hybrids, methylation levels increased at loci that showed large differences in siRNA levels between parents and where hybrids had mid parent levels. Presumably this increase is caused through transchromosomal methylation affecting the low-methylated parental alleles in the hybrids. The increase in methylation of the low parent allele was not absolute, as high parent average values were not attained. This may be due to variable penetrance of transchromosomal methylation (20) and/or to the requirement of other factors, such as responsive chromatin states, needed for the recruitment of the RdDM machinery (15).
Methylation levels are clearly affected at loci with reduced siRNA production. The most severely affected methylation state was CHH methylation, which requires siRNAs for its establishment and continued maintenance (18). Even though siRNAs play a role in maintenance of CHG methylation, it was affected to a lesser extent than CHH methylation, presumably through continued maintenance via CHROMOMETHYLASE 3 (CMT3) (18, 24, 25), which is not dependent on siRNAs. The hybrids retained and exhibited some transchromosomal CG methylation despite the reduced siRNA levels at these loci. This transchromosomal methylation was most likely predicated by low levels of de novo CG methylation by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) through the RdDM pathway, which was then efficiently maintained by METHYLTRANSFERASE1 (MET1) (15, 25–27).
Potential Contribution of Altered Hybrid Epigenome to Heterosis.
No one common cause for improved hybrid performance has yet been reported; however, along with this study, a decreased level of 24-nt siRNA has now been reported in three different hybrid systems showing positive heterosis (13, 22). In addition, rice hybrids show significant changes in DNA methylation levels (13). However, mutants compromising the RdDM pathway do not confer an obvious growth advantage over WT, and in some cases actually cause abnormal dwarfed development (27–30). Possible heterosis-promoting epialleles generated in these mutant backgrounds may be masked by broader, indiscriminate, genome-wide changes, activating genes potentially detrimental to development (29). Changes to epialleles that severely affect development would presumably be infrequent in hybrids, as such genes are more than likely to have been silenced in both parents and therefore are likely to be silenced in the hybrids. Conversely, genes with positive influences on development and/or environmental responses are more likely to be differentially expressed between parents adapted to different niches. Epiallelic variants of such genes with parent-specific benefits would be reinforced through selective pressures. This could account for the enrichment of variable 24-nt siRNA loci between C24 and Ler in genic regions. The prediction is that those epi-loci that differ greatly between parents and are altered in the hybrids would be more likely to be associated with genes that could positively influence development and adaptation. Epi-allelic variants of such genes may contribute significantly to hybrid vigor.
Alterations to epigenomes, whether genome-wide (31–34) or at a single locus (20, 35–37), can lead to widespread changes in gene expression. Of particular interest were epi-loci in the hybrids with altered methylation patterns in upstream sequences, as differences in promoter methylation affect gene expression levels more than does gene body methylation (31).
The fact that higher-order regulatory genes (i.e., transcriptional and signal transduction regulators) are well represented within the population of genes with altered siRNA levels in the hybrids suggests that changes in siRNA and methylation levels may be among the first steps in a cascade of altered gene activities contributing to the final observed heterotic phenotype. The concept of early changes in key regulatory genes suggests that the activity of even a few genes affected by the initial epigenetic alterations can lead to widespread effects on downstream regulatory networks involved in the production of the mature phenotype of the plant.
Materials and Methods
Plant material and experimental designs are described in SI Materials and Methods.
Small RNAs were prepared from 2-wk-old seedlings, with libraries and sequencing performed by Geneworks on the Genome Analyzer I (Illumina). MethylC-seq was carried out as described elsewhere (38) and sequenced on the Genome Analyzer II (Illumina). mRNA-seq was carried as per the Illumina protocol and sequenced on the Genome Analyzer II. Processing of sRNA, methylation, and transcriptome sequences, validation of miRNAs, and methylated regions, are described in SI Materials and Methods. Raw and processed mapped sRNA sequences are deposited in GEO (accession no. GSE25022).
Supplementary Material
Acknowledgments
Prof. Jay Hollick provided critical and important comments, as did Prof. Thomas Altmann. This work was supported by National Collaborative Research Infrastructure Strategy (NCRIS) and Science and Industry Endowment Fund (SIEF) funded by the Australian Government, and by the Agricultural Genomics Centre, New South Wales Government BioFirst initiative.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE25022).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019217108/-/DCSupplemental.
References
- 1.Shull GH. Beginnings of the heterosis concept. In: Gowan JW, editor. Heterosis. Ames, IA: Iowa State College Press; 1952. pp. 14–48. [Google Scholar]
- 2.Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochholdinger F, Hoecker N. Towards the molecular basis of heterosis. Trends Plant Sci. 2007;12:427–432. doi: 10.1016/j.tplants.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Lippman ZB, Zamir D. Heterosis: Revisiting the magic. Trends Genet. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Meyer RC, et al. QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor Appl Genet. 2010;120:227–237. doi: 10.1007/s00122-009-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. Plant Cell. 2010;22:2105–2112. doi: 10.1105/tpc.110.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frascaroli E, et al. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics. 2007;176:625–644. doi: 10.1534/genetics.106.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radoev M, Becker HC, Ecke W. Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics. 2008;179:1547–1558. doi: 10.1534/genetics.108.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semel Y, et al. Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 11.Melchinger AE. Genetic diversity and heterosis. In: Coors JG, Pandey S, editors. Genetics and Exploitation of Heterosis in Crops. Mexico City: International Symposium on the Genetics and Exploitation of Heterosis in Crops; 1999. pp. 99–118. [Google Scholar]
- 12.Meyer RC, Törjék O, Becher M, Altmann T. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 2004;134:1813–1823. doi: 10.1104/pp.103.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He GM, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matzinger DF, Wernsman EA. Genetic diversity and heterosis in Nicotiana. Zuchter. 1967;37:188–191. [Google Scholar]
- 15.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasschau KD, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosher RA, Melnyk CW. siRNAs and DNA methylation: Seedy epigenetics. Trends Plant Sci. 2010;15:204–210. doi: 10.1016/j.tplants.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 19.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Zhai J, et al. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000056. doi: 10.1371/journal.pgen.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughn MW, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha MS, et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teixeira FK, Colot V. Repeat elements and the Arabidopsis DNA methylation landscape. Heredity. 2010;105:14–23. doi: 10.1038/hdy.2010.52. [DOI] [PubMed] [Google Scholar]
- 24.Cao XF, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 25.Chan SWL, et al. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 26.Cao XF, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao XF, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 28.Chan SWL, et al. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2:e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He XJ, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XY, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Zhang XY, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia Y, et al. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5:e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baev V, et al. Identification of RNA-dependent DNA-methylation regulated promoters in Arabidopsis. Plant Physiol Biochem. 2010;48:393–400. doi: 10.1016/j.plaphy.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Soppe WJJ, et al. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 36.Shibukawa T, Yazawa K, Kikuchi A, Kamada H. Possible involvement of DNA methylation on expression regulation of carrot LEC1 gene in its 5′-upstream region. Gene. 2009;437:22–31. doi: 10.1016/j.gene.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Shibuya K, Fukushima S, Takatsuji H. RNA-directed DNA methylation induces transcriptional activation in plants. Proc Natl Acad Sci USA. 2009;106:1660–1665. doi: 10.1073/pnas.0809294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.