Abstract
C1q-like genes (C1ql1–C1ql4) encode small, secreted proteins that are expressed in differential patterns in the brain but whose receptors and functions remain unknown. BAI3 protein, in contrast, is a member of the cell-adhesion class of G protein-coupled receptors that are expressed at high levels in the brain but whose ligands have thus far escaped identification. Using a biochemical approach, we show that all four C1ql proteins bind to the extracellular thrombospondin-repeat domain of BAI3 with high affinity, and that this binding is mediated by the globular C1q domains of the C1ql proteins. Moreover, we demonstrate that addition of submicromolar concentrations of C1ql proteins to cultured neurons causes a significant decrease in synapse density, and that this decrease was prevented by simultaneous addition of the thrombospondin-repeat fragment of BAI3, which binds to C1ql proteins. Our data suggest that C1ql proteins are secreted signaling molecules that bind to BAI3 and act, at least in part, to regulate synapse formation and/or maintenance.
Among the G protein-coupled receptors (GPCRs), the cell-adhesion class of GPCRs is the second-largest subfamily, comprising 33 members (1–3). Cell-adhesion–type GPCRs contain a relatively large N-terminal extracellular sequence that is composed of multiple independent domains and is linked to the typical seven-transmembrane region architecture of GPCRs. In all cell-adhesion–type GPCRs (except GPR123, which may not be a true member of this class), an autoproteolytic sequence known as the GPCR proteolysis site (GPS) is located N-terminal to the first transmembrane region (4). Cell-adhesion GPCRs appear to be generally cleaved at the GPS, and, at least for those proteins in which this has been investigated, the two fragments of adhesion GPCRs produced by autoproteolysis do not separate, but rather remain noncovalently bound (4–6). Adhesion GPCRs are evolutionarily conserved, and members of this class of GPCRs are found in arthropods and nematodes (7, 8). Although a direct ligand-dependent activation of any adhesion GPCR has not yet been demonstrated, genetic evidence in zebrafish indicates that a member of this class of GPCRs functions as a true GPCR and mediates an intracellular signal transduction event (9).
Brain-specific angiogenesis inhibitor (BAI) proteins are cell-adhesion GPCRs that are encoded by three genes, BAI1, BAI2, and BAI3. The extracellular sequences of BAI proteins are composed of an N-terminal CUB domain, five (BAI1) or four (BAI2 and BAI3) thrombospondin type 1 repeats, a hormone-binding domain, and the GPS (10, 11). RNA quantification and in situ hybridization have indicated that in adult mice, BAI proteins are expressed primarily in the brain, with low-level expression in other tissues (12–15). The BAI1 gene, originally identified in a screen for p53-inducible genes, is thought to inhibit neovascularization, a process required for tumor growth (10, 13, 16–19). Human BAI1 expression is down-regulated in glioblastoma and is inversely correlated with neovascularization in colorectal and lung cancers, consistent with an antiangiogenesis function (20–22). The antiangiogenic activity of BAI1 is thought to be mediated by a fragment of its N-terminal extracellular sequence, a fragment known as vasculostatin (17, 23, 24). BAI2 and BAI3 expression are not p53-inducible, but both proteins also act as inhibitors of angiogenesis (11, 14, 15, 25). Moreover, BAI1 functions as a surface receptor for phosphatidylserine that is exposed on the surface of apoptotic cells, suggesting that BAI1 is an engulfment receptor that can initiate phagocytosis (26). Consistent with a role in neovascularization and/or engulfment of apoptotic cells, systematic cancer genomics identified all three BAI genes as targets of mutations in several types of cancer (27). Thus, a wealth of evidence implicates BAI proteins in oncogenic transformation, neovascularization, and phagocytosis of apoptotic cells; however, whether (and if so, how) BAI proteins function as GPCRs remains unclear.
BAI proteins exhibit only limited sequence identity with each other (e.g., mouse BAI1 and BAI3 are 48.1% identical), but are highly conserved evolutionarily (e.g., human and mouse BAI3 sequences are 98.4% identical). In mice, the largely brain-specific expression of BAI3 peaks during neonatal development but persists throughout adult life at lower levels (15). Interestingly, two SNPs within the human BAI3 gene have been significantly associated with schizophrenia in genome-wide association studies (28). An explanatory model for the development of schizophrenia is that the disease is a consequence of aberrant brain development before symptoms become manifest (29). The temporal and spatial BAI3 expression pattern is consistent with its role in a pathogenic process like schizophrenia.
C1q-like (C1ql) proteins are small, secreted proteins of unknown function that are synthesized from four genes in mammals, expressed almost exclusively in brain similar to BAI proteins, and produced in differential patterns by specific types of neurons (30, 31). C1ql proteins belong to a large family of proteins containing a globular complement 1q (gC1q) domain that associates into homotrimers or heterotrimers (32–34). Besides the eponymic C1q complement factor, the gC1q-domain protein family contains small signaling molecules containing short N-terminal sequences and a C-terminal gC1q domain (e.g., cerebellins, adiponectin, C1ql proteins), as well as larger proteins, including collagens containing C-terminal gC1q domains. C1ql proteins are composed of an N-terminal signal peptide followed by a short conserved sequence (∼15 residues) with two closely spaced cysteine residues, a spacer (15–35 residues), a collagen-like sequence (∼50 residues), and a C-terminal gC1q domain (∼140 residues) that accounts for approximately half of the total C1ql sequence (238–287 residues). Structurally, C1ql proteins resemble a combination of cerebellins and adiponectin, both of which contain C-terminal gC1q domains, but with either a short conserved N-terminal cysteine-rich sequence (cerebellin) or an N-terminal collagen-like sequence (adiponectin) (35, 36). Notably, in cerebellins, the N-terminal sequence multimerizes the C-terminal trimeric gC1q domains (37); this sequence likely does the same in C1ql proteins (31).
The present study was initiated to identify possible ligands for BAI3, based on the hypothesis that the neuronal expression and structure of this cell-adhesion GPCR suggests a possible role in neuronal signaling. Using affinity chromatography, we identified C1ql proteins as BAI3 ligands and found that the presence of C1ql proteins causes a decrease in synapse numbers in cultured neurons in a manner that can be inhibited by the C1ql-binding fragment of BAI3.
Results
Identification of C1ql Proteins as BAI3 Ligands.
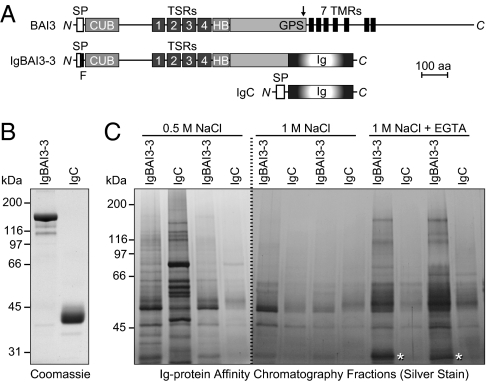
We produced a recombinant Ig-fusion protein composed of the extracellular domains of BAI3 fused to the Fc region of human IgG (IgBAI3-3), along with a control Ig-fusion protein composed of only the Fc region (IgC; Figs. 1 A and B). We then used the Ig fusion proteins in affinity chromatography experiments with mouse brain proteins extracted in 1% Triton X-100. Proteins were bound in the presence of Ca2+ and Mg2+ and then sequentially eluted with solutions containing 0.5 M or 1 M NaCl with Ca2+ and Mg2+ and finally with a solution containing 1 M NaCl and EGTA to chelate the Ca2+ ions. Two sequential fractions for each elution condition were analyzed by SDS/PAGE and silver staining (Fig. 1C).
Fig. 1.
Identification of C1ql proteins as BAI3 ligands. (A) Schematic depiction of the domain organization of BAI3 (1,522 amino acids), of the BAI3 Ig-fusion protein IgBAI3-3, and of the control Ig-fusion protein IgC. SP, signal peptide; F, FLAG tag; CUB, complement C1r/C1s-Uegf-Bmp1 domain; TSRs, thrombospondin repeats; HB, hormone-binding domain; GPS, GPCR proteolysis sequence (proteolytic site denoted by an arrow); 7 TMR, seven-transmembrane region; Ig, Fc domain of human IgG. (B) Coomassie blue–stained SDS gel of Ig-fusion proteins produced in HEK293 cells and purified by protein A–Sepharose affinity chromatography. (C) Silver-stained SDS gel of a representative pull-down experiment. Mouse brain homogenates were incubated with protein A–Sepharose beads containing either IgBAI3-3 or IgC. After the beads were washed, bound proteins were sequentially eluted with buffers containing 0.5 M NaCl, 1 M NaCl, and 1 M NaCl with 5 mM EGTA. Two fractions per buffer were collected and analyzed. Mass spectrometry analysis identified C1ql2 and C1ql3 in the band marked by an asterisk.
Multiple proteins were bound to BAI3 and eluted by chelation of Ca2+ with EGTA. Mass spectroscopy analysis of these proteins showed that most proteins constituted abundant intracellular molecules, such as tubulin, actin, heat-shock proteins, and GAPDH, which are not likely to be true ligands. Only two bona fide extracellular proteins were identified, C1ql2 and C1ql3, with multiple peptides unequivocally assigned to both (24 peptides out of a total of 215 in two independent experiments).
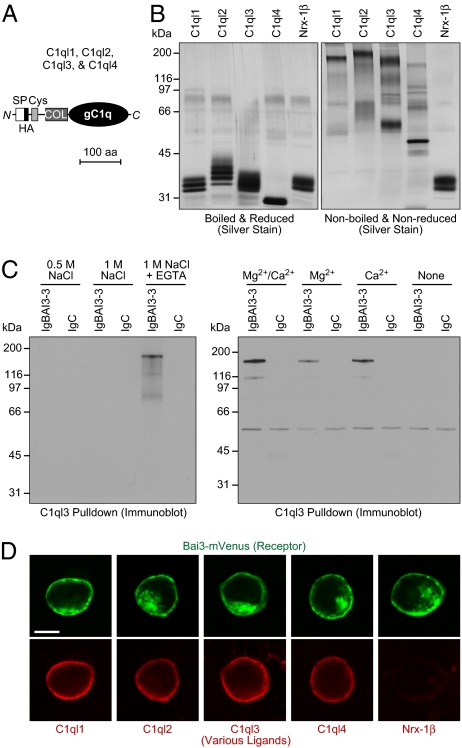
Four different C1ql isoforms are expressed in the mammalian brain with an identical domain structure, comprising an N-terminal short cysteine loop domain, a central collagen-like sequence, and a C-terminal gC1q domain (Fig. 2A) (30, 31). We expressed all four C1ql isoforms as HA-tagged proteins by transfection into HEK293 cells, purified them from the culture medium by immunoaffinity chromatography with immobilized anti-HA antibody, and examined the recombinant C1ql proteins by SDS/PAGE. Under reducing conditions in boiled samples, the C1ql proteins appeared slightly larger than expected with considerable heterogeneity, presumably due to differential glycosylation (Fig. 2B). However, when we analyzed the same proteins by SDS/PAGE without reducing agents or boiling, none of the proteins migrated at their expected monomer size, and most exhibited a very high apparent molecular weight (Fig. 2B). In contrast, recombinant neurexin-1β protein composed of only the neurexin-1β extracellular sequences exhibited no difference in migration as a function of boiling and disulfide bond reduction. These findings suggest that C1ql proteins, similar to C1q complement factor, form higher-order multimers via their interactions within the gC1q domains and/or their collagenous and cysteine loop domains, as suggested previously (31).
Fig. 2.
Characterization of BAI3 binding by C1ql proteins. (A) Domain structure of C1ql proteins C1ql1–C1ql4 (238–287 amino acids). SP, signal peptide; Cys, cysteine loop domain; COL, collagen-like domain; gC1q, globular C1q domain. An HA epitope was inserted immediately following the signal peptide. (B) Silver-stained SDS gel of purified HA-tagged full-length C1ql proteins and of a recombinant protein containing the extracellular neurexin-1β sequences loaded onto SDS gels with (Left) or without (Right) reducing agents and boiling. Proteins were expressed in HEK293 cells and purified from the medium by anti-HA agarose. (C) Analyses of C1ql3-mediated pull-downs of recombinant IgBAI3-3 protein, using IgC as a negative control. HA-tagged C1ql3 immobilized on an anti-HA column was incubated with IgBAI3-3 or IgC, and bound proteins were analyzed by immunoblot analysis for human Ig. (Left) Eluates obtained after binding in Ca2+/Mg2+-containing buffer and elution with the indicated solutions. (Right) Proteins bound to immobilized C1ql3 in buffers containing the indicated divalent cations. (D) Cell-surface labeling assay demonstrating that all four C1ql isoforms bind to BAI3. HEK293 cells transfected with a vector encoding mVenus-tagged BAI3 (green) were incubated with purified HA-tagged C1ql1–C1ql4 and neurexin-1β (extracellular sequences). Bound proteins were visualized by immunofluorescence labeling (red). (Scale bar: 10 μm.)
We next incubated recombinant C1ql3 with BAI3 Ig-fusion protein (IgBAI3-3) or with IgC control protein under various conditions, and measured the interactions by immunoprecipitation of C1ql3 via its HA epitope (Fig. 2C). We found that C1ql3 and IgBAI3-3 specifically interacted with each other, and that this interaction required divalent cations, most likely Ca2+ given EGTA's binding preference.
To examine whether all four C1ql proteins bind to BAI3, and to assess this binding with an independent assay, we measured the binding of C1ql proteins to HEK293 cells expressing full-length BAI3 (Fig. 2D). In this assay, mVenus-tagged BAI3 was expressed by transient transfection, and its localization to the plasma membrane was confirmed by fluorescence microscopy. Purified recombinant C1ql proteins (Fig. 2B) were then added to the medium. Cells were washed, fixed, and analyzed for C1ql protein binding to the cell surface by indirect immunofluorescence for the HA epitope included in the recombinant proteins without cell permeabilization. Recombinant HA-tagged neurexin-1β protein served as a control. All four C1ql isoforms, but not the neurexin-1β extracellular domain, exhibited binding to BAI3-expressing HEK293 cells, suggesting that all four C1ql proteins interact with BAI3 on the cell surface (Fig. 2D). No binding of C1ql proteins to cells that did not express BAI3 was observed.
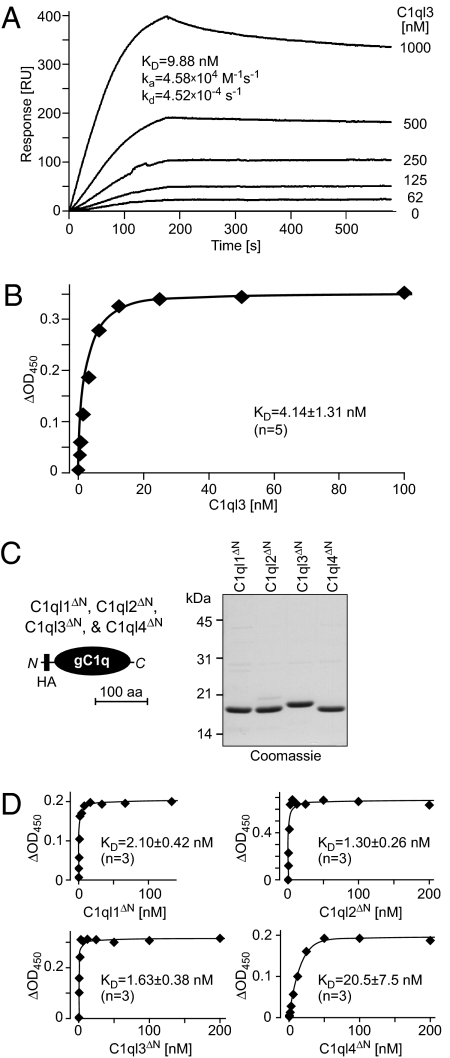
Measurement of Binding Affinities.
We next strove to determine the binding affinities of the C1ql proteins for BAI3. For this purpose, we first performed surface plasmon resonance experiments with recombinant full-length C1ql3. We observed fast on-rates but slow off-rates, suggesting an overall dissociation constant (KD) of ∼10 nM (Fig. 3A). We confirmed this estimate of the C1ql3–BAI3 binding affinity with a quantitative cell-surface binding assay in which we incubated HEK293 cells expressing BAI3 with different concentrations of C1ql3, using mock-transfected cells as a control. We then fixed the cells and quantified the amount of bound C1ql3 on the surface using an HRP-coupled anti-HA antibody. Affinities were calculated after subtraction of background binding observed with control cells. In five independent experiments, we observed an apparent dissociation constant of ∼4 nM for full-length C1ql3 and BAI3, affirming a high-affinity interaction (Fig. 3B). It should be noted that the dissociation constant estimated by this method might underestimate the true binding affinity, because the assay also relies on the interaction between the HA epitope and the anti-HA antibody.
Fig. 3.
Nanomolar affinity of C1ql1, C1ql2, C1ql3, and C1ql4 for BAI3. (A) Surface plasmon resonance measurements. Full-length C1ql3 was injected at the indicated concentrations onto chips containing immobilized IgBAI3-3 or IgC for 180 s, followed by injection of buffer alone. Deduced binding constants are listed in the graph. RU, resonance units. (B) Cell-surface binding measurements with full-length C1ql3 as a ligand. HEK293 cells were transfected with a BAI3 cDNA or mock-transfected, incubated with purified HA-tagged full-length C1ql3 at the indicated concentrations, and fixed. Bound protein was detected with a peroxidase-conjugated antibody against the HA epitope. The signal shown was subtracted for background observed with mock-transfected cells. Data are from a representative experiment independently repeated five times, with Scatchard fits resulting in the indicated binding constant (mean ± SEM). (C) Schematic diagram (Left) and Coomassie blue–stained SDS gel (Right) of recombinant gC1q domains of all four C1ql proteins. gC1q domains were expressed with an N-terminal HA tag as GST-fusion proteins in Escherichia coli, and GST was removed by thrombin cleavage after purification with glutathione-Sepharose. gC1q domains are referred to as C1ql1ΔN–C1ql4ΔN because the N-terminal cysteine sequence and collagen domain were deleted from the C1ql proteins to generate these recombinant domains. (D) Binding curves generated as described in B, but with HA-tagged N-terminally truncated C1ql molecules consisting only of the HA-tagged gC1q domains of the respective proteins. Data shown are from a representative experiment independently repeated three times, with Scatchard fits resulting in the indicated binding constants (mean ± SEM).
To measure the affinities of all C1ql proteins for BAI3, we needed to produce sufficient quantities of recombinant proteins, which proved difficult in transiently transfected HEK293 cells. Thus, based on studies of other gC1q domain proteins (32–34), we speculated that the binding of C1ql proteins to BAI3 might be mediated by the gC1q domain. We produced the isolated gC1q domains of all C1ql proteins as HA-tagged recombinant proteins in bacteria by first generating GST-fusion proteins, and then cleaving off the C1ql moiety with thrombin (Fig. 3C).
We then used the recombinant bacterial gC1q domains of the C1ql proteins for quantitative cell-surface binding assays (Fig. 3D). Strikingly, the bacterially produced gC1q domains of all C1ql proteins were specifically and saturably bound to BAI3-expressing HEK293 cells. As determined in three independent experiments, the gC1q domains of C1ql1, C1ql2, and C1ql3 bound to BAI3 with low nanomolar affinities (KD = 1.3–2.1 nM), whereas the gC1q domain of C1ql4 exhibited an ∼10-fold lower affinity (KD = 20.5 nM) (Fig. 3D). Thus, all C1ql proteins are high-affinity ligands for BAI3.
Characterization of Binding Domains.
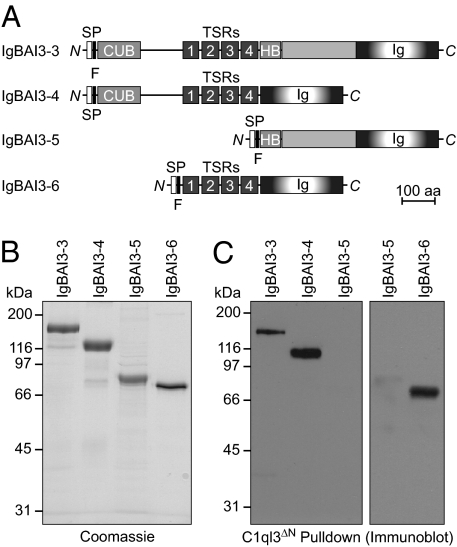
The extracellular sequence of BAI3 contains a CUB domain, four thrombospondin repeats, a hormone-binding domain, and a GPS motif preceded by a long uncharacterized sequence (Fig. 1A). To examine which of these sequences constitutes the binding site for C1ql proteins, we produced four BAI3 Ig-fusion proteins containing different extracellular BAI3 domains (Figs. 4 A and B). We then used these proteins in pull-down experiments with purified C1ql3 protein. These experiments revealed that the thrombospondin repeats of BAI3 were both necessary and sufficient to bind to C1ql3 (Fig. 4C). Given that thrombospondin repeats are known to adopt an autonomously folded, disulfide-bonded conformation composed of three β-strands (38), it is possible that fewer than four thrombospondin repeats are required for binding.
Fig. 4.
BAI3 fragment containing thrombospondin repeats binds to the gC1q domain of C1ql3. (A) Schematic diagram of BAI3 deletion constructs. Abbreviations are as in Fig. 1. (B) Coomassie blue–stained SDS gels of BAI3 Ig-fusion proteins expressed in HEK293 cells and purified from the medium by anti-FLAG agarose. (C) Pull-down experiments with the HA-tagged gC1q domain of C1ql3 that was immobilized on an HA-antibody column and used as an affinity matrix for the indicated purified BAI3 Ig-fusion proteins. Bound proteins were detected by immunoblot analysis for the FLAG epitopes. Data show representative experiments independently repeated three times.
Effect of C1ql Proteins on Synapse Density.
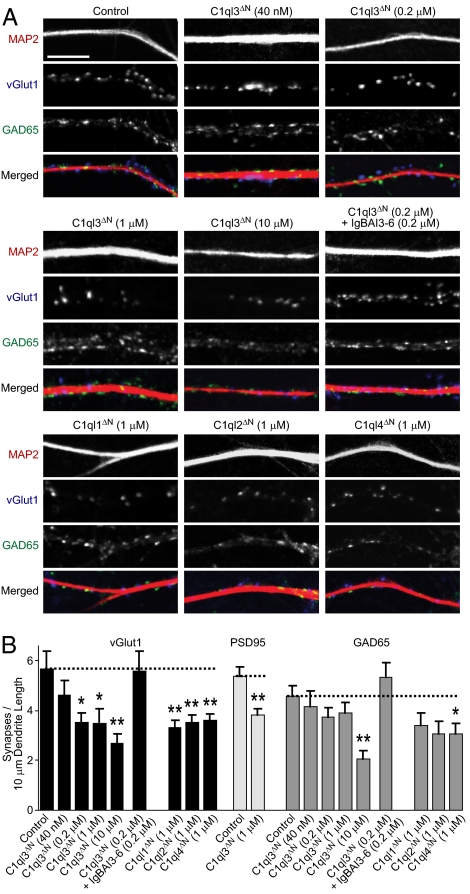
Because BAI3 and C1ql proteins are both expressed at high levels in neurons, and cerebellin-1—which is related to C1ql proteins—regulates synapse formation in the brain (39, 40), we hypothesized that C1ql proteins and BAI3 might have a function in synapses. As a first test of this hypothesis, we added to cultured hippocampal neurons at 10 d in vitro (DIV10) increasing concentrations of recombinant bacterial C1ql3 protein (composed of only the gC1q domain), and determined the density of excitatory and inhibitory synapses on pyramidal neuronal dendrites on DIV14 (Fig. 5). Synapse densities were measured by indirect immunofluorescence with antibodies to excitatory (vGlut1, the vesicular glutamate transporter, and PSD95, a postsynaptic density protein) and inhibitory markers (GAD65, a 65-kDa glutamic acid decarboxylase). Simultaneously, we also quantified morphological parameters of the neurons, including soma size, dendrite length, and dendritic branching (Fig. 6).
Fig. 5.
Incubation of primary hippocampal neurons with recombinant gC1q domains from C1ql1–C1ql4 decreases synapse density. (A) Representative images of dendrites from cultured hippocampal neurons that were incubated with recombinant HA-tagged gC1q domains of the indicated C1ql proteins at DIV10 and analyzed at DIV14 by immunofluorescence staining with antibodies to MAP2 (to label dendrites) and vGlut1 or GAD65 as markers for excitatory and inhibitory synapses, respectively. (Scale bar: 10 μm.) (B) Summary graphs of excitatory and inhibitory synapse densities measured by staining with antibodies to vGlut1 and PSD95 (for excitatory synapses) or to GAD65 (for inhibitory synapses). Synapse densities were determined in neurons treated as described in A, with the incubation conditions indicated below each bar. Graphs show mean ± SEM of three independent experiments (two-tailed Student's t test; *P < 0.05; **P < 0.01).
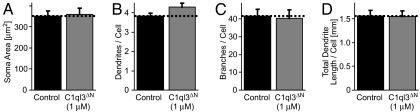
Fig. 6.
Treatment of cultured hippocampal neurons with the C1ql3 gC1q domain does not alter overall neuron size, dendrite length, or dendritic branching. Hippocampal neurons were treated as described in Fig. 5 with recombinant C1ql3 gC1q domain (1 μM). Graphs show the mean size of the neuronal soma (A), number of dendritic processes per soma (B), number of dendritic branches per cell (C), and total length of dendrites per cell (D). Data are mean ± SEM of three independent experiments.
As measured with staining for vGlut1, C1ql3 concentrations as low as 0.2 μM significantly (∼40%) decreased the excitatory synapse density in cultured hippocampal neurons, with a maximal decrease of ∼55% obtained with 10 μM C1ql3 (Fig. 5B). The decrease in excitatory synapse density was also detected by staining excitatory synapses for PSD95, whereas C1ql3 had no statistically significant effect on inhibitory synapse density except at high concentrations (i.e., 10 μM). Strikingly, the effect of C1ql3 on synapse density was occluded by addition of an equimolar concentration of the recombinant BAI3 fragment that binds to C1ql3 (IgBAI3-6) (Fig. 5). Examination of the other C1ql proteins indicated that they also decreased the density of synapses, with similar potencies. As a control, we measured the effects of C1ql3 applied at 1 μM on the length and branching of dendrites in the cultured neurons, and found no significant change (Fig. 6). Taken together, our findings suggest that the C1ql proteins selectively affect synapse density in cultured neurons.
Discussion
In a search for endogenous ligands for the cell-adhesion–type GPCR BAI3, we have identified C1ql proteins as endogenous BAI3 ligands. C1ql proteins are small, secreted proteins containing a gC1q domain preceded by two short multimerization sequences (a cysteine loop motif and a collagen-like domain; Fig. 2A), whereas BAI3 is composed of a large extracellular sequence containing a CUB domain, four thrombospondin repeats, and a GPS sequence, followed by a typical GPCR architecture composed of seven transmembrane regions and a cytoplasmic C-terminal tail (Fig. 1A). We found that the gC1q domain of all four C1ql proteins binds to the thrombospondin repeats of BAI3 (Figs. 2–4). The high affinity of this binding (KD = 1–20 nM), along with the nature of C1ql proteins as small secreted proteins and of BAI3 as a membrane-bound GPCR, suggest that BAI3 is a genuine receptor for C1ql proteins in the brain.
Both C1ql and BAI3 proteins are expressed in adult animals almost exclusively in the brain and appear to be enriched in neurons (11, 12, 31). Strikingly, the addition of the recombinant gC1q domains of C1ql proteins to the medium of cultured hippocampal neurons caused significant decreases in synapse density without affecting other measured morphological parameters (Figs. 5 and 6). This effect was observed for all four C1ql proteins and was blocked in the case of C1ql3 by the addition of the recombinant thrombospondin repeats of BAI3 (Fig. 5). These findings suggest that C1ql proteins regulate synapses and thus are functionally analogous to cerebellins, which are homologous to C1ql proteins, have a well-documented function in synapse formation, and bind to the orphan glutamate receptor δ2 (39, 40).
In summary, our data indicate that C1ql proteins represent a family of signaling molecules, analogous to cerebellins (39, 40) and adiponectin (36, 41, 42), and that they act, at least in part, by binding to BAI3 as a receptor. Our study raises many additional questions, however. Whether C1ql proteins actually activate BAI3 as a GPCR to initiate an intracellular signaling cascade is unclear. One possibility is that C1ql proteins, as large multimers, link multiple BAI3 molecules into complexes, thereby activating them. Moreover, our experiments do not demonstrate whether C1ql proteins decrease synapse density through a direct effect on synapse formation, on synapse stability, or on other properties of the neurons. This question remains to be addressed in future experiments. Furthermore, although the BAI3 fragment binding to C1ql3 blocked the effect of C1ql3 on synapse density, it is possible that C1ql3 decreases synapse density by binding to a protein other than BAI3 on the neuronal surface. Nonetheless, the high affinity and specificity of C1ql protein binding to BAI3, along with the strong expression of BAI3 in pyramidal hippocampal neurons (15), strongly support the notion that their interaction underlies the effect of C1ql proteins on synapse density. Finally, some basic questions— such as that of the localization of BAI3—also remain to be addressed. These questions are difficult to answer without either overexpressing tagged proteins, which often gives erroneous results, or generating outstanding antibodies to the endogenous protein, which is often difficult. Thus, our study is an initial exploration of C1ql and BAI3 proteins that suggests an unexpected relationship between these fascinating molecules and raises the possibility that together they might regulate synapses, and that the association of human BAI3 SNPs with schizophrenia (28) may reflect, at least in part, aberrant synapse formation and/or maintenance in the carriers of these mutations. Our work here will hopefully initiate future studies investigating a potential association between C1ql mutations and schizophrenia.
Materials and Methods
Antibodies.
The following antibodies were used: rabbit anti-FLAG, rabbit anti-HA, and mouse anti-MAP2 (clone HM-2) antibodies (Sigma-Aldrich); rabbit anti-MAP2 and guinea pig anti-vGlut1 antibodies (Millipore); mouse anti-PSD95 (clone 7E3-1B8) antibodies (Thermo Scientific); mouse anti-GAD65 (clone GAD-6) antibodies (Developmental Studies Hybridoma Bank); secondary peroxidase-conjugated antibodies (Cappel Biomedical and MP Biomedicals); and secondary antibodies conjugated with Alexa Fluor 488, 546, and 633 (Molecular Probes).
Expression Plasmids.
All BAI3 constructs include the preprotrypsin signal peptide followed by the FLAG epitope from vector pFLAG-CMV (Sigma-Aldrich). Ig-fusion proteins contain the Fc fragment of human IgG (43). Full-length C1ql proteins were expressed with the Ig κ-chain signal sequence and HA epitope from the pDisplay vector (Invitrogen). gC1q domain GST-fusion proteins were expressed in pGEX-KG (American Type Culture Collection); an HA epitope was ligated into the XmaI site of the linker region following the thrombin cleavage site. The following plasmids were used in this study: pCAGGS/FLAG-BAI3-mVenus (residues 20–1522 of human BAI3, with a monomeric Venus inserted into the BstXI site), pCMV/FLAG-BAI3-3-Ig (residues 20–799 of human BAI3), pCMV/FLAG-BAI3-4-Ig (residues 20–509 of human BAI3), pCMV/FLAG-BAI3-5-Ig (residues 510–799 of human BAI3), pCMV/FLAG-BAI3-6-Ig (residues 287–509 of human BAI3), pCMV/IgC (residues 1–48 of rat neurexin-1α; ref. 43), pDisplay/HA-C1ql1 (residues 21–258 of mouse C1ql1), pDisplay/HA-C1ql2 (residues 22–287 of mouse C1ql2), pDisplay/HA-C1ql3 (residues 21–255 of mouse C1ql3), pDisplay/HA-C1ql4 (residues 21–238 of mouse C1ql4), pGEX/GST-HA-C1Q1 (residues 122–258 of mouse C1ql1), pGEX/GST-HA-C1Q2 (residues 152–287 of mouse C1ql2), pGEX/GST-HA-C1Q3 (residues 119–255 of mouse C1ql3), and pGEX/GST-HA-C1Q4 (residues 104–238 of mouse C1ql4).
Biochemical Procedures.
GST-fusion proteins expressed in bacteria and Flag-tagged or Ig-fusion proteins expressed in transfected HEK293 cells were purifed essentially as described previously using glutathione agarose (44) or anti-Flag or protein A-Sepharose affinity columns (43). SDS/PAGE and immunblot analysis were performed using standard methods. For pull-down assays and affinity chromatography experiments, brain proteins solubilized in 20 mM Hepes-NaOH (pH 7.4), 0.1 M NaCl, 2 mM CaCl2, 2 mM MgCl2, 1% Triton X-100, 1 mM EGTA, and protease inhibitors were used in standard procedures (45), and bound proteins were analyzed by SDS/PAGE, visualized by silver staining, and identified by mass spectrometry at the Stanford University Mass Spectrometry Facility. To test the binding of C1ql3 to various BAI3 Ig-fusion proteins, HA-tagged C1ql3 attached to anti-HA agarose was incubated with BAI3 Ig-fusion proteins [either purified proteins in 10 mM Hepes-NaOH (pH 7.4), 0.15 M NaCl, 2 mM CaCl2, and 2 mM MgCl2 or medium from transfected HEK293 cells] overnight at 4 °C under agitation. Beads were washed, boiled for 4 min in SDS sample buffer, and analyzed by SDS/PAGE and immunoblot analysis.
Cell-Surface Assays.
Cell-surface labeling assays were performed as described previously (46). For cell-surface binding affinity determinations, BAI3-transfected HEK293 cells were incubated with HA-tagged ligands [in DMEM containing 50 mM Hepes-NaOH (pH 7.4), 0.1% BSA, 2 mM CaCl2, 2 mM MgCl2] in a serial dilution (typically 11 dilutions: 400 nM, 200 nM, 100 nM, etc., and without protein, in duplicate) for 16 h at 4 °C under gentle agitation. Cells were washed, fixed with 4% paraformaldehyde for 10 min on ice, washed with PBS, and incubated in blocking solution (3% BSA in PBS) for 30 min at room temperature. Peroxidase-conjugated anti-HA antibodies (diluted 1:50,000; Sigma-Aldrich) were added in blocking solution for 60 min. After washing with PBS, cells were incubated in TMB peroxidase EIA substrate solution (Bio-Rad) for 5–20 min under vigorous agitation. The reaction was stopped with an equal volume of 1N sulphuric acid, and absorbance at 450 nm was measured in a 96-well plate using an Apollo-8 LB912 plate reader (Berthold Technologies). The ligand concentration was plotted versus the absorbance difference between transfected and mock-transfected cells, and the KD value was calculated using Scatchard analysis.
Surface Plasmon Resonance Analysis.
Binding of C1ql3 to IgBAI3-3 was measured on a Biacore 3000 system. IgBAI3-3 was covalently bound to the carboxymethylated dextran matrix of a research-grade CM5 biosensor chip using EDC/NHS amine-coupling chemistry. A separate reference surface contained IgC protein. C1ql3 was diluted in running buffer [107 mM Hepes-NaOH (pH 7.1), 89 mM glycine, 2 mM CaCl2, 2 mM MgCl2], and injected over the two surfaces at different concentrations with a flow rate of 30 μL/min at 10 °C. The injection interval was 180 s, followed by a decay interval of 400 s in which running buffer alone was run through the surface. The surfaces were regenerated by two 30-s injections of a regeneration solution [107 mM Hepes-NaOH (pH 7.1), 89 mM glycine, 5 mM EGTA]. Reference data were subtracted from the sample flow channel to obtain specific C1ql3 binding.
Neuronal Culture Experiments.
Primary hippocampal neurons were cultured from newborn mice. Neurons were dissociated by papain treatment (20 min at 37 °C), triturated, plated onto coverslips coated with Matrigel (BD Biosciences) in 24-well plates and cultured in MEM (Gibco), 5 g/L of glucose, 100 mg/L of transferrin, 200 mg/L of NaHCO3, 0.5 mM l-glutamine, 5% FBS, 2% B-27 supplement, and 2 μM cytosine arabinoside. Purified C1ql proteins were added at the indicated concentrations, either alone or together with IgBAI3-6, to the cultured neurons on DIV10, and neurons were analyzed on DIV14 by double-immunofluorescence staining using methanol fixation for vGlut1, GAD65, and MAP2 or fixation in cold 4% paraformaldehyde/2% sucrose for PSD95 and MAP2. Images were acquired with a Leica TCS2 confocal microscope and analyzed with MetaMorph software (Molecular Devices). For each independent experiment, the first ∼50 μm of a basal dendrite (starting from the soma) from 15 pyramidal neurons were imaged, and synapse densities were averaged. The calculations shown in Fig. 6 were performed using the MetaMorph neurite analysis application.
Acknowledgments
We thank Dr. Tong Zang (University of Texas Southwestern Medical Center) for help with the initial cloning of the BAI3 expression vector. This study was supported by National Institute of Mental Health Grant R37 MH052804-17 (to T.C.S.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Yona S, Lin HH, Siu WO, Gordon S, Stacey M. Adhesion-GPCRs: Emerging roles for novel receptors. Trends Biochem Sci. 2008;33:491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnadóttir TK, et al. The human and mouse repertoire of the adhesion family of G protein-coupled receptors. Genomics. 2004;84:23–33. doi: 10.1016/j.ygeno.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Krasnoperov V, et al. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor: Role of the G protein-coupled receptor proteolysis site (GPS) motif. J Biol Chem. 2002;277:46518–46526. doi: 10.1074/jbc.M206415200. [DOI] [PubMed] [Google Scholar]
- 5.Lin HH, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 6.Krasnoperov V, et al. Dissociation of the subunits of the calcium-independent receptor of alpha-latrotoxin as a result of two-step proteolysis. Biochemistry. 2009;48:3230–3238. doi: 10.1021/bi802163p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das G, Reynolds-Kenneally J, Mlodzik M. The atypical cadherin Flamingo links Frizzled and Notch signaling in planar polarity establishment in the Drosophila eye. Dev Cell. 2002;2:655–666. doi: 10.1016/s1534-5807(02)00147-8. [DOI] [PubMed] [Google Scholar]
- 8.Langenhan T, et al. Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Dev Cell. 2009;17:494–504. doi: 10.1016/j.devcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monk KR, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–1405. doi: 10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimori H, et al. A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene. 1997;15:2145–2150. doi: 10.1038/sj.onc.1201542. [DOI] [PubMed] [Google Scholar]
- 11.Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T. Cloning and characterization of BAI2 and BAI3, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1) Cytogenet Cell Genet. 1997;79:103–108. doi: 10.1159/000134693. [DOI] [PubMed] [Google Scholar]
- 12.Haitina T, et al. Expression profile of the entire family of adhesion G protein-coupled receptors in mouse and rat. BMC Neurosci. 2008;9:43. doi: 10.1186/1471-2202-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh JT, et al. Characterization of mouse brain-specific angiogenesis inhibitor 1 (BAI1) and phytanoyl-CoA alpha-hydroxylase–associated protein 1, a novel BAI1-binding protein. Brain Res Mol Brain Res. 2001;87:223–237. doi: 10.1016/s0169-328x(01)00004-3. [DOI] [PubMed] [Google Scholar]
- 14.Kee HJ, et al. Expression of brain-specific angiogenesis inhibitor 2 (BAI2) in normal and ischemic brain: Involvement of BAI2 in the ischemia-induced brain angiogenesis. J Cereb Blood Flow Metab. 2002;22:1054–1067. doi: 10.1097/00004647-200209000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Kee HJ, et al. Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett. 2004;569:307–316. doi: 10.1016/j.febslet.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Duda DG, et al. Overexpression of the p53-inducible brain-specific angiogenesis inhibitor 1 suppresses efficiently tumour angiogenesis. Br J Cancer. 2002;86:490–496. doi: 10.1038/sj.bjc.6600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24:3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 18.Kang X, et al. Antiangiogenic activity of BAI1 in vivo: Implications for gene therapy of human glioblastomas. Cancer Gene Ther. 2006;13:385–392. doi: 10.1038/sj.cgt.7700898. [DOI] [PubMed] [Google Scholar]
- 19.Kudo S, et al. Inhibition of tumor growth through suppression of angiogenesis by brain-specific angiogenesis inhibitor 1 gene transfer in murine renal cell carcinoma. Oncol Rep. 2007;18:785–791. [PubMed] [Google Scholar]
- 20.Fukushima Y, et al. Brain-specific angiogenesis inhibitor 1 expression is inversely correlated with vascularity and distant metastasis of colorectal cancer. Int J Oncol. 1998;13:967–970. doi: 10.3892/ijo.13.5.967. [DOI] [PubMed] [Google Scholar]
- 21.Hatanaka H, et al. Vascularization is decreased in pulmonary adenocarcinoma expressing brain-specific angiogenesis inhibitor 1 (BAI1) Int J Mol Med. 2000;5:181–183. doi: 10.3892/ijmm.5.2.181. [DOI] [PubMed] [Google Scholar]
- 22.Kaur B, Brat DJ, Calkins CC, Van Meir EG. Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am J Pathol. 2003;162:19–27. doi: 10.1016/S0002-9440(10)63794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardcastle J, et al. Enhanced antitumor efficacy of vasculostatin (Vstat120) expressing oncolytic HSV-1. Mol Ther. 2010;18:285–294. doi: 10.1038/mt.2009.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh JT, et al. Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Exp Cell Res. 2004;294:172–184. doi: 10.1016/j.yexcr.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Jeong BC, et al. Brain-specific angiogenesis inhibitor 2 regulates VEGF through GABP that acts as a transcriptional repressor. FEBS Lett. 2006;580:669–676. doi: 10.1016/j.febslet.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 26.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 27.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 28.DeRosse P, et al. The genetics of symptom-based phenotypes: Toward a molecular classification of schizophrenia. Schizophr Bull. 2008;34:1047–1053. doi: 10.1093/schbul/sbn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: Update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 30.Bérubé NG, et al. Cloning and characterization of CRF, a novel C1q-related factor, expressed in areas of the brain involved in motor function. Brain Res Mol Brain Res. 1999;63:233–240. doi: 10.1016/s0169-328x(98)00278-2. [DOI] [PubMed] [Google Scholar]
- 31.Iijima T, Miura E, Watanabe M, Yuzaki M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur J Neurosci. 2010;31:1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 32.Kishore U, et al. C1q and tumor necrosis factor superfamily: Modularity and versatility. Trends Immunol. 2004;25:551–561. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Carland TM, Gerwick L. The C1q domain containing proteins: Where do they come from and what do they do? Dev Comp Immunol. 2010;34:785–790. doi: 10.1016/j.dci.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Ghai R, et al. C1q and its growing family. Immunobiology. 2007;212:253–266. doi: 10.1016/j.imbio.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Urade Y, Oberdick J, Molinar-Rode R, Morgan JI. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc Natl Acad Sci USA. 1991;88:1069–1073. doi: 10.1073/pnas.88.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 37.Bao D, Pang Z, Morgan JI. The structure and proteolytic processing of Cbln1 complexes. J Neurochem. 2005;95:618–629. doi: 10.1111/j.1471-4159.2005.03385.x. [DOI] [PubMed] [Google Scholar]
- 38.Tan K, et al. Crystal structure of the TSP-1 type 1 repeats: A novel layered fold and its biological implication. J Cell Biol. 2002;159:373–382. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda K, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- 40.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 41.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 42.Maeda K, et al. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (adipose most abundant gene transcript 1) Biochem Biophys Res Commun. 1996;221:286–289. doi: 10.1006/bbrc.1996.0587. [DOI] [PubMed] [Google Scholar]
- 43.Ushkaryov YA, et al. Conserved domain structure of beta-neurexins: Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 1994;269:11987–11992. [PubMed] [Google Scholar]
- 44.Burré J, et al. α-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ichtchenko K, et al. Neuroligin 1: A splice site-specific ligand for β-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 46.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice-code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]