Abstract
Angiosperms exhibit staggering diversity in floral form, and evolution of floral morphology is often correlated with changes in pollination syndrome. The showy, bilaterally symmetrical flowers of the model species Antirrhinum majus (Plantaginaceae) are highly specialized for bee pollination. In A. majus, CYCLOIDEA (CYC), DICHOTOMA (DICH), RADIALIS (RAD), and DIVARICATA (DIV) specify the development of floral bilateral symmetry. However, it is unclear to what extent evolution of these genes has resulted in flower morphological divergence among closely related members of Plantaginaceae differing in pollination syndrome. We compared floral symmetry genes from insect-pollinated Digitalis purpurea, which has bilaterally symmetrical flowers, with those from closely related Aragoa abietina and wind-pollinated Plantago major, both of which have radially symmetrical flowers. We demonstrate that Plantago, but not Aragoa, species have lost a dorsally expressed CYC-like gene and downstream targets RAD and DIV. Furthermore, the single P. major CYC-like gene is expressed across all regions of the flower, similar to expression of its ortholog in closely related Veronica serpyllifolia. We propose that changes in the expression of duplicated CYC-like genes led to the evolution of radial flower symmetry in Aragoa/Plantago, and that further disintegration of the symmetry gene pathway resulted in the wind-pollination syndrome of Plantago. This model underscores the potential importance of gene loss in the evolution of ecologically important traits.
Evolution of flower symmetry is tightly correlated with pollinator shifts and has been proposed as a key innovation within the angiosperms (1–6). In the plantain family (Plantaginaceae, Lamiales), morphological and phylogenetic analyses suggest that the ancestral flower was five-parted, bilaterally symmetrical (monosymmetrical and zygomorphic), and insect-pollinated (7, 8). For example, in Antirrhinum majus (snapdragon, Antirrhineae) and Digitalis purpurea (foxglove, Digitalideae), large bee-pollinated flowers are five-parted and establish bilateral symmetry due to differential growth rates along the dorsiventral axis. The dorsal (adaxial), lateral, and ventral (abaxial) petals and stamens are easily distinguishable based on differences in shape and/or ornamentation, further reinforcing a single plane of symmetry within mature flowers (Fig. 1; Fig. S1) (8–13).
Fig. 1.
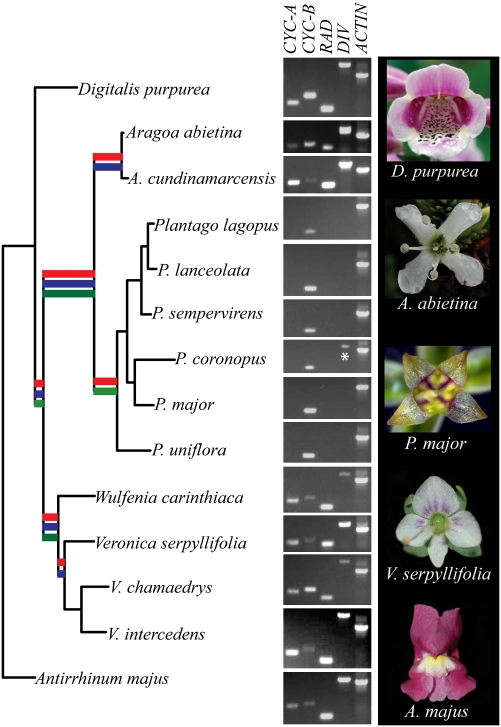
Maximum-likelihood phylogram based on combined nuclear markers showing evolution of flower symmetry and its underlying genetic pathway in Plantaginaceae. Bold branches indicate supported (>70% bootstrap and/or >90% posterior probability) generic relationships based on the chloroplast markers rps2, rbcL, ndhF, rps16, and matK-trnK (blue branch) (7, 15, 20), combined nuclear markers DEL, ITS, and DIV intron (red branch), and ACT (green branch). Morphological transitions include a reduction from five large (Antirrhinum and Digitalis) to four small (Veronica, Plantago, and Aragoa) petals, and a shift from bilateral (Antirrhinum, Digitalis, and Veronica) to radial flower symmetry (Aragoa and Plantago). PCR analyses using conserved primers on genomic DNA demonstrate that the loss of showy petals, but not radial flower symmetry, is correlated with a loss of the A-clade CYC-like gene and its downstream targets RAD and DIV in six Plantago species, representing four of the five monophyletic subgenera (21). Note: Our sequence analysis confirmed that the band amplified by the DIV primers in P. coronopus (indicated with an asterisk) is the DIV-like paralog PcDVL (Fig. 2C).
Veroniceae is the sister tribe to Digitalideae, and includes the large cosmopolitan genera Veronica (ca. 450 species) and Plantago (ca. 200 species) and the small páramo endemic genus Aragoa (19 species), each of which is monophyletic based on phylogenetic analyses (14–19). In contrast to the ancestral flower form found in Digitalideae, several Veroniceae species have flowers that are four-parted, radially symmetrical (polysymmetrical and actinomorphic), and/or wind-pollinated (Fig. 1) (8, 15, 20–22). The small flowers of Veronica are primarily fly- and/or self-pollinated, and develop four sepals and four petals (23, 24). The enlarged dorsal petal is inferred to develop from two dorsal petals that fuse early in development (11, 25) and, consequently, Veronica flowers have retained bilateral symmetry within the petal whorl (11, 25). Whereas the radially symmetrical showy flowers of Aragoa have retained the ancestral number of five sepals but have four identical petals and stamens (11, 12, 21), the diminutive radially symmetrical flowers of wind-pollinated Plantago are four-merous in all three outer organ whorls (Fig. 1). In early development, both Aragoa and Plantago flowers are bilaterally symmetrical due to the unidirectional emergence of organs in a ventral-to-dorsal sequence (21). However, flowers at mid to late stages of development in both genera are fully radially symmetrical (21, 26).
It is hypothesized that changes in the expression or biochemical function of genes that establish the ancestral condition of floral bilateral symmetry underlie the evolution of radial symmetry in the Aragoa/Plantago clade (8, 21, 27). The floral symmetry genetic pathway is best understood in A. majus, and involves the action of three dorsal identity genes, CYCLOIDEA (CYC), DICHOTOMA (DICH), and RADIALIS (RAD), and one ventral identity gene, DIVARICATA (DIV) (13, 28–32). CYC and DICH are recently duplicated type II TCP transcription factors that establish bilateral symmetry in both early and late flower development, partly through their positive regulation of the MYB transcription factor RAD in the dorsal petals and staminode (13, 29, 31, 33). Although not expressed outside the dorsal petals and staminode, RAD appears to negatively regulate the ventral identity protein DIV in the dorsal and lateral regions of the flower (29, 30). Thus, ventral identity is confined to the ventral region of the flower (28, 30).
Unlike close relatives Veronica serpyllifolia and D. purpurea, which have two and three CYC-like genes, respectively, only one CYC-like gene has been recovered from Plantago lanceolata (ribwort plantain; subgenus Psyllium sensu lato) and P. major (common plantain; subgenus Plantago) genomes, suggesting a recent gene loss event in the Plantago lineage (14, 18, 27, 33–35). Furthermore, Reardon et al. recently demonstrated that expression of this gene does not appear to be confined to the dorsal region of the P. lanceolata flower (27). Instead, evidence suggests expression in the region below the flower, correlating with the loss of differentiated dorsal flower identity and a shift to radial symmetry. Interestingly, a nondorsal pattern of expression for the V. serpyllifolia ortholog to the single Plantago CYC-like gene was also recently demonstrated (34). The aim of this study is to better understand the role of floral symmetry gene network evolution in the shift from bilateral to radial symmetry in the Veroniceae. Here we test three nonexclusive hypotheses: (i) loss of a dorsally expressed CYC-like paralog predates evolution of radial symmetry in the Aragoa/Plantago lineage; (ii) the nondorsal expression pattern of the single Plantago CYC-like gene evolved before the loss of its sister paralog; and (iii) the loss of a dorsally expressed CYC-like paralog resulted in loss, or change in expression, of its downstream targets.
Results
Plantago and Aragoa Are More Closely Related to Veronica than to Digitalis.
Although the sister relationship of Aragoa and Plantago is well-supported based on multiple markers (7, 15, 20) (Fig. 1), previous phylogenetic analyses have been equivocal regarding the relationship between the monophyletic genera Aragoa/Plantago, Veronica, and Digitalis (7, 14–20). Analyses based on combined rbcL, ndhF, and rps2, as well as trnL-F chloroplast sequences, support a close relationship between Aragoa/Plantago and Veronica, with Digitalis sister to these genera together (7, 15, 20) (Fig. 1). However, analyses based on the matK-trnK and rps16 introns and the internal transcribed spacer (ITS) give different topologies with varying levels of support (20).
To better resolve these relationships with the ultimate aim of elucidating the direction of evolutionary change within the floral symmetry gene network, we combined ITS sequences generated from multiple studies (15, 18, 20, 36) and generated partial sequences of the nuclear DELILA (DEL) and ACTIN (ACT) genes and the full DIV intron for A. majus, D. purpurea, two Aragoa species, six Plantago species representing four of the five monophyletic subgenera (except the DIV intron), three Veronica species representing three of the twelve subgenera, and the Veronica outgroup Wulfenia carinthiaca (14–19) (Fig. 1; Fig. S2). DEL, ACT, and the DIV intron have been successfully used to resolve species relationships in eukaryotes and other plant groups (37–39).
With the exception of ACT, individual analyses of aligned nucleotides strongly supported the monophyly of Veronica, Plantago, and Aragoa, as previously suggested (14–19) (Fig. S2). Furthermore, consistent with combined analyses based on chloroplast gene markers (7, 20), all trees from individual analyses resolved a sister relationship between Aragoa/Plantago and Wulfenia/Veronica genes, with the genes from D. purpurea being more distantly related (Fig. S2). Results were concordant between maximum-likelihood and Bayesian reconstructions, and were supported by likelihood bootstrap (LB) and/or posterior probability (PP) values >70% and >90%, respectively (Fig. S2). Based on concordance between gene trees, DEL, ITS, and DIV intron sequences were concatenated for further analysis. The best trees based on combined analyses were not significantly different from independent analyses; the monophyly of each genus and the sister relationship between Aragoa/Plantago and Wulfenia/Veronica was supported by LB (100% and 76%, respectively) and/or PP (100% and 86%, respectively) values (Fig. 1; Fig. S2).
Floral Symmetry Gene Degeneration in Representative Species of Plantago but Not Aragoa.
To determine copy-number and orthology/paralogy relationships among floral symmetry genes, multiple clones derived from PCR using various CYC-, RAD-, and DIV-like primer sets were sequenced from D. purpurea, V. serpyllifolia, P. major, P. lanceolata, and A. abietina. Consistent with previous studies, only one CYC-like gene was isolated from P. major (PmTCP1) and P. lanceolata (PlTCP1), three were isolated from D. purpurea (DpCYC1, DpCYC2, and DpCYC3), and two were isolated from V. serpyllifolia (VsCYC1 and VsCYC2) (14, 27, 33–35). Similar to V. serpyllifolia, two putatively functional CYC-like genes were also isolated from A. abietina (AaCYC1 and AaCYC2).
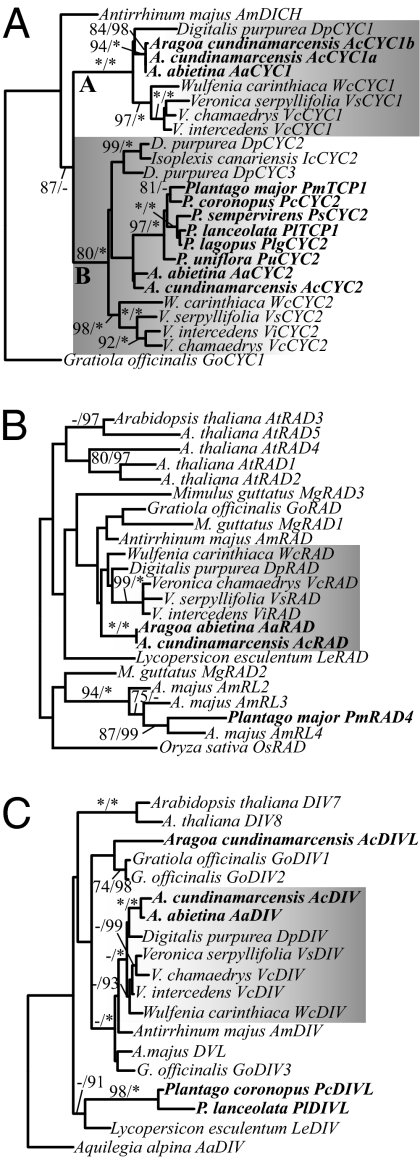
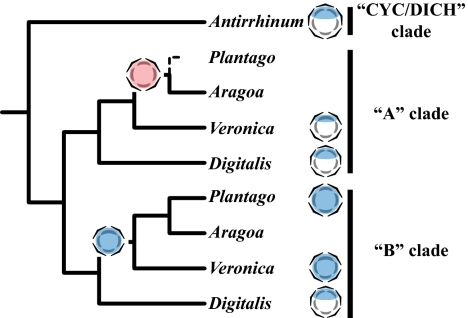
Phylogenetic analyses of these sequences, supplemented with sequences from additional species (see below), revealed a well-supported (LB of 100%, PP of 100%) sister relationship between DpCYC1, VsCYC1, and AaCYC1 (“A” clade in Fig. 2A) and among the recently duplicated D. purpurea genes DpCYC2 and DpCYC3, VsCYC2, AaCYC2, PmTCP1, and PlTCP1 (“B” clade in Fig. 2A; LB of 80%, PP of 100%). Although inferred gene relationships within the A and B clades did not completely track the species phylogeny (Fig. 1 versus Fig. 2A), there was limited support from the CYC dataset for generic relationships within either the A or B clade. Thus, together with support for the species tree topology, the CYC-like gene tree suggests a single gene duplication event at the base of Digitalideae + Veroniceae, followed by A lineage gene loss at the base of Plantago. Comparison of maximum-likelihood models found no evidence of positive or relaxed selection on branches leading to AaCYC1, AaCYC2/PmTCP1/PlTCP1, or PmTCP1/PlTCP1 (Table S1).
Fig. 2.
Maximum-likelihood phylograms showing best estimates of floral symmetry gene relationships. (A) CYC-like genes were duplicated following the divergence of Antirrhineae (represented by Antirrhinum majus) and Digitalideae (represented by Digitalis purpurea) + Veroniceae (represented by Wulfenia, Veronica, Aragoa, and Plantago), giving rise to two gene clades designated A and B (shaded). Plantago species have a single B-clade CYC-like gene (bold), but no genes were found within the CYC A clade, suggesting loss of this gene at the base of Plantago. (B) Plantago genomes have RAD-like paralogs (bold) but lack RAD orthologs. (C) Plantago genomes have DIV-like (DIVL) paralogs (bold) but lack DIV orthologs. Maximum-likelihood bootstrap values >70% (Left) and Bayesian posterior probabilities >90% (Right) are shown. Asterisks denote support values of 100%.
Sequencing of multiple clones, in combination with phylogenetic analyses, revealed single RAD and DIV gene orthologs in A. majus, D. purpurea, V. serpyllifolia, and A. abietina (Fig. 2 B and C). Again, comparison of maximum-likelihood models found no evidence of positive or relaxed selection on branches leading to the A. abietina genes (Table S1). By contrast, despite the isolation of more distantly related paralogs in Plantago (PmRAD4 and PlDIVL; Fig. 2 B and C), no RAD or DIV gene orthologs were found in inflorescence cDNA pools of P. major or P. lanceolata (Fig. 2 B and C).
To further verify that the genomes of all Plantago species lack A-clade CYC, RAD, and DIV orthologs, and to test whether symmetry gene copy number is consistent across genera, highly conserved CYC, RAD, and DIV primers were designed (Table S2). These primers amplified A-clade CYC and B-clade CYC, RAD, and DIV fragments from genomic DNA of A. majus, D. purpurea, W. carinthiaca, and Veronica and Aragoa species (Figs. 1 and 2; Fig. S2). However, only B-clade CYC genes were amplified from genomic DNA of representative Plantago species (Figs. 1 and 2; Fig. S2), confirming the loss of A-clade CYC, RAD, and DIV before the diversification of Plantago.
PmTCP1 Expression Is Not Restricted to the Dorsal Domain.
To test predictions that CYC-like and RAD-like genes are expressed dorsally throughout D. purpurea, but not P. major, flower development, quantitative (q)RT-PCR and in situ hybridization were carried out (Figs. 3 and 4). Unfortunately, because Aragoa is endemic to the páramo of Colombia and Venezuela, similar material was not available for determining gene expression patterns in this genus.
Fig. 3.
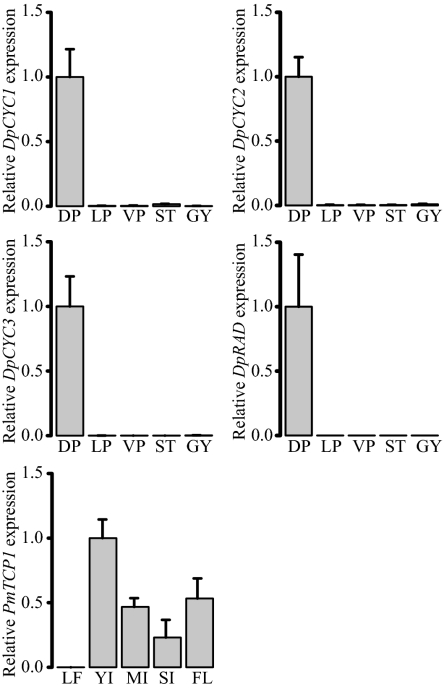
Quantitative RT-PCR analyses showing relative symmetry gene expression levels in different tissues of Digitalis purpurea and Plantago major. Expression levels are relative to EF1alpha (D. purpurea) or UBQ-5 (P. major), with SDs for four technical replicates. DP, dorsal petals; FL, flowers from proximal half of the SI (see below) inflorescence; GY, gynoecia; LF, leaf; LP, lateral petals; MI, midstage (11 mm long) inflorescence; SI, proximal half of young inflorescence with flowers partially removed; ST, stamens; VP, ventral petal; YI, young (7 mm long) inflorescence.
Fig. 4.
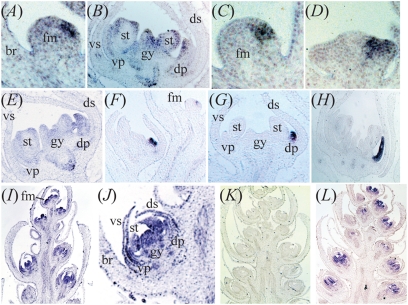
In situ hybridization analyses showing spatiotemporal patterns of symmetry genes in Digitalis purpurea and Plantago major. DpCYC1 (A and B), DpCYC3 (C–E), and DpRAD (F–H) are expressed asymmetrically in floral meristems and flowers, whereas PmTCP1 (I and J) is expressed symmetrically in floral meristems and flowers. (K) No staining is detectable in P. major inflorescences probed with the PmTCP1 sense probe. (L) PmHistone4 is strongly expressed in actively dividing cells of P. major flowers. br, bract; dp, dorsal petal; ds, dorsal sepal; fm, floral meristem; gy, gynoecium; st, stamen; vp, ventral petal; vs, ventral sepal.
Quantitative RT-PCR analyses on dissected D. purpurea flowers revealed that the transcript levels for all three CYC-like genes, and the single RAD-like gene, were at least 62-fold higher in dorsal petals than in lateral petals, ventral petals, stamens, and gynoecia (Fig. 3). Thus, unlike the B-clade V. serpyllifolia ortholog VsCYC2, which is expressed across dorsal, lateral, and ventral petals (34), petal expression of the B-clade genes DpCYC2 and DpCYC3 was characteristic of CYC-like genes from other species with bilaterally symmetrical flowers (13, 34, 40–48).
For in situ hybridization in D. purpurea, one recent duplicate in the B clade (DpCYC3), DpCYC1, and DpRAD were selected for further analysis (Fig. 2 A and B). Similar to qRT-PCR analyses, mRNA of all three genes was most strongly detectable in the dorsal region of the floral meristem and in later-stage flowers (Fig. 4 A–H). At later stages of flower development, during petal fusion, all three genes were expressed most strongly in the dorsal petals, with a lower level of expression in stamens, gynoecia, and lateroventral petals for the CYC-like genes (Fig. 4 B, E, and H). Staining was undetectable on sections hybridized with sense control probes for each gene.
Flowers of P. major cannot be easily dissected due to their small size. Instead, cDNA from differently staged inflorescences and vegetative tissues was prepared for qRT-PCR. PmTCP1 expression was highest in inflorescence tissues, with no detectable expression in leaves (Fig. 3). Within inflorescence tissues, PmTCP1 expression was over 1.5-fold higher in young (7 mm) versus midstage (11 mm) whole inflorescences. Furthermore, expression was over 2-fold higher in flowers separated from the proximal half of a young inflorescence compared with the proximal half of the same young inflorescence with most of its flowers removed (Fig. 3).
Similar to the qRT-PCR data, in situ hybridization revealed expression of PmTCP1 in P. major inflorescences (Fig. 4 I and J). In early development, PmTCP1 was expressed throughout floral meristems, and in mid- to late-stage development, PmTCP1 expression was evident in all organs of the flower, in the dorsal, lateral, and ventral domains (Fig. 4 I and J). Expression throughout the flowers was similar to that of Histone4, a marker of cell proliferation (Fig. 4L), and appeared to be specific, based on the lack of staining in sections probed with a sense control (Fig. 4K). The wide expression pattern of PmTCP1 across the flower was similar to that previously found for its V. serpyllifolia ortholog, VsCYC2, but different from the dorsal expression patterns of orthologs DpCYC2 and DpCYC3 and that reported for P. lanceolata (27, 34).
Discussion
Our findings support the hypothesis that radially symmetrical flowers in the Aragoa/Plantago lineage evolved gradually through gene duplication, expression diversification, and degeneration in the symmetry developmental genetic pathway (21, 27, 34, 35) (this study) (Fig. 5). Specifically, we have shown that at least two species of Aragoa have maintained apparently functional copies of both dorsal (CYC and RAD) and ventral (DIV) identity genes, which may be important for the development of its attractive corolla (Fig. 1). By contrast, degeneration of both the dorsal and ventral identity programs in six representative species of Plantago implicates the lateral floral organ identity program in the development of all four diminutive petals (Fig. 1), the proposed default pathway in A. majus (13, 31). Given all available evidence, we hypothesize a single origin of floral radial symmetry in the ancestor of Aragoa/Plantago due to the expansion of gene expression in the CYC-like A and B clades, followed by further disintegration of the symmetry gene pathway, including the loss of a dorsally expressed A-clade CYC paralog, as well as RAD and DIV orthologs, resulting in the wind-pollination syndrome of Plantago species (Fig. 5).
Fig. 5.
Hypothesis of floral symmetry evolution based on phylogenetic analyses and expression of CYC-, RAD-, and DIV-like genes. The ancestral CYC-like gene at the base of tribes Antirrhineae, Digitalideae, and Veroniceae was probably expressed asymmetrically on the dorsal side of the floral meristem and flower. The ancestral CYC-like gene was maintained through speciation events and duplicated independently at the base of the Antirrhineae (represented by Antirrhinum), and in the ancestor of Digitalideae + Veroniceae (represented by Digitalis, Veronica, Aragoa, and Plantago). At the base of Veroniceae, regulatory evolution occurred within the B clade, from dorsally restricted expression to expression across the flower; expression data from Veronica suggest that the Veronica B-clade CYC gene product no longer regulates RAD expression (34). Because both Aragoa and Plantago flowers are radially symmetrical and there is no evidence of relaxed selection on AaCYC1 (or on AaRAD or AaDIV), it is hypothesized that there was an expansion of the A-clade CYC-like gene in the ancestor of these genera (shaded flower on branch). In the Plantago lineage, the A-clade CYC-like gene is lost (dotted line). Loss of the A-clade CYC-like gene coincides with loss of downstream genes RAD and DIV, and with the evolutionary change from a showy to a nonpigmented, diminutive corolla, and from insect to wind pollination. Expression patterns illustrated in blue are based on empirical data (13, 31, 34); patterns illustrated in red are hypothesized as described above.
Gradual disintegration of the floral symmetry pathway in the Aragoa/Plantago lineage is supported by two lines of evidence: phylogenetic and gene expression. Phylogenetic analyses strongly support a CYC-like gene duplication event at the base of the tribes Digitalideae + Veroniceae, followed by a more recent duplication event within Digitalideae (27, 33, 35) (this study). Both paralogs generated from the first duplication event were maintained in D. purpurea, W. carinthiaca, and numerous species of Veronica and Aragoa. However, independent studies demonstrate the loss of the A-clade CYC gene along the lineage leading to Plantago, represented by distantly related P. major, P. coronopus, P. uniflora, P. lanceolata, P. lagopus, and P. sempervirens (27, 35) (this study) (dotted line in Fig. 5).
Gene expression analyses of A-clade CYC genes from D. purpurea and V. serpyllifolia, B-clade CYC genes from D. purpurea, and CYC and DICH from A. majus suggest that the ancestral Digitalideae + Veroniceae CYC-like gene was expressed dorsally within the flower (13, 31, 34) (this study) (Fig. 5). This is similar to expression patterns of other CYC-like genes within the Lamiales and, in combination with the observed dorsal coexpression of DpRAD and VsRAD, is consistent with an ancestral function in dorsal petal identity specification (13, 31, 34, 40, 44–46). By contrast, expression of V. serpyllifolia and P. major B-clade CYC-like genes is not restricted to the dorsal domain of the flower (34) (this study), and our data suggest that this condition is derived relative to dorsally restricted expression of closely related CYC-like genes. Both VsCYC2 and PmTCP1 are expressed ubiquitously across floral meristems and petals; expression varies within the stamen whorl. These data demonstrate an expansion of the B-clade CYC-like gene expression domain within Veroniceae, suggesting a loss (nonfunctionalization) or change (neofunctionalization) of developmental function following gene duplication (Fig. 5). Our working hypothesis is that the Veroniceae B-clade CYC-like gene has acquired a novel developmental function in regulating flower size, correlating with a significant reduction of flower size in this lineage.
Codon models suggest that the A. abietina floral symmetry genes are functionally constrained, implicating expression changes in the evolution of floral radial symmetry. Based on evidence from VsCYC2 and PmTCP1, we infer that AaCYC2 is also expressed throughout the flower (34) (this study). Furthermore, given that AaRAD and AaDIV appear to be functionally constrained, we predict a similar expansion in expression of the A-clade CYC-like gene in the ancestor of Aragoa and Plantago, resulting in a single origin of floral radial symmetry. Assuming that this most parsimonious interpretation is correct, we suggest two ways in which AaRAD and AaDIV might have been maintained in the genus Aragoa. One scenario is that AaRAD no longer negatively regulates AaDIV, and therefore all four symmetry genes contribute to development of the entire corolla whorl. Alternatively, AaRAD and AaDIV may have evolved novel functions in flower development. Regardless of which hypothesis is correct, our results are similar to those from previous studies showing expansion or contraction of CYC-like gene expression correlated with independent shifts from bilateral to radial symmetry in the showy corollas of Cadia purpurea (Leguminoseae), Bournea leiophylla (Gesneriaceae), and disk flowers of Gerbera hybrida (40, 42, 49).
The final step in the transition from insect to wind pollination in the Plantago genus was likely due to the loss of the A-clade CYC-like gene (Fig. 5) and the correlated loss of RAD and DIV. In A. majus, RAD expression is positively regulated by CYC in the dorsal region of the flower. In turn, RAD negatively regulates the DIV protein in the dorsal and lateral regions of the flower, leading to differential petal and stamen identity along the dorsiventral axis of the flower (28–30, 50). Unlike A. majus, there is no intrawhorl organ differentiation in Plantago flowers (21). Assuming that our search for RAD and DIV orthologs was exhaustive, the absence of these genes from representative Plantago genomes indicates the loss of both the dorsal and ventral identity programs. This would implicate the same “lateral” (sensu A. majus) identity program in the development of dorsal, lateral, and ventral floral organs of Plantago, highlighting the potential importance of gene duplication and loss in the evolution of form.
Materials and Methods
Plant Material.
Seeds of Digitalis purpurea (“Candy Mountain”) and Veronica serpyllifolia were obtained from Thompson and Morgan Seed; seeds of Plantago major were obtained from the University of Washington Medicinal Herb Garden. Plants were grown in the greenhouse under standard long-day (16 h light/8 h dark) conditions. Fresh material of P. lanceolata was collected from wild populations in Lawrence, KS. DNA of Aragoa abietina (reference no. 12226), A. cundinamarcensis (reference no. 11177), P. coronopus (reference no. 2763), P. lagopus (reference no. 9433), P. sempervirens (reference no. 9426), P. (formerly Littorella) uniflora (reference no. 2798), V. chamaedrys (reference no. 7419), V. intercedens (reference no. 9211), and W. carinthiaca (reference no. 8940) was obtained from Kew Gardens DNA Bank.
Isolation of Nuclear Markers.
DEL, ACT, and the DIV intron were amplified from genomic DNA using the primers MYCF1/R2, ACT1/2, and DIV-55-F/403-R, respectively, based on their phylogenetic utility in other plant groups (34, 37–39) (Table S2). Although ACT genes are multicopy in angiosperms, ACT1/2 selectively amplified genes belonging to the REP clade, with closest homology to the Solanum tuberosum (potato, Solanaceae) genes Ac46, Ac71, and Ac75 (51). PCR bands were individually gel-extracted (Qiagen) and cloned into the pGEM-T vector (Invitrogen). At least four clones were sampled per PCR band for sequencing on both strands.
Floral Symmetry Gene Isolation.
Multiple CYC-like clones were sequenced from inflorescence cDNA synthesized from total mRNA or genomic DNA, following amplification with a set of degenerate forward primers (CYCF1, CYC73aaF, CYC73aF, and CYC73bF) in combination with the reverse primers CYC-R2 and/or poly-TQT (34, 52). These primers have been used previously to successfully amplify CYC-like genes from across Plantaginaceae (34, 52). To isolate RAD and DIV orthologs, the degenerate forward primers RAD.deg.F, RAD.deg (2).F, DIV.deg.F, and DIV.deg (2).F were used in combination with the reverse primer poly-TQT, and multiple clones were sequenced as previously described (Table S2) (34). To verify the presence/absence of CYC, RAD, and DIV genes, conserved primers were designed based on aligned A. majus and V. serpyllifolia sequences. The primer pairs Arag.CYC.A.F/R, Arag.CYC.B.F/R, RAD-70-F/240-R, and DIV-55-F/403-R were used to amplify CYC A, CYC B, RAD, and DIV orthologs, respectively, from genomic DNA (Table S2).
Phylogenetic Analysis.
DEL, ACT, ITS, and floral symmetry genes were separately aligned using MAFFT, with readjustment by eye in MacClade (53–55). Phylogenetic relationships were estimated using maximum-likelihood methods in GARLI 0.951 and Bayesian methods in MrBayes 3.1.2 (56, 57). For both types of analysis, MrModeltest 2.3 was used to estimate the best model of molecular evolution (GTR + I + Γ for the DIV intron, ACT, and ITS; HKY + I + Γ for CYC, DIV, and DEL; SYM I + Γ for RAD) (58). Maximum-likelihood analyses were run using 10 random addition sequences, and Bayesian analyses were run twice for 1 million generations, sampling every 1,000 generations. LB values were obtained using 200 replicates in GARLI. Combined analyses were partitioned in GARLI-Part-0.97 and MrBayes based on different model selection by MrModeltest.
Tests for Selection.
To test for positive or relaxed selection along specific branches of the symmetry gene trees, maximum-likelihood branch (one- and two-ratio) and branch-site (M1a, Null, and model A) models were run on an alignment of the longest sequences from A. majus, D. purpurea, V. serpyllifolia, P. major, P. lanceolata, and A. abietina using the codeml program in PAML Version 4.3 (59). Foreground branches were specified as those ancestral to clades containing AaCYC1 only, PmTCP1, PlTCP1, and AaCYC2 together, PmTCP1 and PlTCP1 together, AaRAD only, and AaDIV only. Nested models were compared against a χ2 distribution of trees using a likelihood ratio test. To test for elevated ω (synonymous-to-nonsynonymous site ratio) on the foreground branch(es), the one- and two-ratio models were compared (60). To incorporate heterogeneity of parameters over both sites and branches, M1a was compared with model A and, when significant at the 0.05 level, null model A was compared with model A (61).
Quantitative RT-PCR.
Total RNA was extracted from vegetative tissues and dissected floral tissues of D. purpurea and P. major using TriReagent (Ambion) according to the manufacturer's instructions. Extracted RNA was treated with rDNaseI (Ambion), and cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was carried out as previously described (62). The gene-specific primers pairs DpCYC1qrtF/R, DpCYC2qrtF/R, DpCYC3qrtF/R, PmTCP1qrtF/R, and DpRADqrtF/R were designed in Primer3 (63) (Table S2). EF1alpha and UBQ-5 showed little transcriptional variation across different tissues, and were therefore selected as the reference genes. Cycle threshold values were corrected for transcriptional stability and normalized against EF1alpha (D. purpurea) or UBQ-5 (P. major). For each gene, the mean and SD were determined for four technical replicates.
In Situ Hybridization.
Inflorescences of D. purpurea and P. major were fixed in formaldehyde-acetic acid-alcohol (FAA), dehydrated in an alcohol series, and embedded in paraffin wax blocks. Antisense and sense gene-specific probes of DpCYC1, DpCYC3, DpRAD, and PmTCP1 spanning the C-terminal end of the coding regions and the 3′-UTRs were generated using T7 and T3 RNA Taq polymerase (Roche) according to the manufacturer's instructions. As an experimental control, an antisense probe was generated for PmHistone4 as described previously (34). In situ hybridization was performed as in refs. 64 and 65.
Supplementary Material
Acknowledgments
We thank Felipe Zapata for help with phylogenetic analyses and two anonymous reviewers for comments on an early version of the manuscript. DNA material was used with the permission of the Board of Trustees of the Royal Botanic Gardens, Kew. C.C.M. was supported by National Institute of General Medical Science Grant 5R25GM078441 through the Post-Baccalaureate Research Education Program at the University of Kansas. This work was supported by National Science Foundation Grant IOS-0616025 (to L.C.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ853598–HQ853667.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011361108/-/DCSupplemental.
References
- 1.Donoghue MJ, Ree RH, Baum DA. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 1998;3:311–317. [Google Scholar]
- 2.Endress PK. Symmetry in flowers: Diversity and evolution. Int J Plant Sci. 1999;160(Suppl 6):S3–S23. doi: 10.1086/314211. [DOI] [PubMed] [Google Scholar]
- 3.Knapp S. On ‘various contrivances’: Pollination, phylogeny and flower form in the Solanaceae. Philos Trans R Soc Lond B Biol Sci. 2010;365:449–460. doi: 10.1098/rstb.2009.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ree RH, Donoghue MJ. Inferring rates of change in flower symmetry in asterid angiosperms. Syst Biol. 1999;48:633–641. [Google Scholar]
- 5.Sargent RD. Floral symmetry affects speciation rates in angiosperms. Proc Biol Sci. 2004;271:603–608. doi: 10.1098/rspb.2003.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vamosi JC, Vamosi SM. Key innovations within a geographical context in flowering plants: Towards resolving Darwin's abominable mystery. Ecol Lett. 2010;13:1270–1279. doi: 10.1111/j.1461-0248.2010.01521.x. [DOI] [PubMed] [Google Scholar]
- 7.Olmstead RG, et al. Disintegration of the Scrophulariaceae. Am J Bot. 2001;88:348–361. [PubMed] [Google Scholar]
- 8.Reeves PA, Olmstead RG. Evolution of novel morphological and reproductive traits in a clade containing Antirrhinum majus (Scrophulariaceae) Am J Bot. 1998;85:1047–1056. [PubMed] [Google Scholar]
- 9.Arber A. Studies on flower structure. I. On a peloria of Digitalis purpurea L. Ann Bot. 1932;46:929–939. [Google Scholar]
- 10.Coen ES, et al. Evolution of floral symmetry. Philos Trans R Soc Lond B Biol Sci. 1995;350:35–38. [Google Scholar]
- 11.Endress PK. Evolution of floral symmetry. Curr Opin Plant Biol. 2001;4:86–91. doi: 10.1016/s1369-5266(00)00140-0. [DOI] [PubMed] [Google Scholar]
- 12.Kampny CM, Dengler NG. Evolution of flower shape in Veroniceae (Scrophulariaceae) Plant Syst Evol. 1997;205:1–25. [Google Scholar]
- 13.Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 14.Albach DC, Meudt HM. Phylogeny of Veronica in the Southern and Northern Hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Mol Phylogenet Evol. 2010;54:457–471. doi: 10.1016/j.ympev.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 15.Bello MA, Chase MW, Olmstead RG, Ronsted N, Albach D. The páramo endemic Aragoa is the sister genus of Plantago (Plantaginaceae; Lamiales): Evidence from plastid rbcL and nuclear ribosomal ITS sequence data. Kew Bull. 2002;57:585–597. [Google Scholar]
- 16.Cho Y, Mower JP, Qiu Y-L, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci USA. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahn K. A phylogenetic study of Plantaginaceae. Bot J Linn Soc. 1996;120:145–198. [Google Scholar]
- 18.Ronsted N, Chase MW, Albach DC, Bello MA. Phylogenetic relationships within Plantago (Plantaginaceae): Evidence from nuclear ribosomal ITS and plastid trnL-F sequence data. Bot J Linn Soc. 2002;139:323–338. [Google Scholar]
- 19.Tay ML, Meudt HM, Garnock-Jones PJ, Ritchie PA. DNA sequences from three genomes reveal multiple long-distance dispersals and non-monophyly of sections in Australasian Plantago (Plantaginaceae) Aust J Bot. 2010;23:47–68. [Google Scholar]
- 20.Albach DC, Meudt HM, Oxelman B. Piecing together the “new” Plantaginaceae. Am J Bot. 2005;92:297–315. doi: 10.3732/ajb.92.2.297. [DOI] [PubMed] [Google Scholar]
- 21.Bello MA, Rudall PJ, González F, Fernández-Alonso JL. Floral morphology and development in Aragoa (Plantaginaceae) and related members of the order Lamiales. Int J Plant Sci. 2004;165:723–738. [Google Scholar]
- 22.Oxelman B, Backlund M, Bremer B. Relationships of the Buddlejaceae s. l. investigated using parsimony jackknife and branch support analysis of chloroplast ndhF and rbcL sequence data. Syst Bot. 1999;24:164–182. [Google Scholar]
- 23.Grime JP, Hodgson JG, Hunt R. Comparative Plant Ecology. London: Unwin Hyman; 1988. [Google Scholar]
- 24.Pennell FW. The Scrophulariaceae of Eastern Temperate North America. Philadelphia: Acad Nat Sci; 1935. Monograph 1. [Google Scholar]
- 25.Kampny CM, Dickinson TA, Dengler NG. Quantitative comparison of floral development in Veronica chamaedrys and Veronicastrum virginicum (Scrophulariaceae) Am J Bot. 1993;80:449–460. [Google Scholar]
- 26.Hong D-Y. Taxonomy and evolution of the Veroniceae (Scrophulariaceae) with special reference to palynology. Opera Bot. 1984;75:1–60. [Google Scholar]
- 27.Reardon W, Fitzpatrick DA, Fares MA, Nugent JM. Evolution of flower shape in Plantago lanceolata. Plant Mol Biol. 2009;71:241–250. doi: 10.1007/s11103-009-9520-z. [DOI] [PubMed] [Google Scholar]
- 28.Almeida J, Rocheta M, Galego L. Genetic control of flower shape in Antirrhinum majus. Development. 1997;124:1387–1392. doi: 10.1242/dev.124.7.1387. [DOI] [PubMed] [Google Scholar]
- 29.Corley SB, Carpenter R, Copsey L, Coen E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc Natl Acad Sci USA. 2005;102:5068–5073. doi: 10.1073/pnas.0501340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galego L, Almeida J. Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 2002;16:880–891. doi: 10.1101/gad.221002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo D, et al. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/s0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- 32.Preston JC, Hileman LC. Developmental genetics of floral symmetry evolution. Trends Plant Sci. 2009;14:147–154. doi: 10.1016/j.tplants.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Hileman LC, Baum DA. Why do paralogs persist? Molecular evolution of CYCLOIDEA and related floral symmetry genes in Antirrhineae (Veronicaceae) Mol Biol Evol. 2003;20:591–600. doi: 10.1093/molbev/msg063. [DOI] [PubMed] [Google Scholar]
- 34.Preston JC, Kost MA, Hileman LC. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol. 2009;182:751–762. doi: 10.1111/j.1469-8137.2009.02794.x. [DOI] [PubMed] [Google Scholar]
- 35.Reeves PA, Olmstead RG. Evolution of the TCP gene family in Asteridae: Cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol Biol Evol. 2003;20:1997–2009. doi: 10.1093/molbev/msg211. [DOI] [PubMed] [Google Scholar]
- 36.Albach DC, Chase MW. Paraphyly of Veronica (Veroniceae; Scrophulariaceae): Evidence from the internal transcribed spacer (ITS) sequences of nuclear ribosomal DNA. J Plant Res. 2001;114:9–18. [Google Scholar]
- 37.Fan C, Purugganan MD, Thomas DT, Wiegmann BM, Xiang JQ. Heterogeneous evolution of the myc-like anthocyanin regulatory gene and its phylogenetic utility in Cornus L. (Cornaceae) Mol Phylogenet Evol. 2004;33:580–594. doi: 10.1016/j.ympev.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Howarth DG, Donoghue MJ. Duplications and expression of DIVARICATA-like genes in Dipsacales. Mol Biol Evol. 2009;26:1245–1258. doi: 10.1093/molbev/msp051. [DOI] [PubMed] [Google Scholar]
- 39.Keeling PJ. Foraminifera and Cercozoa are related in actin phylogeny: Two orphans find a home? Mol Biol Evol. 2001;18:1551–1557. doi: 10.1093/oxfordjournals.molbev.a003941. [DOI] [PubMed] [Google Scholar]
- 40.Broholm SK, et al. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busch A, Zachgo S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci USA. 2007;104:16714–16719. doi: 10.1073/pnas.0705338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Citerne HL, Pennington RT, Cronk QCB. An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci USA. 2006;103:12017–12020. doi: 10.1073/pnas.0600986103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng X, et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA. 2006;103:4970–4975. doi: 10.1073/pnas.0600681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Q, Tao JH, Yan D, Wang YZ, Li ZY. Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae) Dev Genes Evol. 2008;218:341–351. doi: 10.1007/s00427-008-0227-y. [DOI] [PubMed] [Google Scholar]
- 45.Hileman LC, Kramer EM, Baum DA. Differential regulation of symmetry genes and the evolution of floral morphologies. Proc Natl Acad Sci USA. 2003;100:12814–12819. doi: 10.1073/pnas.1835725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song CF, Lin QB, Liang RH, Wang YZ. Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae) BMC Evol Biol. 2009;9:244. doi: 10.1186/1471-2148-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, et al. Genetic control of floral zygomorphy in pea (Pisum sativum L.) Proc Natl Acad Sci USA. 2008;105:10414–10419. doi: 10.1073/pnas.0803291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Kramer EM, Davis CC. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc Natl Acad Sci USA. 2010;107:6388–6393. doi: 10.1073/pnas.0910155107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X-R, Wang Y-Z, Smith JF, Chen R. Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae) New Phytol. 2008;178:532–543. doi: 10.1111/j.1469-8137.2008.02384.x. [DOI] [PubMed] [Google Scholar]
- 50.Costa MMR, Fox S, Hanna AI, Baxter C, Coen E. Evolution of regulatory interactions controlling floral asymmetry. Development. 2005;132:5093–5101. doi: 10.1242/dev.02085. [DOI] [PubMed] [Google Scholar]
- 51.An SS, Möpps B, Weber K, Bhattacharya D. The origin and evolution of green algal and plant actins. Mol Biol Evol. 1999;16:275–285. doi: 10.1093/oxfordjournals.molbev.a026109. [DOI] [PubMed] [Google Scholar]
- 52.Howarth DG, Donoghue MJ. Duplications in CYC-like genes from Dipsacales correlate with floral form. Int J Plant Sci. 2005;166:357–370. [Google Scholar]
- 53.Katoh T, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddison DR, Maddison WP. MacClade 4: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 2003. Version 4.0.6. [DOI] [PubMed] [Google Scholar]
- 56.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 57.Zwickl D. Austin, TX: University of Texas at Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis. [Google Scholar]
- 58.Nylander JAA. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest. Version 2. [Google Scholar]
- 59.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 60.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 62.Preston JC, Hileman LC. SQUAMOSA-PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J. 2010;62:704–712. doi: 10.1111/j.1365-313X.2010.04184.x. [DOI] [PubMed] [Google Scholar]
- 63.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 64.Jackson D. In situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPherson M, editors. Molecular Plant Pathology: A Practical Approach. Oxford: Oxford Univ Press; 1991. pp. 163–174. [Google Scholar]
- 65.Preston JC, Kellogg EA. Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: Evidence from gene expression analyses. Plant J. 2007;52:69–81. doi: 10.1111/j.1365-313X.2007.03209.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.