Abstract
Lassa fever virus, a member of the family Arenaviridae, is a highly endemic category A pathogen that causes 300,000–500,000 infections per year in Western Africa. The arenaviral nucleoprotein NP has been implicated in suppression of the host innate immune system, but the mechanism by which this occurs has remained elusive. Here we present the crystal structure at 1.5 Å of the immunosuppressive C-terminal portion of Lassa virus NP and illustrate that, unexpectedly, its 3D fold closely mimics that of the DEDDh family of exonucleases. Accompanying biochemical experiments illustrate that NP indeed has a previously unknown, bona fide exonuclease activity, with strict specificity for double-stranded RNA substrates. We further demonstrate that this exonuclease activity is essential for the ability of NP to suppress translocation of IFN regulatory factor 3 and block activation of the innate immune system. Thus, the nucleoprotein is a viral exonuclease with anti-immune activity, and this work provides a unique opportunity to combat arenaviral infections.
Keywords: immunology, structural biology, virology, arenavirus
Lassa fever virus (LASV) causes severe hemorrhagic fever and is highly endemic in Western Africa, causing 300,000–500,000 infections per year (1). In its most severe form, LASV infection is marked by severe immunosuppression and a rapidly progressive febrile illness, culminating in a septic shock-like syndrome and multisystem failure. It is also the hemorrhagic fever most frequently transported out of Africa to the United States and Europe (2, 3). Development of specific treatments and therapeutics is a high priority of public health and biodefense efforts.
LASV belongs to the family Arenaviridae. The arenaviruses have a world-wide distribution and include other significant human pathogens, such as the hemorrhagic fever viruses Machupo, Junin, and Lujo (1, 4) as well as the widely studied lymphocytic choriomeningitis virus (LCMV), a reemerging obstetric pathogen (5). All arenaviruses have a bisegmented single-stranded RNA (ssRNA) genome with a unique ambisense coding strategy that produces just four known proteins: a nucleoprotein NP, a matrix protein Z, a polymerase L, and a glycoprotein GP (1). Of these, NP is the most abundant in an infected cell. NP associates with L to form the ribonucleoprotein (RNP) core for RNA replication and transcription (6) and with the matrix protein Z for viral assembly (7–10).
A key hallmark of virus infection is the presence of double-stranded RNA (dsRNA) in the infected cell. Sensing of dsRNA by cellular immune sentry proteins, such as retinoic acid-inducible I (RIG-I) (11) and melanoma differentiation-associated 5 (MDA-5) (12), initiates signaling pathways that result in the translocation of IFN regulatory factor 3 (IRF-3) to the nucleus. Once in the nucleus, IRF-3 activates expression of IFN-α/β, which initiates the antiviral response in the infected cells and primes neighboring cells for a rapid response to viral invasion. The NP proteins of LASV, Junin, Machupo, and LCMV have all been implicated in suppression of the innate IFN response, resulting in the inhibition of nuclear translocation of IRF-3 (13). This inhibition can result in unchecked viral replication, failure to initiate an adaptive immune response, and increased morbidity and mortality (14–19). Mutagenesis throughout LCMV NP, before the availability of a structure, identified key residues within the C-terminal half of NP critical for immunosuppressive function (20, 21). However, the role of these residues, and the specific mechanism by which arenavirus NPs cause immunosuppression, remained unknown.
Results
Structure of LASV NPΔ340.
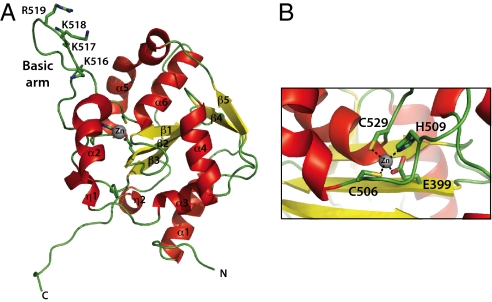
To gain insight into the mechanism by which NP suppresses the innate immune system, we determined the crystal structure of the C-terminal half of LASV NP (residues 341–569, termed NPΔ340) to 1.5 Å by SIRAS (22) using the inherent zinc atom of NP and eight introduced selenium atoms from selenomethionine incorporation for phasing. NPΔ340 consists of a mixed, five-stranded β-sheet with one antiparallel strand (β2) and six α-helices connected by a series of loops, with one particularly long loop between α5 and α6 forming a basic “arm” off one side (Fig. 1A). The α-helices surround the β-sheet and are grouped into three sets of one (α2), two (α5, α6), and three (α1, α3, α4) helices each. The zinc atom is coordinated by E399, C506, H509, and C529, which are located at the bases of β2, the protruding basic arm, and α6, respectively (Fig. 1B). The first 20 residues of the construct are disordered and are contained within a likely flexible linker between the N- and C-terminal domains of NP.
Fig. 1.
Structure of the C-terminal, immunosuppressive domain of LASV NP. (A) Cartoon representation of LASV NPΔ340. The basic arm includes residues Lys516, Lys517, Lys518, and Arg519. (B) A single zinc is coordinated by Glu399, His506, Cys509, and Cys529.
Similarity to DEDDh Exonucleases.
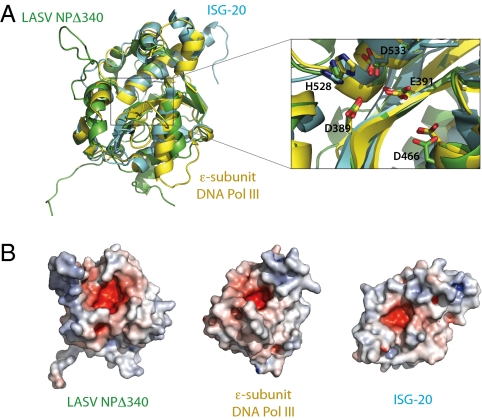
A BLAST (23) search indicates no similarity in primary amino acid sequence for LASV NP outside the arenavirus family. However, an HHpred search, which searches alignment databases rather than sequence databases (24), and a DALI (25) search for 3D folds similar to that of LASV NP surprisingly return numerous hits, all of which are members of the DEDD superfamily of exonucleases. The DEDD superfamily contains strictly conserved Asp-Glu-Asp-Asp catalytic residues in the active site and can further be subdivided into DEDDh or DEDDy groups based on the presence of a histidine or tyrosine, respectively, in close apposition to the DEDD core (26). Alignment of NPΔ340 with members of this family demonstrates that the 3D arrangement of their secondary structure elements is strikingly similar, with an average rmsd between LASV NP and known exonucleases of ∼3 Å (Fig. 2A). Furthermore, both main-chain and side-chain positions of D389, E391, D533, and H528 of LASV NP align with D1, E2, D4, and h in active sites of known DEDDh enzymes, such as IFN-stimulated gene 20 (ISG-20) (27) and the ε subunit of Escherichia coli DNA polymerase III (DNA pol IIIε) (28). D466 of LASV NP, which corresponds to D3 of the DEDDh active site, resides on an α-helix that is shifted slightly from the D3 of ISG-20 and DNA pol IIIε but adopts an alternate rotamer in LASV NP such that its carboxylate oxygens approach the equivalent positions in other exonucleases (Fig. 2A Inset).
Fig. 2.
Superimposition of LASV NP and known DEDD exonucleases. (A) Structural comparison of NPΔ340 and two known DEDDh exonucleases. NPΔ340 is colored green, ISG-20 (PDB ID 1WLJ; ref. 27) is colored cyan, and the E. coli DNA pol IIIε (PDB ID 2GUI; ref. 28) is colored yellow. Inset shows a close-up view of the superimposed DEDDh residues of the active site. Numbered residues reflect those of LASV NP. (B) Electrostatic surface potential calculated with APBS (the Adaptive Poisson-Boltzmann Solver) software (49) shows that each exonuclease has an acidic active site and highlights the basic arm of LASV NP. Positive surface is colored blue; negative surface is colored red with limits ± 10 kT/e.
The DEDD-containing active sites of known exonucleases form a negatively charged cavity. Similarly, D389, E391, D466, D533, and H528 of LASV NP also are located in a negatively charged cavity (Fig. 2B). At the entrance to this cavity is a positively charged arm formed by the long loop that connects α5 to α6.
Exoribonucleases in the DEDD superfamily share a common catalytic mechanism that depends on two metal ions (29). Known exonucleases have been crystallized in complex with divalent ions as well as mononucleotides, and, in these structures, the ions coordinate to the DEDD residues (27, 28, 30, 31). Our initial crystal structure, crystallized in the absence of added divalent ions and nucleotides, shows positive Fo − Fc density in the active site (Fig. S1 A1 and A2). A subsequent structure, in which NPΔ340 was cocrystallized with 5 mM MnCl2 and 5 mM UMP (Table S1) demonstrates the presence of a single, partially occupied Mn2+ ion in the active site coordinated to D389, E391, and D533 (Fig. S1 B1 and B2). We do not observe density for a second Mn2+ or the UMP, indicating that NPΔ340 may not bind mononucleotides in isolation and that coordination of the second ion may require the presence of nucleic acid oligomers. A similar phenomenon is observed with RNase D, ExoI, and E. coli DNA pol I, in which only one of the two active site ions is bound in the absence of a nucleotide substrate (26, 32, 33).
LASV NPΔ340 Has Exonuclease Activity.
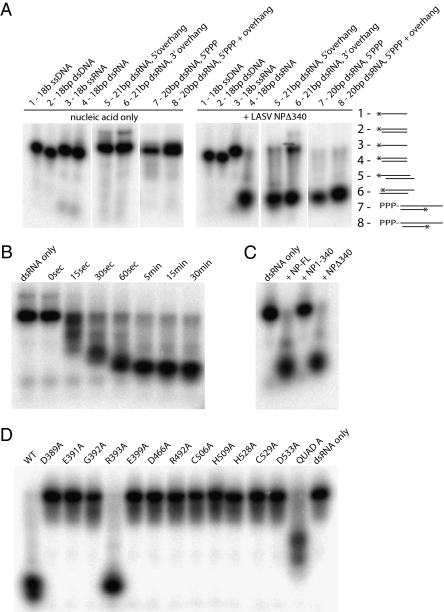
LASV NP appears structurally to be an exonuclease. To determine whether NPΔ340 indeed has this enzymatic activity, NPΔ340 was tested in exonuclease assays. First, to gain insight into the presence, specificity, and directionality of the exonuclease activity, a range of nucleic acid substrates were incubated with NPΔ340 and then analyzed by denaturing PAGE and autoradiography. NPΔ340 readily hydrolyzes an 18-bp dsRNA oligonucleotide to less than 6 bases but has no effect on ssRNA, ssDNA, or dsDNA (Fig. 3A and Fig. S2). Subsequent experiments further elucidate NPΔ340 specificity for different dsRNA substrates. dsRNA with a 3-nt overhang on either the 5′ or the 3′ end is digested to approximately the same size as blunt-ended dsRNA. However, the 3′ overhang somewhat reduces the efficiency of the reaction. dsRNA containing a 5′ triphosphate is also digested regardless of whether the ends are blunt or contain a single 5′ nucleotide overhang (34). Lastly, in a time-course experiment, we found that a 5′-labeled, blunt-ended 18-bp dsRNA substrate was hydrolyzed to increasingly smaller products that still carried the 5′ label, leading us to conclude that NPΔ340 operates in a 3′ to 5′ direction (Fig. 3B). In summary, NPΔ340 specifically digests dsRNA in a 3′ to 5′ direction and can process blunt-ended, overhang, or 5′ triphosphate-labeled dsRNA.
Fig. 3.
Ribonucleolytic activity of LASV NP. (A) Substrate specificity. NPΔ340 was incubated with different 32P-labeled substrates, and the reaction products were analyzed by denaturing PAGE and autoradiography. (Left) Migration patterns of each nucleic acid substrate alone (in the absence of NPΔ340 exonuclease). (Right) Migration of each substrate when incubated with NPΔ340 for 15 min. A schematic for each substrate is also shown, with the location of the 32P label indicated by an asterisk. Sequences for each substrate can be found in Table S2. (B) Time course of exonuclease activity. The 18-bp blunt-ended dsRNA is increasingly digested by wild-type NPΔ340 from 0 to 15 min. (C) Comparison of ribonucleolytic activity of the N terminus of NP (NP1–340) and full-length NP (NP-FL). The 18-bp blunt-ended dsRNA is digested by full-length NP and NPΔ340, whereas NP1–340 does not have exonuclease activity. (D) Effects of mutations to active site and proximal residues on ribonucleolytic activity. Wild-type and NPΔ340 point mutants were incubated with 18-bp blunt-ended dsRNA for 15 min, and products were analyzed by PAGE. QuadA designates a quadruple alanine mutation in residues K516K517K518R519. Note that all point mutations to residues in or near the exonuclease active site and the Zn coordination site, save R393, abrogate exonuclease activity, leaving the dsRNA undigested. The QuadA mutation to the basic arm partially diminishes exonuclease activity.
In addition, NPΔ340 demonstrates exonuclease activity equivalent to that of full-length NP (NP-FL) on the 18-bp blunt-ended dsRNA. By contrast, the N-terminal half of NP (NP1–340) has no ribonucleolytic activity in the same assay, indicating that NPΔ340 is sufficient for the exonuclease activity of the complete LASV NP (Fig. 3C). Alanine scanning mutagenesis of NPΔ340 was performed to test the catalytic and/or structural roles of conserved residues in and proximal to the active site, with each mutant incubated with blunt-ended 18-bp dsRNA (data are given in Fig. 3D, and residues are illustrated in Fig. 4). Mutations at any of the DEDDh active site residues abolish exonuclease activity. Furthermore, mutations of nearby residues and residues that coordinate the Zn atom also abolish exonuclease activity. An R393A mutation has no effect on exonuclease activity, suggesting that R393, although proximal to the active site, is not critical for enzymatic function. A four-site mutation targeting the stretch of basic residues in the α5–α6 arm (K516K517K518R519, termed “QuadA”) shows partial activity. The structure of the E. coli DNA polymerase I Klenow fragment bound to dsDNA revealed the presence of a basic arm that makes contact with the primer strand of the duplex DNA (35). Furthermore, structures of several other DEDDh exonucleases that process dsDNA or structured RNA substrates also reveal the presence of either a basic arm or a basic groove proximal to the active site (28, 31, 36). Thus, is it is possible that the side chains located in the basic arm region of LASV NP may also interact with the phosphodiester backbone of nucleic acid strands as they enter the acidic cavity (Fig. S3).
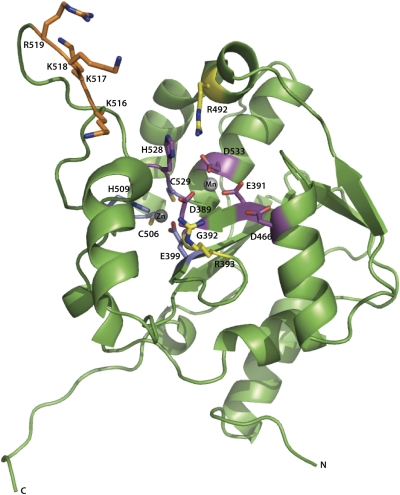
Fig. 4.
Cartoon representation of NPΔ340 and location of residues important for the exonuclease and anti-IFN activities. Residues corresponding to the DEDDh motif are colored magenta, two Arg residues proximal to the active site are colored yellow, residues involved in coordination of the Zn2+ atom are colored blue, and residues corresponding to the basic arm are colored orange.
Assessment of the metal ion requirements of NPΔ340 revealed a strict dependence on divalent cations. The protein was active in the presence of Mg2+, Mn2+, Co2+, and Zn2+ ions (Fig. S4). Similar to other DEDD exonucleases, LASV NPΔ340 was inactive in the presence of EDTA and Ca2+ and Cu2+ ions (37, 38).
Exonuclease Activity of LASV NPΔ340 Is Essential for Innate Immune Suppression.
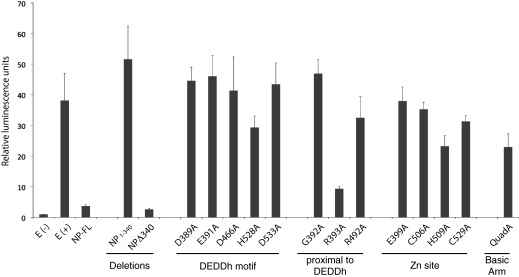
Previous studies have established that the NPs from many arenavirus species are able to suppress the innate immune system by preventing the translocation of IRF-3 into the nucleus (20, 39). In addition, residues D382 and G385 of LCMV NP as well as the equivalent residues of LASV NP (D389 and G392) have been implicated in the suppression of the innate immune system (21). To expand on these results and further assess role of individual amino acids and domains of LASV NP in counteracting the IFN-α/β response, a panel of single-residue mutations to the C-terminal region of full-length LASV NP, as well as the N-terminal and C-terminal portions of NP expressed individually (NP1–340 and NPΔ340), were tested for their ability to suppress translocation of IRF-3 into the nucleus, as measured by the expression of firefly luciferase under the control of an IRF-3–dependent promoter (Fig. 5). The ability of each mutant to suppress IRF-3 translocation mirrors its respective exonuclease activity: wild-type full-length NP and R393A NP suppress IRF-3 translocation. By contrast, NP point mutants that abrogate exonuclease activity permit IRF-3 translocation, indicating that proper exonuclease function is critical to innate immune suppression in vitro. All mutants are expressed to similar levels as wild-type NP (Fig. S5), indicating the failure to inhibit the IRF-3–dependent promoter was not caused by a defect in protein synthesis. Furthermore, the N-terminal portion of NP (NP1–340) has no effect on IRF-3 translocation, whereas NPΔ340 suppresses translocation to a similar level as that of wild-type NP, indicating NPΔ340 is necessary and sufficient for immune suppression in vitro.
Fig. 5.
Sendai virus-mediated activation of an IRF-3 promoter. Full-length NP (NP-FL), the N-terminal domain of NP (NP1–340), the C-terminal domain of NP (NPΔ340), and amino acid substitutions of full-length NP were assayed for inhibition of Sendai virus-mediated activation of an IRF-3–dependent promoter. Values displayed reflect relative luminescence units corrected to an uninfected, empty vector control. E(−) and E(+) indicate an uninfected empty vector control and an infected empty vector control, respectively. Note that the wild-type function of NP is to block IRF-3 translocation and therefore block reporter luminescence.
Discussion
Although no particular enzymatic activity was previously known for LASV NP, the 1.5-Å crystal structure presented here demonstrates that the immunosuppressive domain of LASV NP is extremely similar in 3D fold and active site residue placement to the DEDDh family of exonucleases. In addition to structurally resembling an exonuclease, NP functions like an exonuclease, with high specificity for dsRNA-specific degradation in the 3′ to 5′ direction. This exonuclease activity is likely required for the immunosuppression caused by LASV NP, as residues previously identified as essential for arenaviral anti-IFN activity are now shown to lie within the exonuclease active site (21). Arenavirus NPs are 60–85% similar in the immunosuppressive domain, and residues in the DEDDh active site are completely conserved in the virus family (Fig. S6), suggesting that this activity may be a shared feature of arenavirus NPs.
Might the exonuclease activity play a role in replication? In LCMV NP, mutations that abolish replication (I373A, W380A, C518A, I524A, and L544A) are largely distinct from those that abolish IFN suppression (D382, G385, and C518) (21). The only mutations that consistently abolish both functions are those that likely disrupt the tertiary structure of NP: C518A, which coordinates a Zn atom critical to the structure, and excisions of two or more adjacent residues contained within an α-helix or within the exonuclease active site (as indicated by the LASV NP structure). Residues in LCMV NP that showed defects in viral replication but not anti-IFN activity did not appear to map to regions in or proximal to the exonuclease active site of LASV NP. Thus, it is likely that the C-terminal domain of the arenavirus NP has separate but perhaps complementary functions, whereby the anti-IFN activity may be closely tied with the overall replicative capacity of the virus. Elimination of dsRNA generated during viral replication by the exonuclease NP would remove a key signature of viral infection and allow the virus to replicate unchecked. For example, Martínez-Sobrido et al. (21) demonstrated that mutation of D382 in LCMV NP (equivalent to LASV NP D389) had only a slight effect on viral titers when LCMV was grown in IFN-deficient cell lines but a marked effect on titers when the virus was grown in IFN-competent cell lines. Hence, mutations that affect the anti-IFN capabilities of the NP may also, consequently, affect the ability of the virus to replicate efficiently and without interference by the immune system.
Although the DEDDh motif is conserved among arenaviruses, there are a few differences elsewhere in NP between LASV and the other arenaviruses, and it is possible that the exact residues critical for exonuclease and immune suppression functions vary. For LASV NP, we find that E391 is important for blocking IRF-3, whereas R393 is not (Fig. 5). By contrast, a previous report (21) indicates that the reverse is true for LCMV. For LCMV NP, E384 (E391 equivalent) is not important for suppression of the innate immune response, whereas R386 (R393 equivalent) is important (21). Furthermore, Tacaribe virus (TACV), a nonpathogenic arenavirus, does not effectively suppress the innate immune system (13), although it is not immediately clear why because TACV shares the identical DEDDh and Zn-coordination residues. The deposited sequence for TACV NP (accession no. AAA47903) contains an Asp at position 392, which lies in the exonuclease active site, rather than a Gly as in other arenavirus nucleoproteins, suggesting that the G to D change might contribute to the decreased anti-IFN function of TACV. However, by sequencing multiple independent clones of TACV, Martínez-Sobrido et al. (21) found that all sequenced TACV NP clones actually contained Gly-392 rather than Asp-392. Thus it is unlikely that the decreased anti-IFN function of TACV is related to this particular residue. It is possible that other variations within the TACV genome contribute to its altered immunosuppression phenotype, and it is not yet clear whether TACV NP possesses the same exonuclease activity as LASV does.
Beyond LASV, only one other mammalian RNA virus is known to encode a protein with exonuclease activity. The nsp14 protein of severe acute respiratory syndrome (SARS) coronavirus is a member of the DEDDh family and therefore also has 3′ to 5′ activity (40). Nsp14 shows activity against both ssRNA and dsRNA and has been implicated in the control of RNA synthesis and genome fidelity (41). However, no IFN suppressive activity is known for nsp14. Another DEDD exonuclease, TREX1, has recently been shown to digest HIV reverse transcripts found in the cytosol (42). However, TREX1 is encoded by the host rather than the virus, and is specific for ssDNA rather than any form of RNA. Regardless of this difference, it is likely LASV NP serves the same general function as Trex-1: to remove viral replication intermediates generated during the virus life cycle and prevent them from being recognized by immune sensor molecules.
This mechanism of suppression of dsRNA-mediated immune responses appears to be unique. Several proteins of other viruses are known to suppress dsRNA-mediated responses by binding to and physically shielding dsRNA from recognition by immune sensors such as RIG-I and MDA-5 or by RNA interference machinery. Example viral proteins with this activity include Ebolavirus VP35 (43, 44), Flock House virus B2 (32), and Tombusvirus P19 (33). However, rather than coating dsRNA generated during replication and transcription, the arenavirus NP instead digests the dsRNA.
In summary, the exonuclease activity of LASV NP presents a mechanism by which arenaviruses subvert the innate immune system. This work thus provides an additional avenue for development of therapeutics for LASV and other arenaviral threats to human health as well as the high-resolution structural template necessary for their design and analysis. It also opens the possibility that other viruses may encode proteins with similar activity critical for host immune suppression and opens the door to novel lines of inquiry in the field of viral immunosuppression.
Materials and Methods
Plasmids.
Full-length LASV NP (Josiah strain), the N-terminal domain (residues 1–340), and the C-terminal domain (NPΔ340, residues 341–569) were cloned into the pET46-ek/LIC vector (Novagen), resulting in NP_pET46, NP1-340_pET46, and NPΔ340_pET46, respectively. To generate mammalian NP expression vectors, full-length NP, the N-terminal portion of NP (residues 1–340), or NPΔ340 were cloned into phCMV, which contains a C-terminal HA tag, to generate NP-HA_phCMV, NP1-340-HA_phCMV, and NPΔ340-HA_phCMV. Point mutants were introduced via site-directed mutagenesis with Phusion Hot Start Polymerase (Finnzymes).
Expression and Purification.
Native LASV NP constructs and all mutants were expressed in Rosetta 2(DE3)pLysS E. coli cells (Novagen) at 37 °C and grown in 1-L cultures (supplemented with 100 μM ZnCl) to an OD600 of ∼0.4. Cells were then induced with 500 μM isopropyl-β-d-thiogalactopyranoside and incubated overnight at 25 °C with shaking at 300 rpm (New Brunswick Scientific Innova 4430). Selenomethionine-derivatized NPΔ340 was expressed in prB834 (DE3) cells in M9 minimal media, supplemented with 40 μg/mL l-amino acids (all amino acids except l-methionine), 0.4% (wt/vol) glucose, 2 mM MgSO4, 25 μg/mL FeSO40.7H2O, 1 μg/mL riboflavin, 1 μg/mL niacinamide, 1 μg/mL pyradoxine monohydrochloride, 1 μg/mL thiamine, 100 μg/mL ampicillin, and 60 μg/mL d/l-selenomethionine.
Expression cultures were harvested via centrifugation and lysed in 50 mM Tris, pH 8.0, 300 mM NaCl, 20 mM imidazole, 0.1 mg/mL lysozyme, 0.1 mg/mL DNase, and one EDTA-free Complete Protease Inhibitor tablet (Roche) using an M-110L Laboratory Microfluidizer (Microfluidics). The lysate was cleared by centrifugation, and His-tagged NP was purified from the supernatant via immobilized metal affinity chromatography using Ni-NTA beads (Qiagen). Beads were washed three times with 20 mL of wash buffer (50 mM Tris, pH 8.5, 300 mM NaCl, and 20 mM imidazole) and eluted with 20 mL of wash buffer supplemented with 500 mM imidazole. The protein was then purified on a Superdex 75 GL 10/300 (GE Healthcare) column equilibrated with 10 mM Tris, pH 8.0, and 300 mM NaCl. Fractions from the single major peak, corresponding to a monomer, were pooled and used in crystallization trials (Fig. S7). The final yield of pure NPΔ340 protein was 75 mg/L of culture. The molecular mass (27,163 Da) was confirmed by MALDI-TOF mass spectrometry.
Crystallization and Data Collection.
Initial crystal hits were obtained with the Hampton sparse-matrix screens PegRx, Crystal Screen 2, and Peg/Ion 1 and 2 in a 96-well format using a 2:1 ratio of well solution to protein at 10–12 mg/mL. Larger crystals were obtained by hanging drop vapor diffusion using a 1:1 ratio of well solution to protein at 10–12 mg/mL in 0.4 M potassium phosphate and 20% PEG 3350. Native and selenomethionine-derivatized crystals were grown in identical conditions. Crystals were harvested and cryoprotected in well solution containing 15% glycerol and flash-cooled in liquid nitrogen. Data for native and selenomethionine-derivatized NPΔ340 crystals were collected in-house on a Rigaku 007 generator and a mar345 detector (Marresearch). High-resolution data for native NPΔ340 and NPΔ340 crystals grown in the presence of UMP/MnCl2 were obtained at the Advanced Light Source, Beamline 8.3.1.
Data Processing and Structure Determination.
Data were indexed, integrated, and scaled in space group P212121 by using d*TREK (Rigaku) (45). Structure determination of LASV NPΔ340 via SIRAS was carried out by AutoRickshaw (46). Nine possible heavy-atom sites were identified, with occupancies ranging from 1.0 to 0.473. Molecular replacement for the high-resolution structures of native NPΔ340 and Mn2+-soaked native NPΔ340 was performed with PHENIX (47) using the initial model derived from the 2.2-Å native data. Iterative cycles of model building were performed by using Coot (48), and subsequent rounds of refinement were carried out with PHENIX. Five percent of the reflections were set aside for Rfree calculations. The final native NPΔ340 structure has an Rfree of 24.90% and R of 19.17% using all data to 1.7-Å resolution, and the Mn2+-soaked NPΔ340 had an Rfree of 21.57% and R of 19.15% using all data to 1.5-Å resolution (Table S1).
RNA.
For experiments involving 18-bp blunt-ended dsDNA or dsRNA or 3-nt–overhang dsRNA without 5′ triphosphate, deprotected, desalted, duplex synthetic DNA and RNA oligos were purchased from Integrated DNA Technologies. For all experiments involving these nucleic acids, the sense strand was end-labeled with [γ32P]ATP and T4 polynucleotide kinase. For experiments involving 20-bp blunt-ended or 1-nt–overhang dsRNA with a 5′ triphosphate, 5′ triphosphate-containing RNA oligos were synthesized via in vitro transcription using T7 polymerase and DNA templates. Templates contained a T7 promoter and the desired RNA sequence. Transcription products were chloroform-extracted, precipitated, and purified by denaturing PAGE to obtain the desired 54-base RNA oligo with 5′ triphosphate. In these experiments, the antisense strand was end-labeled as noted above. Sequences for all nucleic acid substrates are outlined in Table S2.
Exonuclease Activity Assays.
Standard reactions contained 500 nM wild-type or mutant NPΔ340 and 2 nM (labeled) and 100 nM (unlabeled) oligo(ribo)nucleotide. For reactions with dsDNA/dsRNA, an excess of unlabeled antisense DNA/RNA was added to labeled sense DNA/RNA and annealed at 65 °C for 5 min. In vitro transcription was used to generate a 54-nt 5′ triphosphate-capped RNA oligo. Excess, unlabeled, in vitro-transcribed sense RNA was annealed to labeled antisense oligomers corresponding to either nucleotides 1–20 or 2–21 to create 5′ triphosphate-containing 20-bp dsRNA with either blunt ends or with a single 5′ unpaired nucleotide, respectively. Reactions were performed in 20 mM Tris, pH 7.5, 150 mM NaCl, and 5 mM MgCl2. After incubation for 15 min, reactions were stopped with the addition of an equal volume of formamide loading buffer and boiled. For time-course experiments, reactions were stopped at intervals between 0 and 30 min. The products were then analyzed in 18% polyacrylamide gels containing 8 M urea and buffered with 0.5× Tris-borate-EDTA. Gels were exposed overnight to a phosphor screen (Amersham) and then imaged with a Storm 860 PhosphorImager (GE Healthcare).
IFN Induction Reporter Assays.
293T cells were seeded into a 96-well plate (1 × 104 cells per well) and cotransfected with FuGene HD (Roche Diagnostics) with 30 ng of the IRF-3 promoter reporter plasmid p55C1B-FF (a gift from J. C. de la Torre, The Scripps Research Institute) and 100 ng of LASV NP expression vectors together with 10 ng of a SV40 Renilla luciferase (SV40-RL) expression plasmid to normalize transfection efficiencies. Cells were infected with Sendai virus (multiplicity of infection of 3) at 24 h posttransfection. At 16–20 h postinfection, cells were lysed and luciferase activities were measured with the dual-luciferase kit as recommended by the manufacturer (Promega). Reporter gene activation is expressed as induction over an empty vector control.
Western Blot Analysis.
Proteins were separated by SDS/PAGE and then transferred to Immobilon-P membranes (Millipore). After blocking with 10% milk, membranes were probed with anti-HA 16B12 (Covance) for 1 h. After three washes in PBS containing 0.5% Tween 20 (PBS/Tween), membranes were incubated with secondary HRP-conjugated anti-mouse Ig antibodies for 1 h. After three washes with PBS/Tween, membranes were developed with SIGMAFAST BCIP/NBT (Sigma-Aldrich) according to the manufacturer's protocol.
Note Added in Proof.
While this manuscript was under consideration, Qi et al. published “Cap binding and immune evasion revealed by Lassa nucleoprotein structure” (50). In experiments using various ssRNA oligonucleotides, which could potentially form duplexes or hairpins, and RNA and DNA molecular weight markers as substrates, Qi et al conclude that LASV NP has no specificity for any nucleic acid type, leaving open the question of how the exonuclease activity could suppress an antiviral immune response in particular. Here we illustrate that the exonuclease activity of LASV NP is indeed specific for dsRNA, such as the products produced during viral infection.
Supplementary Material
Acknowledgments
We thank Dr. Juan Carlos de la Torre (The Scripps Research Institute) for the gift of the p55C1B-FF plasmid and the Sendai virus, Marin Gantner (The Scripps Research Institute) for assistance with IRF-3 translocation assays, and Dr. Robert Garry (Tulane University, New Orleans, LA) for valuable discussions. We also acknowledge the Viral Hemorrhagic Fever Research Consortium and Contract HHSN272200900049C (BAA-NIAID-DAIT-NIHAI2008031), an Investigators in Pathogenesis of Infectious Diseases award from the Burroughs Wellcome Fund, and The Skaggs Institute for Chemical Biology (to E.O.S.) for support as well as Beamline 8.31 of the Advanced Light Source (Berkeley, CA) for data collection.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3Q7B (native) and 3Q7C (Mn2+ soaked)].
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016404108/-/DCSupplemental.
References
- 1.Buchmeier MJ, de la Torre JC, Peters CJ. Arenaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1791–1827. [Google Scholar]
- 2.Haas WH, et al. Imported Lassa fever in Germany: Surveillance and management of contact persons. Clin Infect Dis. 2003;36:1254–1258. doi: 10.1086/374853. [DOI] [PubMed] [Google Scholar]
- 3.Holmes GP, et al. Lassa fever in the United States. Investigation of a case and new guidelines for management. N Engl J Med. 1990;323:1120–1123. doi: 10.1056/NEJM199010183231607. [DOI] [PubMed] [Google Scholar]
- 4.Briese T, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamieson DJ, Kourtis AP, Bell M, Rasmussen SA. Lymphocytic choriomeningitis virus: An emerging obstetric pathogen? Am J Obstet Gynecol. 2006;194:1532–1536. doi: 10.1016/j.ajog.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Pinschewer DD, Perez M, de la Torre JC. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol. 2003;77:3882–3887. doi: 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichler R, et al. Characterization of the Lassa virus matrix protein Z: Electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP) Virus Res. 2004;100:249–255. doi: 10.1016/j.virusres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Groseth A, Wolff S, Strecker T, Hoenen T, Becker S. Efficient budding of the Tacaribe virus matrix protein Z requires the nucleoprotein. J Virol. 2010;84:3603–3611. doi: 10.1128/JVI.02429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvato MS, Schweighofer KJ, Burns J, Shimomaye EM. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 1992;22:185–198. doi: 10.1016/0168-1702(92)90050-j. [DOI] [PubMed] [Google Scholar]
- 10.Shtanko O, et al. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J Virol. 2010;84:5415–5422. doi: 10.1128/JVI.02417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 12.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Sobrido L, Giannakas P, Cubitt B, García-Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baize S, et al. Early and strong immune responses are associated with control of viral replication and recovery in Lassa virus-infected cynomolgus monkeys. J Virol. 2009;83:5890–5903. doi: 10.1128/JVI.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrion R, Jr., et al. Lassa virus infection in experimentally infected marmosets: Liver pathology and immunophenotypic alterations in target tissues. J Virol. 2007;81:6482–6490. doi: 10.1128/JVI.02876-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djavani MM, et al. Early blood profiles of virus infection in a monkey model for Lassa fever. J Virol. 2007;81:7960–7973. doi: 10.1128/JVI.00536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher-Hoch SP, et al. Physiological and immunologic disturbances associated with shock in a primate model of Lassa fever. J Infect Dis. 1987;155:465–474. doi: 10.1093/infdis/155.3.465. [DOI] [PubMed] [Google Scholar]
- 18.Walker DH, et al. Pathologic and virologic study of fatal Lassa fever in man. Am J Pathol. 1982;107:349–356. [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson KM, et al. Clinical virology of Lassa fever in hospitalized patients. J Infect Dis. 1987;155:456–464. doi: 10.1093/infdis/155.3.456. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Sobrido L, Zúñiga EI, Rosario D, García-Sastre A, de la Torre JC. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2006;80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Sobrido L, et al. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2009;83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Söding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 26.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horio T, et al. Crystal structure of human ISG20, an interferon-induced antiviral ribonuclease. FEBS Lett. 2004;577:111–116. doi: 10.1016/j.febslet.2004.09.074. [DOI] [PubMed] [Google Scholar]
- 28.Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 29.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brautigam CA, Sun S, Piccirilli JA, Steitz TA. Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′-5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry. 1999;38:696–704. doi: 10.1021/bi981537g. [DOI] [PubMed] [Google Scholar]
- 31.Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005;13:973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 34.Garcin D, Kolakofsky D. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J Virol. 1990;64:6196–6203. doi: 10.1128/jvi.64.12.6196-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 36.Hsiao YY, et al. Crystal structure of CRN-4: Implications for domain function in apoptotic DNA degradation. Mol Cell Biol. 2009;29:448–457. doi: 10.1128/MCB.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derbyshire V, et al. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 38.Deutscher MP, Marlor CW. Purification and characterization of Escherichia coli RNase T. J Biol Chem. 1985;260:7067–7071. [PubMed] [Google Scholar]
- 39.Fischer SA, et al. LCMV in Transplant Recipients Investigation Team. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med. 2006;354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 40.Minskaia E, et al. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;107:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimberlin CR, et al. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci USA. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung DW, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 46.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-Rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr. 2005;61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.