Abstract
The cell surfaces of Gram-negative bacteria are composed of lipopolysaccharide (LPS). This glycolipid is found exclusively in the outer leaflet of the asymmetric outer membrane (OM), where it forms a barrier to the entry of toxic hydrophobic molecules into the cell. LPS typically contains six fatty acyl chains and up to several hundred sugar residues. It is biosynthesized in the cytosol and must then be transported across two membranes and an aqueous intermembrane space to the cell surface. These processes are required for the viability of most Gram-negative organisms. The integral membrane β-barrel LptD and the lipoprotein LptE form an essential complex in the OM, which is necessary for LPS assembly. It is not known how this complex translocates large, amphipathic LPS molecules across the OM to the outer leaflet. Here, we show that LptE resides within the LptD β-barrel both in vitro and in vivo. LptD/E associate via an extensive interface; in one specific interaction, LptE contacts a predicted extracellular loop of LptD through the lumen of the β-barrel. Disrupting this interaction site compromises the biogenesis of LptD. This unprecedented two-protein plug-and-barrel architecture suggests how LptD/E can insert LPS from the periplasm directly into the outer leaflet of the OM to establish the asymmetry of the bilayer.
Keywords: lipopolysaccharide assembly, outer membrane protein complex
The outer membrane (OM) of Gram-negative bacteria is an asymmetric bilayer (1–3), with an inner leaflet composed of phospholipids and an outer leaflet consisting mainly of lipopolysaccharide (LPS). In Escherichia coli, the LPS molecule typically contains six fatty acyl chains and as many as several hundred sugars. In the presence of divalent cations, LPS is proposed to form a gel of relatively low fluidity, which impedes the passage of hydrophobic molecules that can readily cross other biological membranes (4). This unique barrier enables Gram-negative bacteria to survive in harsh environments and to exclude several antibiotics effective against Gram-positive organisms. Not surprisingly, given its important function, LPS is essential in most Gram-negative bacteria.
LPS biogenesis involves synthesis at the inner membrane (IM), transport across the periplasmic space, and insertion into the outer leaflet of the outer membrane (OM). The LPS biosynthetic pathway has been well characterized (5); however, it is not clear how the cell transports this large amphiphilic molecule across the aqueous periplasmic compartment and to its final location at the cell surface. This process requires seven essential Lpt (lipopolysaccharide transport) proteins that form a transenvelope complex (6, 7). Two of these proteins, LptD and LptE (formerly Imp and RlpB, resp.), reside in the OM and are responsible for the translocation of LPS across the OM (to the cell surface) in the final stages of assembly (8–10).
LptD, an integral membrane β-barrel protein, and the lipoprotein LptE form a stable complex (9, 10), whose structure remains uncharacterized. LptD consists of a periplasmic N-terminal domain and a C-terminal transmembrane β-barrel domain (10), which are linked by two disulfide bonds (11). The C-terminal β-barrel domain of LptD is sufficient for the tight interaction with LptE and protects LptE from proteolytic digestion in vitro; these observations led us to propose that LptE may form a plug within the LptD β-barrel (10). LptE is required for LptD biogenesis (10); proper disulfide bond formation in LptD depends on the presence of LptE (11), but how LptE facilitates this process is unknown. Interestingly, LptE also binds specifically to LPS in vitro, suggesting that it directly handles LPS during transport (10). The mechanism by which LptD/E work together to translocate LPS across the OM and establish the asymmetry of the bilayer is unknown.
To address this question, we sought to characterize the architecture of this two-protein complex in a physiological environment. Here, we have used unnatural amino acid mutagenesis and photocrosslinking in vivo to show that LptD and LptE interact via an extensive interface spanning multiple faces of LptE. Mass spectrometric analysis of crosslinked LptD/E complexes identified a ten-residue region of LptD that interacts with LptE, as confirmed in vivo. Remarkably, this region is found in a putative extracellular loop of LptD. Deletion of this interaction site compromises the assembly of the LptD/E complex in vivo and leads to OM defects. Taken together, these results establish that LptE resides within the lumen of the transmembrane β-barrel of LptD and suggest how the LptD/E complex assembles LPS at the cell surface. As highlighted by the recent report of antibiotics that target LptD in Pseudomonas spp. (12), understanding the mechanism by which LptD/E function is critical for developing new drugs against clinically important Gram-negative pathogens.
Results
Specific Regions of LptE Directly Contact LptD in Vivo.
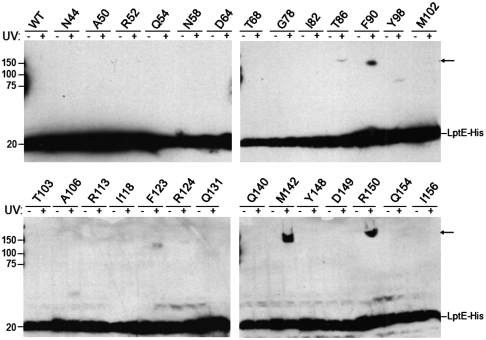
Recently, we described the isolation and characterization of LptD/E, the two-protein complex responsible for assembling lipopolysaccharide in the outer membrane of E. coli (10). Here, to develop a more detailed understanding of LptD/E structure, we have carried out in vivo photocrosslinking (13) to map intermolecular interactions within the complex. Guided by the three-dimensional structures of three LptE orthologs, we introduced the UV-photocrosslinker para-benzoyl-L-phenylalanine (pBPA) at 27 positions throughout oligohistidine-tagged LptE (LptE-His) via unnatural amino acid mutagenesis (14; Fig. 1). Low-level expression of these mutant proteins supports growth in the absence of endogenous LptE, indicating that they are functional (see SI Text).
Fig. 1.
In vivo photocrosslinking of LptE. E. coli expressing low levels of wild-type or pBPA-substituted LptE-His protein, as indicated, in a chromosomal ΔlptE background were either left untreated or irradiated with UV light, followed by Western blot analysis of whole-cell lysates with an anti-His antibody. Arrows, UV-photocrosslinked complexes containing pBPA-substituted LptE-His. The analyzed positions were selected from the 193 residues of full-length LptE (residue numbers include the signal sequence, a.a. 1–18) on the basis of structural conservation with LptE orthologs from other Gram-negative bacteria: no structural information exists for residues 19–35, and residues 166–193 of E. coli LptE constitute a C-terminal extension that is not found in other organisms (see Fig. 2B).
To determine which sites within the protein sequence of LptE are involved in interactions with LptD, we prepared whole-cell lysates from exponentially growing cells expressing pBPA-containing LptE-His, which had been either irradiated with UV light or left untreated. In cells in which pBPA was incorporated at position 86, 90, 123, 124, 142, or 150 of LptE-His (numbering includes the LptE signal sequence, a.a. 1–18), UV-irradiation led to the formation of LptE-His-containing intermolecular complexes with apparent molecular weights above 100 kDa (Fig. 1), consistent with crosslinking between LptE-His (∼20 kDa) and LptD (∼87 kDa).
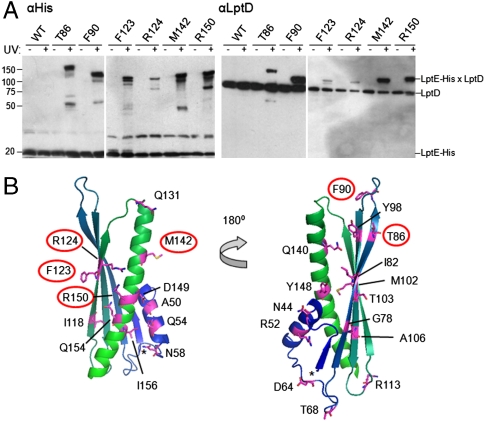
To confirm that LptD was the UV-photocrosslinking partner, we enriched His-tagged species from unirradiated or UV-irradiated cells by nickel affinity chromatography and showed that, for each of the six positions identified above, the high-molecular weight crosslinked bands could be detected by both anti-His and anti-LptD antibodies (Fig. 2A). This result indicates the UV-dependent formation of covalently linked LptD/E complexes in which LptE residue T86, F90, F123, R124, M142, or R150 is replaced with pBPA. These data show that LptD and LptE directly interact in vivo and establish a map of the regions of LptE involved in this interaction.
Fig. 2.
Specific residues of LptE interact directly with LptD in vivo. (A) Live E. coli expressing low levels of the indicated pBPA-substituted LptE–His protein were either left untreated or irradiated with UV light, followed by nickel affinity chromatography and Western blotting using an anti-His antibody or an anti-LptD polyclonal antiserum. (B) Mapping of the tested positions onto a structural model of E. coli LptE, generated on the basis of the three-dimensional structures of three LptE orthologs (RlpB from Shewanella oneidensis, Nitrosomonas europaea, and Neisseria meningitidis; PDB IDs 2R76, 2JXP, and 3BF2, respectively) using HHPred and MODELLER (40). Residues involved in LptD/E interactions, as identified in (A), are circled. The asterisk denotes the N-terminus of the model structure, corresponding to residue I36 of the full-length E. coli LptE. The N-terminal lipidation site is on C19 in this numbering scheme.
This putative LptD-binding interface spans a wide region of LptE. We mapped the LptD-crosslinking positions onto a model LptE structure (Fig. 2B) and found that the LptD/E interaction involves residues from both a conserved β-sheet (T86, F90, F123, and R124) and a conserved C-terminal α-helix (M142 and R150). Notably, residues at both edges (T86 vs. F123/R124) and at both faces (T86/R124 vs. F123) of the β-sheet appear to participate in the interaction with LptD. These findings are consistent with our earlier suggestion that LptE may be localized inside the LptD β-barrel (10; see Discussion).
LptD/E contacts may be critical to the assembly, stability, and/or function of the two-protein complex. To dissect the role of the LptD-crosslinking residues within the native LptE protein, we mutated them to alanine individually and in pairs (Fig. S1). Haploid LptE T86A and M142A mutants, but not alanine mutants at the other four positions, displayed reduced growth on plates containing 0.5% SDS and 1 mM EDTA, indicating defects in OM biogenesis (Fig. S1 A and C). The T86A/M142A double mutant displayed a more severe growth defect than either of the parent strains (Fig. S1A). Notably, neither the T86A and M142A single mutations, nor the combination of the two, affected LptE protein levels or the stability of LptD/E complexes, as judged by LptD pulldown with LptE-His (Fig. S1B). These results suggest that, whereas LptE residues T86 and M142 likely play a subtle role in LptD/E biogenesis and/or function, no residue alone is essential for the stability of the LptD/E complex, consistent with an extensive interface comprising a large number of interactions at widely distributed positions within the two proteins.
Identification of a Specific LptE Interaction Site within LptD.
Little is known about the structure or mechanism of LptD. To obtain a high-resolution map of the LptD/E interface, we used mass spectrometry to determine which residues of LptD are crosslinked to pBPA-containing LptE. Because the natural abundance of LptD/E in vivo is low (15), we turned to an in vitro system based on our established method (10) to overexpress and purify LptD/E complexes. We examined complexes containing the pBPA substitution at four of the six positions that showed UV-photocrosslinking in vivo. As observed in live cells, UV-irradiation of these purified complexes led to the formation of high molecular weight products (Fig. S2A), which, upon trypsin digestion and mass spectrometric analysis, proved to contain both LptD and LptE with high coverage (Fig. S2B). By contrast, UV-irradiation of purified complexes containing pBPA at positions 106 and 113 of LptE, where crosslinking to LptD had not been observed in vivo, did not produce appreciable amounts of crosslinked product (Fig. S2A). Our in vitro crosslinking thus displays the same site-specificity as was observed in vivo.
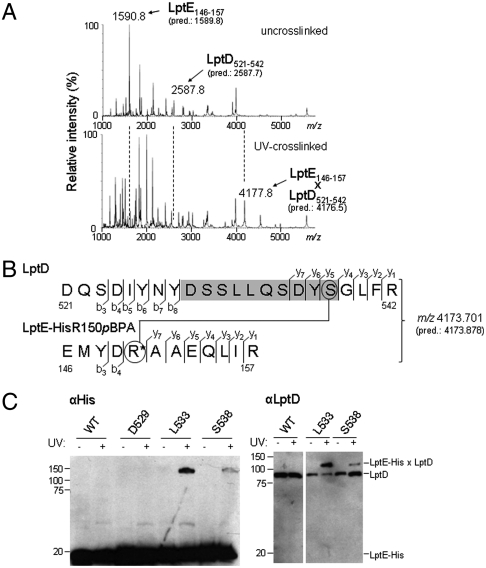
To identify the specific residues of LptD involved in binding LptE, we analyzed tryptic digests from uncrosslinked and crosslinked LptD/E complexes by matrix-assisted laser desorption-ionization mass spectrometry (MALDI-MS). We selected the complexes in which R150 of LptE-His was replaced with pBPA for detailed study because this substitution led to near-quantitative crosslinking in vitro (Fig. S2A). We performed parallel in-gel tryptic digests of the bands corresponding to free LptE-His and LptD from unirradiated LptD/LptE-HisR150pBPA complex, which we combined as the uncrosslinked sample, and of the crosslinked band from the UV-irradiated complex. We reasoned that, relative to uncrosslinked complexes, tryptic digests of crosslinked complexes should contain greatly reduced levels of the pBPA-containing LptE peptide and of any LptD peptide(s) to which it is crosslinked. Indeed, the low-resolution MALDI-MS spectrum of the uncrosslinked sample revealed substantial peaks with mass-to-charge ratios (m/z) around 1,591 and 2,588, which were absent from the spectrum of the crosslinked complex (Fig. 3A). The average [M + H]+ mass of the predicted pBPA-containing tryptic peptide from LptE, EMYD[pBPA]AAEQLIR (spanning residues 146–157), is 1,589.8 Da. Furthermore, an LptD tryptic peptide spanning residues 521–542 has a predicted [M + H]+ mass of 2,587.7 Da, again in agreement with the observed peak at m/z = 2,588. Meanwhile, the MALDI-MS spectrum of the crosslinked complex contained a new peak with an m/z of approximately 4,178, which was not observed in the spectrum of the uncrosslinked complex (Fig. 3A). This mass is consistent with that of a singly charged UV-photocrosslinked species comprised of both of these peptides (1,588.8 + 2,586.7 + 1 = 4,176.5 Da). These results strongly suggested that LptE directly contacts LptD between residues 521 and 542.
Fig. 3.
LptE interacts with a predicted extracellular loop of the LptD β-barrel. (A) LptD/E complexes in which LptE residue R150 was substituted with pBPA (LptE-HisR150pBPA) were overexpressed, purified, and UV-photocrosslinked in vitro (see Fig. S2A). Bands corresponding either to free LptD and LptE protein from uncrosslinked complexes (Top) or to the UV-crosslinked LptD/E complex (Bottom) were excised from an SDS-PAGE gel and subjected to in-gel trypsin digestion, followed by low-resolution MALDI-MS analysis of peptide products. The observed and predicted (pred.) average [M + H]+ masses of the indicated LptD and LptE peptides are shown. (B) High-resolution MS/MS fragmentation analysis of the crosslinked adduct identified in part A. The observed and predicted (pred.) exact [M + H]+ masses of this adduct are shown. Detected fragment ions included those corresponding to the eight N-terminal and four C-terminal residues of the LptD peptide spanning residues 521–542 (denoted b3–b8 and y1–y4, respectively, for the LptD sequence), as well as the four N-terminal and seven C-terminal residues of the pBPA-containing LptE peptide spanning residues 146–157 (denoted b3–b4 and y1–y7, respectively, for the LptE sequence; R* denotes the pBPA-substituted residue). Three additional C-terminal LptD fragments (y5–y7) had masses indicating the presence of a covalent modification on S538 (circled, Top) with molecular weight equal to that of the entire LptE peptide. No unmodified C-terminal LptD fragments of more than four residues were detected. (See Fig. S3 and Table S1.) (C) The indicated residues of LptD were replaced with pBPA in a chromosomal ΔlptD background and in the presence of plasmid-encoded LptE–His. UV-photocrosslinking, nickel affinity chromatography, and Western blotting were performed as in Fig. 2.
To confirm that this 22-residue peptide of LptD was indeed crosslinked to pBPA-containing LptE, and to further resolve this LptE interaction site within LptD, we subjected the same crosslinked LptD/LptE-HisR150pBPA sample to high-resolution two-dimensional MALDI-TOF/TOF spectrometry (Fig. 3B, Fig. S3, and Table S1). Upon fragmentation of the parent ion (m/z 4,173.701; predicted monoisotopic m/z 4,173.871), we detected 19 fragment ions from the N- and C-termini of both the pBPA-containing LptE peptide (residues 146–157) and the LptD peptide spanning residues 521–542 (Fig. 3B). These data establish that LptE-HisR150pBPA is crosslinked to this region of LptD.
Interestingly, automated peptide sequencing identified no ions containing residues from the region of LptD between positions 529–538 (Fig. 3B, shaded) upon fragmentation of the crosslinked parent ion. We hypothesized that this region might contain crosslinks to LptE-His, leading to modified ions that are not recognized by automated analysis. In fact, upon reanalyzing the MS/MS data, we identified three additional C-terminal fragment ions (LptD y5–y7 in Fig. 3B) corresponding to the LptD peptide DYS*GLFR (residues 536–542), where S* is a serine residue bearing a modification of molecular weight 1,587.7, the exact mass of the entire pBPA-containing LptE peptide (Fig. 3B, Fig. S3A, and Table S1A). Although the data do not eliminate the possibility of additional crosslinking sites between residues 529–538 that do not give rise to detectable fragment ions, these results show that LptE-HisR150pBPA is photocrosslinked to residue S538 within the LptD tryptic peptide 521–542. LptD residues 529–538 thus demarcate an LptE interaction site.
Residues within the LptD 529–538 Region Directly Contact LptE in Vivo.
We wished to determine if LptD residues 529–538 constitute an LptE-interacting site in vivo as well as in vitro. To test our hypothesis that LptD residue S538 is a site of direct contact with LptE in the cell, we replaced this residue with pBPA by amber mutagenesis in a chromosomal ΔlptD background and in the presence of wild-type LptE. In fact, UV-irradiation of these cells led to photocrosslinking between LptD and LptE (Fig. 3C), confirming the importance of LptD residue 538 as a site of interaction with LptE. Because we had observed no uncrosslinked fragment ions for the entire region spanning residues 529–538, we wondered if other residues within this region might be crosslinked as well. Indeed, photocrosslinking was also observed when residue L533 of LptD was replaced with pBPA (Fig. 3C). These results confirm that the region of LptD roughly defined by residues 529–538 plays a key role in LptD/E interactions in vivo as well as in vitro.
The LptE Interaction Site Is Found in a Predicted Extracellular Loop of LptD.
The three-dimensional structure of LptD has not been determined. It is noteworthy, however, that the LptE-interacting region between residues 529–538 is found within a predicted extracellular loop spanning residues 515–557 of the LptD β-barrel (Fig. S4), as assigned in silico by the PRED-TMBB method (16, 17) using three distinct algorithms (Viterbi, N-best, and posterior decoding).
In all known OM β-barrel structures, loops of over 15 residues in length are found exclusively between the extracellular ends of transmembrane β-strands. These loops are typically flexible and tolerant of deletion and insertion mutations (18, 19). Consistent with the prediction that the LptE-interacting region at residues 529–538 of LptD is located in such a loop, we constructed an lptDΔ529–538 allele and found that it supported cell viability, indicating that LptDΔ529–538 is functional. We also inserted the seven-residue peptide sequence ENLYFQS (the cleavage site recognized by tobacco etch virus [Tev] protease) at six positions (V517, P518, I525, R544, A553, and S554) within the predicted extracellular loop spanning residues 515–557. All of these constructs also supported cell growth in the absence of the chromosomal lptD allele, indicating that LptD proteins containing insertions at these sites are functional. Taken together, these observations suggest that LptD residues 517–554, and thus the LptE interaction site defined by LptD residues 529–538, are found in a topologically extracellular loop of the LptD β-barrel.
The LptE Interaction Site at Residues 529–538 of LptD Plays a Key Role in LptD/E Biogenesis.
Having identified LptD residues 529–538 as an important site of LptD/E interaction, we asked what role this interaction may play in LptD/E and/or LPS biogenesis in vivo. We found that cells in which the only copy of lptD was encoded by an lptDΔ529–538 allele were viable, indicating that LptDΔ529–538 is functional. In LB media the growth of this deletion strain was unimpaired relative to the wild-type control strain (Fig. 4A and Fig. S5A). By contrast, we found that the lptDΔ529–538 strain was not viable on plates containing 0.5% SDS and 0.75 mM EDTA (Fig. 4A), indicating that deletion of these ten residues of LptD severely compromises OM integrity. As is characteristic for cells with LPS assembly defects (20), MALDI-MS analysis revealed the presence of hepta-acyl and phosphoethanolamine modifications to the lipid A core of LPS in lptDΔ529–538 cells (Fig. S5B). By contrast to cells in which the only copy of lptD lacked residues 529–538, cells harboring a native chromosomal lptD allele as well as plasmid-encoded lptDΔ529–538 had no growth defect in the presence of 0.5% SDS and 0.75 mM EDTA (Fig. S5C). This result rules out the possibility that the OM defects associated with the Δ529–538 deletion are due to nonspecific permeabilization by misfolded, aggregated, and/or pore-forming LptD proteins, and indicates instead that this deletion causes a specific loss of LptD function.
Fig. 4.
The LptE interaction site at residues 529–538 of LptD is required for proper folding of LptD and assembly of the LptD/E complex. (A) Ten-fold serial dilutions of overnight cultures (normalized by optical density) on LB containing 25 μg/mL kanamycin without (Top) or with 0.5% SDS and 0.75 mM EDTA. The WT and Δ529–538 strains harbored a chromosomal ΔlptD2∷kan deletion as well as pET23/42lptD or its derivative pET23/42lptDΔ529–538, respectively. (B) Western blot analysis, using the indicated antibodies, of total membrane extracts or eluates from Ni-NTA chromatography of these extracts from strains expressing low levels of LptE–His and either wild-type LptD or LptDΔ529–538 in a chromosomal ΔlptD background. β-mercaptoethanol (βME) reduces the disulfide bonds found in LptD, decreasing its apparent molecular weight. Membrane extracts were normalized by total protein content; equal volumes of Ni-NTA eluates were loaded. The asterisk denotes a protein that cross-reacts with the anti-LptD antibody and serves as a loading control.
We asked whether impaired OM formation in the lptDΔ529–538 strain results from defects in the assembly of the LptD/E complex, its function in LPS assembly, or both. To determine how the Δ529–538 deletion affects LptD/E assembly, we compared total LptD and LptD/E complexes in cell membranes from the lptDΔ529–538 strain to those in wild-type membranes (Fig. 4B). Cell membranes isolated from the lptDΔ529–538 strain contained lower total levels of LptD protein than membranes from the control strain (Fig. 4B, lanes 1 and 2). Moreover, nearly all of the total LptD protein in the lptDΔ529–538 strain failed to form native disulfide bonds, as judged by its faster mobility on nonreducing SDS-PAGE relative to wild-type LptD protein (Fig. 4B, lanes 3 and 4; ref. 11). Nevertheless, when we isolated LptD/E-His complexes from the lptDΔ529–538 strain by nickel affinity chromatography, we found that only the correctly disulfide-bonded LptD species was associated with LptE (Fig. 4B, lanes 7 and 8); the major LptD species, which did not contain native disulfides, could not be pulled down by LptE. Because we previously found that the native disulfide bonds of LptD are not required for the association of LptD with LptE (10, 11), these results suggest that the Δ529–538 deletion either impairs a step in the assembly of the LptD/E complex or compromises the stability of the fully assembled complex.
To distinguish between these two possibilities, we compared the stability of purified LptD/E complexes containing wild-type LptD or LptDΔ529–538 in vitro using our previously published methods (10). Like wild-type LptD/E complexes, LptDΔ529–538/E complexes eluted as a single peak from a size-exclusion column and migrated as a single band on nondenaturing blue native (BN-)PAGE (Fig. S5D). The overall trypsin susceptibility of LptDΔ529–538/E complexes was comparable to that of wild-type complexes, as shown by the fact that trypsin-digested LptDΔ529–538/E complexes continued to comigrate on BN-PAGE (Fig. S5D). Moreover, trypsin cleavage of LptE within the wild-type and mutant complexes occurred at similar sites and efficiencies, as confirmed by Western blot analysis of trypsin-digested complexes after SDS-PAGE (Fig. S5E). These data indicate that deleting the LptE interaction site between LptD residues 529–538 does not destabilize the mature LptD/E complex. Therefore, this LptD/E interaction site plays an important role in the assembly of the LptD/E complex.
Discussion
We have analyzed specific intermolecular interactions within LptD/E, the essential two-protein complex responsible for assembling LPS into the outer membrane of E. coli. In vivo photocrosslinking indicates that LptE directly contacts LptD at a wide range of positions encompassing multiple surfaces of LptE. Furthermore, we identified a specific LptE interaction site within LptD, which is part of a putative extracellular loop of the LptD β-barrel. Finally, we found that disrupting this interaction site compromises the biogenesis of LptD, as well as that of LPS, producing a defective OM.
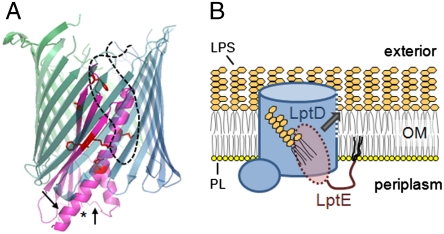
We conclude that a substantial portion of LptE is found inside LptD. We previously proposed that LptE forms a plug within the LptD β-barrel (10). The results presented here, showing that the LptD/E interface is extensive and involves multiple faces of LptE, allow us to build a physical model for how LptE is positioned within LptD (Fig. 5A). Earlier, we observed proteolytic cleavage of LptE within folded LptD/E complexes at specific sites located near the N-terminal lipid anchor of LptE in three-dimensional space (10). That these sites are distant from the LptD/E interface described here explains why they are accessible to proteolytic cleavage in the complex. Given that LptE is anchored on the periplasmic face of the OM, its localization within the lumen of the LptD β-barrel would also account for the prediction that the LptE-interacting site at LptD residues 529–538 is found within an extracellular loop: these loops are often found folded into the lumens of known β-barrel structures, here they can serve as gates or specificity determinants (e.g., in maltoporin, 21). Thus, the interaction between LptE and LptD residues 529–538 may take place inside the β-barrel of LptD, within the OM bilayer.
Fig. 5.
Proposed models for the structure and function of the LptD/E complex. (A) LptE forms a plug within the lumen of the LptD β-barrel. A model LptE structure (as in Fig. 2B) is shown in magenta, with the LptD-interacting residues depicted as red sticks and with an asterisk at the N-terminus of the model structure. The transmembrane portion of LptD is shown as a hypothetical 22-stranded β-barrel in blue-green, with the LptE-interacting extracellular loop as a dashed black line. Arrows denote approximate locations of the trypsin cleavage sites within LptE in LptD/E complexes (10). (B) Proposed model for direct insertion of LPS into the outer leaflet of the OM. LPS is delivered directly from the periplasm to LptE within the LptD pore, and subsequently into the OM lipid bilayer through an opening in the LptD β-barrel wall (dashed blue line), without residing in the inner leaflet. PL, phospholipid. The O-antigen of LPS has been omitted for clarity.
How LPS is transferred from the periplasm to the outer leaflet of the OM is not known; the plug-and-barrel architecture of the LptD/E complex suggests a possible mechanism. Because LptE binds specifically to the lipid portion of LPS (10), its localization within the lumen of LptD may allow it to move the fatty acyl chains of the LPS substrate directly from the periplasm via the lumen of the LptD β-barrel and into the outer leaflet of the OM (Fig. 5B). The oligosaccharide moiety of LPS is presumably threaded through the LptD β-barrel before or during the membrane insertion of the lipid portion of the molecule. Consistent with this direct insertion model, there is no evidence suggesting that LPS ever resides in the inner leaflet of the OM. If LPS did enter the inner leaflet from the periplasm, then maintaining the asymmetry of the OM, in which LPS is found exclusively in the outer leaflet, would pose an additional challenge for the cell. Enclosing the LptE-bound LPS within LptD can prevent LPS insertion into the inner leaflet en route to the cell surface.
Direct insertion of LPS from LptE within the lumen of LptD into the outer leaflet of the OM implies that LPS should diffuse laterally through the LptD β-barrel wall. Such movement of hydrophobic substrates between the OM bilayer and the lumen of a β-barrel has been described in several systems (22), including the long-chain fatty acid transporter FadL (23), the small porins OmpW and OprG (24–26), and the acyltransferase PagP (27). In fact, LPS is a substrate of PagP and is believed to access the active site via a lateral opening in its β-barrel wall (27). Interestingly, the lateral openings of all three of these β-barrels are delimited by proline residues that interrupt hydrogen bonding between adjacent β-strands. An alignment of LptD orthologs from 20 species revealed two pairs of absolutely conserved prolines (P214 and P246; P483 and P510) in adjacent β-strands (Fig. S4). LptD/E may thus utilize a mechanism similar to that of PagP to release LPS substrates via an opening in the β-barrel wall directly into the membrane bilayer (Fig. 5B).
LptD and LptE orthologs are both found in hundreds of Gram-negative genomes, including those of the pathogens Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter baumannii, suggesting a conserved mechanism for LPS assembly. However, some bacterial genomes appear to contain an ortholog of LptD, but not of LptE (28). There are many possible interpretations of this observation. Some proteins that share the fold and function of E. coli LptE may have escaped bioinformatic identification due to low sequence similarity, especially in light of its relatively small size. It is also possible that, in some organisms, the LptD barrel is plugged by a protein of a different fold. We have previously shown that LptE binds LPS, and it would not be surprising if organisms with distinct LPS structures had distinct LPS-binding proteins. Whether the plug-and-barrel architecture represents the only solution for LPS assembly at the cell surface is an interesting question.
It is worth noting that the LptD/E plug-barrel complex consists of two separate proteins. To our knowledge, there are no other known examples of such a two-protein architecture in any organism, as every previously described plug-barrel complex consists of a single polypeptide (29–31). Encoding the plug and barrel as two distinct proteins complicates the assembly of the LPS transport apparatus. We previously showed that LptE is required for the biogenesis of LptD (10, 11). Here, we show that deletion of the LptE interaction site at LptD residues 529–538 impairs a step in LptD biogenesis. In an accompanying paper, Chimalakonda et al. show that a mutation in lptE that also impairs LptD/E interaction leads to assembly defects in LptD (32). Interestingly, like the lptD∆529–538 mutant reported here, two other lptD mutants, lptD4213, and lptD208 (8, 33), harbor deletions within a predicted extracellular loop of LptD (residues 330–352 and 335–359, respectively; see Fig. S4); these mutants also exhibit increased outer-membrane permeability. It remains to be seen whether, as in the case of lptD∆529–538, these deletions compromise LptD/E assembly and whether they do so by impairing the interaction between LptD/E.
It is unclear how LptE can participate in the assembly of LptD, which is folded at the OM by the β-barrel assembly machine (the Bam complex; 34–39). The fact that LptE contacts a region of LptD important for LptD assembly suggests two possible roles for LptE in this process. LptE may associate with and stabilize LptD molecules after their folding is complete; alternatively, because LptE is found inside LptD, it is possible that LptE templates formation of the LptD β-barrel during the folding process. Intriguingly, in the latter case LptD β-barrel folding would require an additional essential protein, LptE, in addition to the general Bam machine.
Our biochemical characterization of the LptD/E interaction has established the plug-and-barrel organization of this two-protein complex, providing a plausible mechanism for how these proteins control the asymmetric placement of LPS in the OM. A detailed understanding of the structure, function and assembly of this essential complex will open the door to antibiotic strategies targeting the outer membrane.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains are described in SI Text. Plasmids are listed in Table S2 and oligonucleotides are listed in Table S3.
In Vivo Photocrosslinking.
For whole-cell lysate analyses, MC4100 ΔlptE∷kan strains harboring pSup-BpaRS-6TRN and pET23/42lptE-His containing the TAG stop codon at the indicated positions were grown overnight, diluted 1∶100 into 2 mL of the same media and grown to midlog phase. After normalization by optical density, cells were pelleted and either used directly or resuspended in 70 μL Tris-buffered saline (TBS; 20 mM Tris pH 8.0, 150 mM NaCl) and irradiated with UV light at 365 nm for 10 min at 4 °C. All samples were finally resuspended in 50 μL of SDS-PAGE buffer, boiled for 5 min, and centrifuged at top speed in a microcentrifuge for 15 min; 15 μL of each sample was analyzed by SDS-PAGE and immunoblotting.
For affinity pulldowns with pBPA-containing LptE-His, overnight cultures of the same strains were diluted 1∶100 into 1.5 L of the same media and grown to midlog phase. Each culture was split in half, and each sample was pelleted and either used directly or resuspended in 1/10 of the original volume in ice-cold TBS pH 8.0 and irradiated with UV light at 365 nm for 15 min. All samples were subsequently kept at 4 °C and protected from light. Samples were resuspended in 15 mL ice-cold TBS containing 1% Anzergent 3–14 (Anatrace), 100 μg/mL lysozyme, 1 mM PMSF, and 50 μg/mL DNase I, lysed by sonication, and centrifuged at 18,500 × g in a table-top centrifuge for 30 min. Nickel affinity purification was performed as described (10). Eluates were concentrated to a final volume of approximately 200 μL using 3,000 Da cut-off Amicon centrifugal concentrators (Millipore) and analyzed by SDS-PAGE and Western blotting. For experiments with pBPA-containing LptD (Fig. 3C), the same procedure was followed with MC4100 ΔlptD2∷kan strains harboring pSup-BpaRS-6TRN, pCLlptE-His, and pET23/42lptD containing the TAG stop codon at the indicated positions.
Overexpression and Purification of pBPA-Containing LptD/E Complexes and in Vitro Photocrosslinking.
The method for overexpression and purification of LptD/E was as previously reported (10), except that each BL21(DE3) overexpression strain harbored pSup-BpaRS-6TRN in addition to pET23/42lptD and amber-mutant pCDFlptE-His. The usual media (10) were supplemented with 15 μg/mL chloramphenicol and 180 μM pBPA.
Other methods are described in SI Text.
Supplementary Material
Acknowledgments.
The authors thank Paul Kowalski and Brian Stall (Bruker Daltonics, Billerica, MA) for generous assistance with MALDI-TOF/TOF analysis. The authors also thank Dr. X. Michelle Li (FAS Center for Systems Biology, Harvard University) and Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard Medical School) for MS assistance. This work was supported by National Institute of Allergy and Infectious Disease Grant AI081059 (D.K.). E.F. is a graduate fellow of the Fannie and John Hertz Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See companion article on page 2492.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015617108/-/DCSupplemental.
References
- 1.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid headgroups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 2.Funahara Y, Nikaido H. Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J Bacteriol. 1980;141:1463–1465. doi: 10.1128/jb.141.3.1463-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mühlradt PF, Golecki JR. Asymmetrical distribution and artefactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur J Biochem. 1975;51:343–352. doi: 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chng SS, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 9.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci USA. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ. Non-consecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci USA. 2010;107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivas N, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 13.Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino acid mutagenesis. ChemBioChem. 2002;3:1135–7. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 15.Takase I, et al. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987;169:5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: A web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32:W400–4. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. A hidden Markov model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinformatics. 2004;5:29–41. doi: 10.1186/1471-2105-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guedin S, et al. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J Biol Chem. 2000;275:30202–30210. doi: 10.1074/jbc.M005515200. [DOI] [PubMed] [Google Scholar]
- 19.Wimley WC. The versatile β-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 20.Raetz CRH, Reynolds CM, Trent SM, Bishop RE. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 22.van den Berg B. Going forward laterally: Transmembrane passage of hydrophobic molecules through protein channel walls. ChemBioChem. 2010;11:1339–1343. doi: 10.1002/cbic.201000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458:367–379. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 25.Neher TM, Lueking DR. Pseudomonas fluorescens ompW: Plasmid localization and requirement for naphthalene uptake. Can J Microbiol. 2009;55:553–563. doi: 10.1139/w09-002. [DOI] [PubMed] [Google Scholar]
- 26.Touw DS, Patel DR, van den Berg B. The crystal structure of OprG from Pseudomonas aeruginosa, a potential channel for transport of hydrophobic molecules across the outer membrane. PLoS ONE. 2010;5:e15016. doi: 10.1371/journal.pone.0015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop RE. The lipid A palmitoyltransferase PagP: Molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 28.Sutcliffe IC. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010;18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Smith BS, Chen LX, Baxter RHG, Deisenhofer J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc Natl Acad Sci USA. 2009;106:7403–7407. doi: 10.1073/pnas.0902789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chimalakonda G, et al. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2011 Jan 21; doi: 10.1073/pnas.1019089108. 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 35.Wu T, et al. Identification of a multi-component complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Malinverni JC, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 37.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 39.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.