Abstract
γδ T cells play important roles in bridging innate and adaptive immunity, but their recognition mechanisms remain poorly understood. Human γδ T cells of the Vδ1 subset predominate in intestinal epithelia and respond to MICA and MICB (MHC class I chain-related, A and B; MIC) self-antigens, mediating responses to tumorigenesis or viral infection. The crystal structure of an MIC-reactive Vδ1 γδ T-cell receptor (TCR) showed expected overall structural homology to antibodies, αβ, and other γδ TCRs, but complementary determining region conformations and conservation of Vδ1 use revealed an uncharacteristically flat potential binding surface. MIC, likewise, serves as a ligand for the activating immunoreceptor natural killer group 2, D (NKG2D), also expressed on γδ T cells. Although MIC recognition drives both the TCR-dependent stimulatory and NKG2D-dependent costimulatory signals necessary for activation, interaction analyses showed that MIC binding by the two receptors was mutually exclusive. Analysis of relative binding kinetics suggested sequential recognition, defining constraints for the temporal organization of γδ T-cell/target cell interfaces.
Keywords: biacore, crystallography
Despite comprising only a small fraction (2–3%) of the total human T-cell population, γδ T cells contribute to immune surveillance by elimination of malignant cells and recognition of mucosal and peripheral blood-borne pathogens (1). However, in contrast to αβ T-cell receptors (TCRs), which require antigen processing and subsequent presentation by MHC molecules, γδ TCRs are believed to recognize antigens directly (2–4). Little is known about the details of γδ TCR ligand recognition mechanisms, because receptor–ligand pairs for this class of immunoreceptors have been difficult to identify. Unlike αβ TCRs, for which there are dozens of 3D structures available that provide a wealth of detailed information (5), there are only three γδ TCR structures currently known: an isolated human Vδ3 domain of unknown ligand specificity [ES204; Protein Data Bank (PDB) code 1TVD], the intact but ligand-free ectodomain of a human Vγ9Vδ2 TCR G115 (the predominant combination in the peripheral blood; PDB code 1HXM), and the murine γδ TCR G8 bound to its nonclassical MHC class I ligand, H-2T22 (PDB 1YPZ) (6–8). These structures are highly informative, showing, for instance, the expected high level of structural homology between αβ and γδ TCRs. However, they provide little detail about how human Vδ1 TCRs recognize ligands or about γδ TCR recognition in general. The murine G8:T22 complex structure, although showing an unusual recognition strategy dominated almost to exclusivity by a single complementary determining region (CDR) CDR3δ, may or may not recapitulate aspects of general γδ TCR ligand recognition. Additional structures of human γδ TCRs, with or without ligand, are, therefore, needed to elucidate molecular recognition mechanisms defining γδ TCR specificity, which are needed to fully understand γδ T cell-mediated immune responses.
Human γδ T cells are classified according to their Vδ gene segment use (Vδ1, Vδ2, or Vδ3) and vary little in terms of sequence among family members (9). T cells bearing Vδ1 receptors are associated with the intraepithelial lymphocyte (IEL) compartment, comprising 70–90% of the γδ T cells in the epithelium, and occur in increased frequencies among tumor-infiltrating lymphocytes (10). They are proposed to play an important role in tumor immunosurveillance, defense against viral infections, and tissue damage repair (10–13). The only specific ligands thus far identified for Vδ1 TCRs are CD1c molecules, independent of the presence of foreign lipid or glycolipid antigens (14), and the MHC class I-homologous MIC self-antigens (10, 15, 16). MIC antigens are induced in response to cellular stress on intestinal epithelium and in epithelially derived tumors (17, 18). Unlike MHC class I proteins, MIC proteins do not associate with β2-microglobin, antigenic peptides, or other ligands.
MIC proteins are also ligands for the stimulatory immunoreceptor NKG2D, broadly expressed on natural killer (NK), γδ T, and CD8 T cells in humans (10, 16, 19). The molecular-level details of the NKG2D:MIC interaction, including multiple MIC, NKG2D, and complex crystal structures, have been studied extensively (20–26). MIC proteins are polymorphic, with over 50 MICA alleles and at least 13 MICB alleles recognized (27). The functional significance of MIC gene polymorphisms is largely unknown, but it maps preferentially to the interface with NKG2D (21) and has been correlated with allelic differences in NKG2D receptor binding strengths (28). How MIC productively interacts with two structurally unrelated immunoreceptors, Vδ1 γδ TCRs and NKG2D, is unclear. The current model holds that MIC delivers both the TCR-dependent signal 1 and the NKG2D-dependent costimulatory signal 2 for activation of MIC-responsive Vδ1 γδ T cells (16). Whether MIC engagement by Vδ1 γδTCR and NKG2D can occur concurrently (and if concurrent, whether binding is cooperative or independent) is not known.
Here, we describe the crystal structure of an MIC-reactive Vδ1 γδ TCR (designated as δ1A/B-3; characterized clonally as Vγ1.4/Jγ2.3 and Vδ1/Jδ1) (10) determined at 3.0 Å resolution as a single-chain Fv (scFv) construct. While maintaining an overall fold similar to other γδ TCR structures, the structure revealed atypical CDR conformations, particularly in the CDR3 regions of both Vγ and Vδ domains. This structure suggests a receptor recognition mode that is substantially different from murine G8:T22 complex interaction, where human δ1A/B-3 likely uses germline-encoded CDR1δ and CDR2δ loops but not variable CDR3δ residues for MIC antigen recognition. Additionally, in sharp contrast to NKG2D:MIC interaction properties, binding between δ1A/B-3 (as an intact ectodomain construct) and MIC ligands was characterized by very low affinity [comparable with peptide MHC (pMHC):CD8 binding] (29) but exceptionally slow on- and off-rates, yielding long complex half-lives. Competitive binding experiments further showed that Vδ1 γδ TCR and NKG2D bind competitively, indicating the use of overlapping binding surfaces on MIC for recognition.
Results
Overall Architecture.
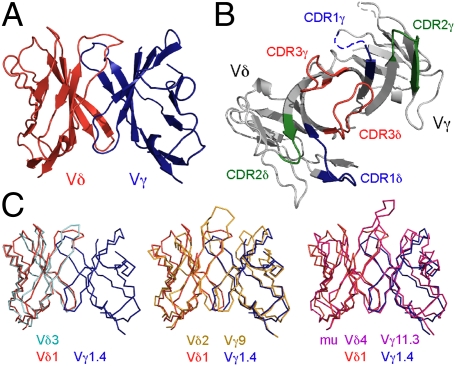
The crystal structure of an scFv construct of the human δ1A/B-3 TCR was determined at 3.0 Å resolution (Rwork = 0.265; Rfree = 0.308), with initial phases generated by multiwavelength anomalous diffraction methods (MAD) (Fig. 1 and Tables S1 and S2). The four molecules (designated A, C, E, and G) in the asymmetric unit of the crystal are nearly identical (pair-wise rmsds range from 0.42 to 0.58 Å on all common Cα atoms), although noncrystallographic symmetry (NCS) restraints were applied during the initial stages of refinement. Only partial models could be built for molecules E and G because of the poor quality of the electron density in various sections of the Fourier syntheses (83%, 81%, 66%, and 40% completeness for models A, C, E, and G, respectively) (Fig. S1); because of this, molecules A and C were used as the focus of the analysis and discussion here, unless otherwise stated. Molecules E and G were likely more disordered because of fewer anchoring contacts with neighboring molecules in the crystal lattice (Fig. S1). For example, density for the δ-chain of molecule G was nearly absent except for three of the β-strands in closest contact with its partner γ-chain. The disordered regions for molecules A and C were confined to the termini, the exogenous 15-residue linker introduced to generate the scFv construct, and the apexes of both the CDR1γ (five residues: 27-AEGST-31) and hypervariable region four (HV4) loops (two residues: 75-TR-76), consistent with conformational flexibility causing static disorder. Partial weak electron density for the missing residues of the CDR1γ and HV4 loops showed that the general positions and orientations of these elements were similar to other γδ TCR structures (Fig. 1B).
Fig. 1.
The overall structure of an MIC-reactive δ1A/B-3 and comparison with other γδ TCR structures. (A and B) Structure of δ1A/B-3 is shown in two orientations, viewing from the side (A) or top (CDR side; B). In B, the color codes are green for CDR1s (residues 26–35 for CDR1γ and 160–169 for CDR1δ), blue for CDR2s (residues 53–59 for CDR2γ and 185–189 for CDR2δ), red for CDR3s (residues 98–106 for CDR3γ and 228–239 for CDR3δ), and light gray for the rest of the molecule. Parts of the CDR1γ and HV4 loops that were not modeled are depicted as dotted lines. (C) Superposition of human Vδ2Vγ9 G115, Vδ3 δ-chain ES204, and murine G8 γδ TCRs onto human Vδ1 δ1A/B-3. Color codes are red for Vδ, blue or light gray for Vγ, orange for Vδ2, cyan for Vδ3 δ-chain, and magenta for G8.
Similar to αβ TCR and antibody structures, the global architecture of δ1A/B-3 scFv comprises two typical V-type Ig-like domains, well-conserved with the other known γδ TCR structures (Fig. 1C), with backbone maximum likelihood rmsdML values (30) of 0.126σ (Vδ3 ES204), 0.157σ (Vδ2Vγ9 G115), and 0.254σ (G8). The Vγ–Vδ interdomain angle between the long axis of the Vγ and the long axis of the Vδ was not significantly different from that observed in structures of other γδ TCRs.
CDR Conformations.
The CDR loops on the membrane-distal face of αβ TCRs comprise the site of ligand recognition; in the only example structurally characterized, the G8 CDR3δ loop dominates contacts with T22. Superposition of the δ1A/B-3 structure with known γδ TCR structures using maximum likelihood-based algorithms reveals dramatic conformational differences in CDR3 loops of both Vγ and Vδ domains (Fig. 2 and Figs. S2 and S3). The CDR3δ loops in both murine G8 and human G115 protrude by over 10 Å from the CDR surface. In the murine G8:T22 complex structure, G8 predominantly uses germline-encoded residues of CDR3δ to bind T22 in an orientation substantially different from that seen in αβ TCR:pMHC or antibody–antigen complexes (8); CDR3δ is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 (31). Such an unusual recognition mode may not be applicable to other γδ TCRs, particularly because CDR3δ regions show the most length variability (32).
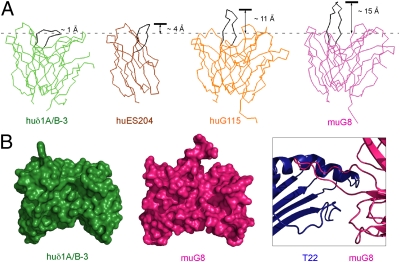
Fig. 2.
Comparison of γδ TCR CDR3δ loop conformations. (A) Three previously determined γδ TCR structures are superimposed onto the δ1A/B-3 structure and displayed in identical orientations. Color (and PDB codes) are green for δ1A/B-3, orange for G115 (1HXM), magenta for murine G8 (1YPZ), and brown for Vδ3 δ-chain ES204 (1TVD). CDR3δ loops are all highlighted in black. (B) Variable domains of human Vδ1 and murine G8 are displayed in the same orientation in a space-filling representation. The protrusion of CDR3δ in G8 relative to δ1A/B-3 is evident. Part of the G8:T22 complex is shown in Right, highlighting the role of CDR3δ in T22 recognition.
In contrast to the extended CDR3δ loop in G8 and G115, the shorter (12 vs. 18 residues) CDR3δ loop of δ1A/B-3 folds over, generating a nearly flat surface (Fig. 2). Examining the neighboring residues that interact with CDR3δ, Tyr33, Tyr103, and Tyr168 form hydrophobic interactions with Arg234, Thr233, and Leu232, respectively. Remaining interactions are less specific, involving the side chains of His35 and the backbone atoms of Arg234, Arg47, Ala235, Trp99, and Asp236, which form hydrogen bonds (Fig. S4). These six neighboring residues are not well-conserved in other classes of γδ TCRs with known structures. Crystal packing does not seem to play a significant role in forming such a compact conformation of CDR3δ, because each CDR3δ is isolated from intermolecular contacts in all four molecules in the asymmetric unit. CDR3γ is also less extended compared with corresponding loops in G8 and G115 receptor structures (Fig. S3). CDR1δ and CDR2δ in this structure project out more than those in the murine G8 and human G115 TCR structures but have similar conformations to those in the human Vδ3 ES204 structure. These CDR conformations together generate a nearly flat surface on the analog of the combining site, even taking into account partially disordered regions (Figs. 1 and 2).
Ig Domain Strand Swapping.
The position of CDR2δ varies among γδ TCR structures (Figs. S2 and S3) partly because of a strand-swapping event in both the human Vδ2 G115 and human Vδ3 ES204 structures, where the C″ strand hydrogen bonds to the C′ strand to form an inner β-sheet five strands wide rather than four strands wide as is seen for the murine G8 and the human δ1A/B-3 structures. In the latter two structures, the C″ strand hydrogen bonds with the D strand in the outer β-sheet. This led us to examine the pattern of strand swapping among Ig folds as CDR2 connects the C′ and C″ strands, and its global position and role in recognition can, thus, be affected by the packing of these β-strands. Although all variable Ig domains have a common structural core defined by a sandwich of antiparallel β-sheets, the positions of the edge strands relative to the common core can be variable (from the outside in, either a 5–4 or 4–5 composition) (33). The human δ1A/B-3 structure exhibits a mixed format such that the γ-chain is a 4–5 and the δ-chain is a 5–4. The majority of αβ TCRs have this same mixed format (α: 5–4, β: 4–5). In antibody structures, the C′ strand is associated with the C″ strand of the same β-sheet through a number of backbone–backbone hydrogen bonds (a 4–5 composition for both heavy- and light-chains), similar to the Vδ2Vγ9 G115. G8 exhibits the opposite 5–4 pairing for both γ- and δ-chains, which is seen in only two of the αβ TCRs (KB5-C20 and BM3.3) (34, 35). Because of this mixed-strand pairing, the position of CDR2 in the δ1A/B-3 is most similar to that seen in most αβ TCRs.
Strength and Stability of γδ TCR:MIC Interaction.
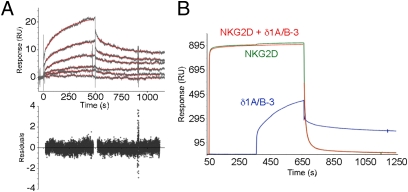
The affinity and kinetics of the δ1A/B-3:MIC interaction were quantified by surface plasmon resonance (SPR) analysis (Table 1) using an intact ectodomain version of δ1A/B-3. MICA was amine-coupled to a standard CM5 chip surface, and six analyte concentrations of δ1A/B-3 were injected in triplicate with randomly interspersed buffer blank injections. Representative sensorgrams and derived residuals are shown in Fig. 3. The dissociation constants (KD) of 110–900 μM at 22 °C for δ1A/B-3:MICA interactions are one to two orders of magnitude weaker than the NKG2D:MICA interaction, more on a par with MHC class I:CD8 interactions (from 11 to ≥1,000 μM) (29). Interactions between murine G8:T22 and human Vδ2Vγ9 γδ TCR (G25):apolipoprotein A–I pairs have much higher affinities of 67–113 nM and 0.81–1.5 μM, respectively (31). Kinetic analyses of δ1A/B-3:MIC interactions at room temperature yielded an association rate constant (kon) of 5–53 M−1·s−1 and a dissociation rate constant (koff) of 0.004–0.005 s−1, the latter corresponding to a complex half-life (τ1/2) of 140–170 s. NKG2D:MICA and TCR:ligand interactions typically have kon values of 1 × 103 to 1 × 105 M−1·s−1 and τ1/2 values of only a few seconds to tens of seconds (21).
Table 1.
Immunoreceptor to ligand interaction parameters
| Receptor | Ligand | kon (M−1·s−1) | koff (s−1) | τ1/2 (s) | KDcalc (μM) | KDeq (μM) | Reference |
| δ1A/B-3 | MICA*001 | 53 | 0.004 | 170 | 110 | — | This study |
| δ1A/B-3 | MICA*001 platform only | 24.3 | 0.004 | 170 | 180 | — | This study |
| δ1A/B-3 | MICA*004 platform only | 9.0 | 0.004 | 170 | 440 | — | This study |
| δ1A/B-3 | MICB*005 | 5.0 | 0.005 | 140 | 900 | — | This study |
| δ1A/B-3 | MICB*005 platform only | 9.0 | 0.004 | 170 | 450 | — | This study |
| Vδ2Vγ9 G25 | Apo A-I | 88 | 0.007 | 97 | 0.81–1.5 | — | 54 |
| G8 γδ TCR | T22 | 2.5 × 105 | 0.017 | 41 | 0.067–0.113 | — | 31 |
| αβ TCR | pMHC Iα | 6 × 102 to 2 × 105 | 0.02–0.2 | 3–35 | — | 0.1–90 | 36 |
| NKG2D | MICA*001 | 4.3 × 104 | 0.013 | 53 | 0.3 | 0.6 | 21 |
| NKG2D | MICA*004 | 1.2 × 104 | 0.24 | 2.9 | 21 | 21 | This study |
Fig. 3.
SPR kinetic analysis of δ1A/B-3:MICA and competitive binding in δ1A/B-3:NKG2D:MICA interactions. (A) SPR sensorgrams of δ1A/B-3:MICA binding are shown, with TCR analyte concentrations of 5, 2.5, 1, 0.5, 0.25, and 0.1 μM. Fitted binding curves are shown as red traces, and corresponding fitting residuals are shown below. (B) SPR-based analysis of the competition between NKG2D and δ1A/B-3 for binding to MICA. Sensorgrams from different channels are colored as blue for δ1A/B-3, green for NKG2D alone, and red for NKG2D (first 5 min) and NKG2D plus δ1A/B-3 (after the first 5 min).
Competitive Binding by TCR and NKG2D to MICA.
The interaction of human intestinal epithelial Vδ1 γδ T cells with MICA- and MICB-expressing target cells involves an epitope or epitopes in the MICA α1α2 platform domain as shown by antibody-blocking studies (10); NKG2D also directly binds the MIC platform domain (21). An SPR-based competition assay was used to determine whether δ1A/B-3 and NKG2D directly compete for binding sites on MIC (Fig. 3B). Saturating concentrations of NKG2D were injected into a flow cell containing amine-coupled MICA and were allowed to come to equilibrium. The analyte was then switched to a mixture of NKG2D (at the same saturating concentration) and the intact ectodomain version of δ1A/B-3. If TCR and NKG2D bound to distinct MICA sites, then an additional binding signal would be observed, comparable with the signal of TCR binding in the absence of NKG2D. If TCR and NKG2D bind to overlapping (or sterically clashing) sites, then the binding response would not change after switching to the mixed analyte. The sensorgrams clearly show the latter behavior, indicating that δ1A/B-3 and NKG2D use overlapping binding surfaces on MICA. These data also provide additional evidence for the specificity of the interaction between δ1A/B-3 and MIC protein; the TCR shows binding responses only to the specific surface on MICA blocked by NKG2D binding and not to random sites left exposed on MICA or NKG2D.
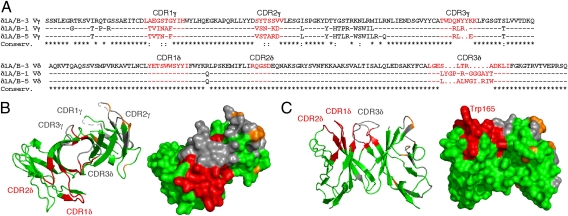
Vδ1 γδ T cells specific for MIC have been shown to recognize a diverse set of nonhuman primate MIC molecules (28), but attempts to use patches of conservation to define potential NKG2D binding sites before the determination of the complex structure were not successful, with the most conserved regions mapping to the opposite face of the MICA platform domain (6, 20). The competition studies argue that these sites are not TCR binding sites, and therefore, use of MIC conservation maps is unlikely to reveal candidate binding sites. MIC-reactive Vδ1 γδ TCR clones have a high degree of sequence conservation overall (83% identity or 86% homology), consistent with conserved use of Vδ1 gene segments. The most variable regions, not surprisingly, are located in the four γ-chain hypervariable regions and CDR3δ in Vδ (Fig. 4A). CDR1δ and CDR2δ are completely conserved, arguing that these loops may play a more important role in MIC recognition (Fig. 4B).
Fig. 4.
MIC-reactive Vδ1 γδ TCR sequences and CDR surface conservation mapped onto the δ1A/B-3 structure. (A) Amino acid sequences of three MIC-responsive Vδ1 γδ TCR clones are shown, designated as δ1A/B-1, δ1A/B-3, and δ1A/B-5 (10), with CDR loops highlighted in red. (B and C) Two views of the δ1A/B-3 structure are shown as ribbons (Left) or molecular surfaces (Right). Identical CDR residues are colored red, identical framework residues are colored green, conservatively substituted residues are colored orange throughout, and nonconserved residues are colored gray throughout.
Discussion
CDR loops, particularly CDR3, are functionally critical for antigen recognition in antibodies and αβ TCRs (reviewed in refs. 5 and 36). In these cases, the relatively straight-on docking mode results in multipoint (i.e., multi-CDR) attachment of the receptor to the ligands. In most cases, such fixed interactions use CDR1 and CDR2 loops to provide a perimeter of contacts surrounding a central region, where CDR3 loops provide major binding interactions. An analysis of the CDR3 length distribution of antibodies and TCRs has revealed that the length profiles of γδ TCRs more closely resemble those of antibodies than αβ TCRs in terms of having long and variable CDR3δs and short and constrained CDR3γs (32). It is conceivable that constrained length distribution of αβ TCR CDR3s may reflect a functional requirement for the αβ TCRs to contact both the MHCs and the presented peptides. The murine G8:T22 structure, the only complex γδ TCR structure available so far, however, presented a very unusual side-on recognition strategy with G8, which almost exclusively uses its CDR3δ loop to bind T22 from the side, with minor contacts from CDR3γ. In human δ1A/B-3, the fold-over conformation of CDR3δ essentially precludes a similar recognition strategy. The relatively flat surface overlying the δ1A/B-3 CDRs suggests that such a functionally important surface may be more likely to accommodate a straight-on docking mode as seen in many examples in antibody–antigen binding, αβ TCR:pMHC recognition, and other protein–protein interaction. However, the ligand docking footprint may be skewed more to the area defined by the surfaces of CDR1δ and CDR2δ based on sequence conservation and away from the central region defined by the surface of the CDR3δ loops. Consistent with this hypothesis, the flatness of the δ1A/B-3 CDR3 loops allows parts of the neighboring CDR1δ and CDR2δ loops to protrude and define a more obvious structural feature for engaging ligands (Fig. 4C). The most protuberant residues include Trp165, Ser166, and Tyr168 of CDR1δ and Asp189 of CDR2δ. Trp165 of CDR1δ, in particular, extends into solvent in all four molecules in the structure. It is possible that this residue plays an important role in MIC recognition, and there are shape- and binding-complementary pockets on the surface of the MIC platform domain. However, in silico docking studies have failed to produce convincing models of a complex, partly because of extensive hard-to-model flexible regions in both the TCR and MIC. Likewise, mutation of Trp165 to test effects on the observed interaction parameters with MIC ligands has also failed because of the solution instability of the mutant. However, a previous SPR-based binding study showed the importance of the δ-chain in MICA binding but also that CDR3δ loops only make minor contributions (37), consistent with our conclusions here. The strength of a Vδ1 γδ TCR:MICA interaction reported here was significantly weaker than estimated by Zhao et al. (37); although the dissociation rates are comparable, the KD values vary by two orders of magnitude (hundreds of micromolar here vs. 2 μM in the previous study) primarily because of differences in association rates of 50- to 500-fold. These on-rate differences could result from different degrees of conformational change during binding, the sequence differences between the two TCRs, or a combination of both.
SPR-based interaction analyses revealed strikingly different binding kinetics and affinities for TCR:MIC vs. NKG2D:MIC interactions. MIC ligand proteins bind to NKG2D receptors with affinities (submicromolar KD) that are nearly 1,000-fold stronger than TCR:MIC interactions (hundreds of micromolar KD), which are explained by kon values for NKG2D:MIC binding that are over two orders of magnitude faster than δ1A/B-3:MIC interactions. However, although weak, TCR:MIC complexes, after they are formed, show unusual stability, with long complex half-lives. We show here that both receptors compete for binding to MIC ligands. Because δ1A/B-3 and NKG2D binding to MIC ligands is completely mutually exclusive, γδ T-cell activation must either (i) be in response to sufficient levels of MIC ligands on the surface of target cells to simultaneously engage both stimulatory and costimulatory receptors or (ii) accommodate sequential engagement of receptors. In the latter case, these data suggest a temporally ordered model for formation of hypothetical T-cell/target cell synapses, with implications for signaling mechanisms: initial interactions at the point of contact may be dominated by NKG2D:MIC binding events, which then give way to longer-lived γδ TCR:MIC complexes.
Materials and Methods
Protein Expression and Purification.
Two soluble forms of an MIC-reactive human Vδ1 γδ TCR (designated as δ1A/B-3) were produced as intact ectodomain and scFv constructs. For crystallization trials, the scFv version of δ1A/B-3 was generated using overlapping PCR, with the variable domains of the γ- and δ-chains [gene access numbers are AF025414 (γ) and AF025423 (δ)] connected through a linker sequence (GSADDAKKDAAKKDG) analogous to constructs used for scFv antibodies and αβ TCRs (38). The scFv construct was expressed in bacteria and refolded in vitro from purified inclusion bodies (20, 23). For binding studies, the extracellular domains of the γ- and δ-chains of δ1A/B-3 were expressed separately in bacteria and corefolded as heterodimer complexes using the same protocol. The intact ectodomain version was used for binding studies, because it represented a more physiological construct and does not undergo monobody/diabody equilibration seen with some antibody and TCR scFv constructs; the scFv version was produced because the intact ectodomain version failed to crystallize. Ectodomains of MICA*001, MICB*005, and MICA*004 and respective platform-only constructs (MICA*001 residues 1–180, MICB*005 residues 1–180, and MICA*004 residues 1–180; numbered as in refs. 20 and 23) were all refolded in vitro (20, 23). Proper folding was confirmed by solution monodispersity of the purified protein after concentration, as determined by size-exclusion chromatography (SEC) and reduced/nonreduced SDS/PAGE analysis. A selenomethionine (SeMet) derivative of the δ1A/B-3 scFv was prepared as described (39) and refolded separately for use in MAD phasing. SeMet incorporation was confirmed by MS.
Crystallization and Crystallography.
The δ1A/B-3 scFv was crystallized by hanging-drop vapor diffusion at 22 °C from a solution containing 17.0 mg/mL protein, 25 mM Pipes (pH 7.0), 1 mM EDTA, and 0.02% NaN3 mixed at a ratio of 1:1 with a well solution of 0.4 M (NH4)2SO4, 1.0 M Li2SO4, 0.1 M sodium citrate, and 0.03 M glycyl-glycyl-glycine. The space group of the crystals was P43 (a = b = 112.4 Å and c = 108.1 Å) with four molecules per asymmetric unit, which resulted in a Vm of 3.0 Å3/Da (40). Diffraction data from native and SeMet crystals were collected at Advanced Light Source (ALS) beamline 5.0.2 or National Synchrotron Light Source (NSLS) beamline X29 and reduced with HKL2000 (41). MAD phases were calculated from 10 of 16 possible selenium sites per asymmetric unit, located using program SHELXD in HKL2MAP (42, 43). Initial phases were significantly improved with NCS electron density averaging using RESOLVE (44) and SHARP (SHARP/autoSHARP; Global Phasing Limited). Using part of known γδ TCR backbone structures (PDB codes 1HXM and 1YPZ) as templates, initial models were built using Molrep (45), Coot (46), and Xfit (47). A series of iterative geometry runs and group B-factor refinements were performed with programs in the Crystallography & NMR System suite (48); NCS restraints were relaxed. The final stages of refinement were carried out with the refinement program refmac5 (45), where individual B factors were refined and NCS restraints were not applied. The four molecules were partitioned into seven tls groups based on analysis by the TLSMD server (49); each scFv consisted of two tls groups except for the fourth molecule, which comprised just one group. The δ-chain of the fourth molecule was too disordered to confidently model except for three of the β-strands in closest contact with the γ-chain. Based on solvent content calculations and the packing of the modeled chains, there may well be a fifth molecule in the asymmetric unit, but if so, it is too disordered to see in the electron density map. Validation of the model was carried out with PROCHECK (50), MolProbity (51), and ADIT (http://deposit.rcsb.org/adit/). Data collection, phasing, and refinement statistics for the model are shown in Tables S1 and S2, and the final model has been deposited in the Research Collaboratory for Structural Bioinformatics PDB (code 3OMZ). Macromolecular structure alignment was performed by using maximum likelihood superposition, as implemented in THESEUS (30); sequence similarity and structural neighborhood searches were performed using the blastp and Dali servers, respectively (52, 53).
Interaction Analysis.
Binding parameters were determined by SPR in 10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% P-20 surfactant (HBS-EP buffer) at 22 °C with a Biacore 3000 or T100 instrument (GE Healthcare Bio-Sciences). δ1A/B-3 analytes were flowed over MIC ligands amine-coupled to CM5 sensor chips following the manufacturer's protocol. The sensor chip was regenerated after each injection with a 20-μL pulse of 10 mM NaOH. The quality of the MIC surface was monitored by determining the NKG2D dissociation constant, ensuring that regeneration was well-tolerated by the ligands. The proteins used in the SPR experiments were purified within 24 h by SEC in HBS-EP buffer. Each experiment was run in triplicate, and each concentration series was randomized independently with interspersed injections of buffer and NKG2D. SPR data were analyzed with BiaEvaluation 3.0 (GE Healthcare Bio-Sciences) or Scrubber 2 (v2.0a; Scrubber 2). All of the sensorgrams were double reference subtracted (54), and the data were fit to a simple 1:1 Langmuir isotherm model. The binding competition analysis used a coinjection procedure where a saturating concentration at 10 μM NKG2D analyte was flowed over an MIC-coupled chip until equilibration followed by an analyte mixture of 10 μM δ1A/B-3 plus 10 μM NKG2D analyte mixture. Flow rates were 20 μM/min; MIC ligands were coupled at a density of 3,000 response units (RU).
Supplementary Material
Acknowledgments
We thank NSLS X29 Beamline staff members, particularly Dr. Annie Heroux, for their expert assistance in MAD data collection and initial phasing. This work was supported in part by National Institutes of Health Grant AI48675 (to R.K.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3OMZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015433108/-/DCSupplemental.
References
- 1.Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Hayday AC. [gamma][delta] cells: A right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 3.Boismenu R, Havran WL. An innate view of gamma delta T cells. Curr Opin Immunol. 1997;9:57–63. doi: 10.1016/s0952-7915(97)80159-8. [DOI] [PubMed] [Google Scholar]
- 4.Chien YH, Konigshofer Y. Antigen recognition by gammadelta T cells. Immunol Rev. 2007;215:46–58. doi: 10.1111/j.1600-065X.2006.00470.x. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 6.Li H, et al. Structure of the Vdelta domain of a human gammadelta T-cell antigen receptor. Nature. 1998;391:502–506. doi: 10.1038/35172. [DOI] [PubMed] [Google Scholar]
- 7.Allison TJ, Winter CC, Fournié JJ, Bonneville M, Garboczi DN. Structure of a human gammadelta T-cell antigen receptor. Nature. 2001;411:820–824. doi: 10.1038/35081115. [DOI] [PubMed] [Google Scholar]
- 8.Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- 9.Thedrez A, et al. Self/non-self discrimination by human gammadelta T cells: Simple solutions for a complex issue? Immunol Rev. 2007;215:123–135. doi: 10.1111/j.1600-065X.2006.00468.x. [DOI] [PubMed] [Google Scholar]
- 10.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 11.Janeway CA, Jr., Jones B, Hayday A. Specificity and function of T cells bearing gamma delta receptors. Immunol Today. 1988;9:73–76. doi: 10.1016/0167-5699(88)91267-4. [DOI] [PubMed] [Google Scholar]
- 12.Halary F, et al. Shared reactivity of Vdelta2(neg) gammadelta T cells against cytomegalovirus-infected cells and tumor intestinal epithelial cells. J Exp Med. 2005;201:1567–1578. doi: 10.1084/jem.20041851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boismenu R, Havran WL. Modulation of epithelial cell growth by intraepithelial gamma delta T cells. Science. 1994;266:1253–1255. doi: 10.1126/science.7973709. [DOI] [PubMed] [Google Scholar]
- 14.Spada FM, et al. Self-recognition of CD1 by gamma/delta T cells: Implications for innate immunity. J Exp Med. 2000;191:937–948. doi: 10.1084/jem.191.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groh V, et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Groh V, Spies T. T cell antigen receptor engagement and specificity in the recognition of stress-inducible MHC class I-related chains by human epithelial gamma delta T cells. J Immunol. 2002;169:1236–1240. doi: 10.4049/jimmunol.169.3.1236. [DOI] [PubMed] [Google Scholar]
- 17.Groh V, et al. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci USA. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diefenbach A, Raulet DH. The innate immune response to tumors and its role in the induction of T-cell immunity. Immunol Rev. 2002;188:9–21. doi: 10.1034/j.1600-065x.2002.18802.x. [DOI] [PubMed] [Google Scholar]
- 19.Cerwenka A, Lanier LL. Ligands for natural killer cell receptors: Redundancy or specificity. Immunol Rev. 2001;181:158–169. doi: 10.1034/j.1600-065x.2001.1810113.x. [DOI] [PubMed] [Google Scholar]
- 20.Li P, et al. Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand. Immunity. 1999;10:577–584. doi: 10.1016/s1074-7613(00)80057-6. [DOI] [PubMed] [Google Scholar]
- 21.Li P, et al. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 22.Steinle A, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 23.Holmes MA, Li P, Petersdorf EW, Strong RK. Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D. J Immunol. 2002;169:1395–1400. doi: 10.4049/jimmunol.169.3.1395. [DOI] [PubMed] [Google Scholar]
- 24.Li P, McDermott G, Strong RK. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity. 2002;16:77–86. doi: 10.1016/s1074-7613(02)00258-3. [DOI] [PubMed] [Google Scholar]
- 25.McFarland BJ, Kortemme T, Yu SF, Baker D, Strong RK. Symmetry recognizing asymmetry: Analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure. 2003;11:411–422. doi: 10.1016/s0969-2126(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 26.McFarland BJ, Strong RK. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: Not induced fit but rigid adaptation. Immunity. 2003;19:803–812. doi: 10.1016/s1074-7613(03)00320-0. [DOI] [PubMed] [Google Scholar]
- 27.Stephens HA. MICA and MICB genes: Can the enigma of their polymorphism be resolved? Trends Immunol. 2001;22:378–385. doi: 10.1016/s1471-4906(01)01960-3. [DOI] [PubMed] [Google Scholar]
- 28.Steinle A, Groh V, Spies T. Diversification, expression, and gamma delta T cell recognition of evolutionarily distant members of the MIC family of major histocompatibility complex class I-related molecules. Proc Natl Acad Sci USA. 1998;95:12510–12515. doi: 10.1073/pnas.95.21.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao GF, Jakobsen BK. Molecular interactions of coreceptor CD8 and MHC class I: The molecular basis for functional coordination with the T-cell receptor. Immunol Today. 2000;21:630–636. doi: 10.1016/s0167-5699(00)01750-3. [DOI] [PubMed] [Google Scholar]
- 30.Theobald DL, Wuttke DS. THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams EJ, Strop P, Shin S, Chien YH, Garcia KC. An autonomous CDR3delta is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by gammadelta T cells. Nat Immunol. 2008;9:777–784. doi: 10.1038/ni.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 34.Housset D, et al. The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain. EMBO J. 1997;16:4205–4216. doi: 10.1093/emboj/16.14.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiser JB, et al. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 36.Davis MM, et al. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 37.Zhao J, Huang J, Chen H, Cui L, He W. Vdelta1 T cell receptor binds specifically to MHC I chain related A: Molecular and biochemical evidences. Biochem Biophys Res Commun. 2006;339:232–240. doi: 10.1016/j.bbrc.2005.10.198. [DOI] [PubMed] [Google Scholar]
- 38.Bird RE, et al. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 39.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 40.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 43.Pape T, Schneider TR. HKL2MAP: A graphical user interface for phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 44.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 48.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 49.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 50.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 51.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 55.Scotet E, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity. 2005;22:71–80. doi: 10.1016/j.immuni.2004.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.