Abstract
Cancer biomarkers are currently the subject of intense research because of their potential utility for diagnosis, prognosis, and targeted therapy. In theory, the gene products resulting from somatic mutations are the ultimate protein biomarkers, being not simply associated with tumors but actually responsible for tumorigenesis. We show here that the altered protein products resulting from somatic mutations can be identified directly and quantified by mass spectrometry. The peptides expressed from normal and mutant alleles were detected by selected reaction monitoring (SRM) of their product ions using a triple-quadrupole mass spectrometer. As a prototypical example of this approach, we demonstrated that it is possible to quantify the number and fraction of mutant Ras protein present in cancer cell lines. There were an average of 1.3 million molecules of Ras protein per cell, and the ratio of mutant to normal Ras proteins ranged from 0.49 to 5.6. Similarly, we found that mutant Ras proteins could be detected and quantified in clinical specimens such as colorectal and pancreatic tumor tissues as well as in premalignant pancreatic cyst fluids. In addition to answering basic questions about the relative levels of genetically abnormal proteins in tumors, this approach could prove useful for diagnostic applications.
Keywords: genetics, proteomics, pancreatic cancer, genetic diagnosis, companion diagnostics
Through genome-wide analysis, it has been shown recently that solid tumors typically contain 20–100 protein-encoding genes that are mutated (1–4). A small fraction of these changes are “drivers” that are responsible for the initiation or progression of the tumors; the remainder are “passengers,” providing no selective growth advantage (5, 6). In principle, these proteins provide unparalleled opportunities for biomarker development. Unlike other protein biomarkers such as carcinoembryonic antigen (CEA) or prostate-specific antigen (PSA), the mutant proteins are produced only by tumor cells. Moreover, they are not simply associated with tumors, as are conventional markers, but in the case of driver gene mutations are directly responsible for tumor generation.
The detection of the proteins encoded by mutated genes (henceforth termed “mutant proteins”) is straightforward when proteins are truncated by a nonsense mutation or fused to other proteins. This detection often can be accomplished simply by Western blotting of cellular extracts. However, a large number of disease-causing mutations are missense mutations that alter the encoded proteins only subtly. For example, in recent studies of the sequences of all protein-encoding genes in human cancers, >80% of the somatic mutations were reported to be missense (1–3). Although it is theoretically possible to detect these abnormal proteins directly with antibodies directed against mutant epitopes, doing so has been difficult to accomplish in practice. For example, although v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) and tumor protein p53 (TP53) are two of the most commonly mutated and intensely studied cancer genes, there still are no antibodies that can reliably distinguish mutant from normal versions of these proteins. Because many different mutations can occur in a single cancer-related gene, it is necessary to develop a specific antibody for each possible mutant epitope, compounding the difficulty of this strategy. Another approach measures the activity of mutant proteins. Although this approach can be useful in special situations, it is not generally applicable because no activity-based assays are available for most proteins, and the proteins resulting from mutated genes often have activities that are only quantitatively, rather than qualitatively, different from their normal counterparts. Thus, there is a critical need for assays that would permit quantification of mutant proteins in a generic fashion.
Recent advances in MS permit sampling of a large fraction of normal and abnormal cellular proteomes in an unbiased and specific fashion (7, 8). MS already has become the method of choice for quantifying protein levels, and a number of quantitative proteomics strategies for this purpose have been described (9–14). Interestingly, MS already has been used to detect and precisely quantify somatic mutations at the DNA level but not at the protein level (15). Indeed, one of the most widely used methods for quantifying such mutations in DNA relies on the measurement of the mass of oligonucleotides differing at a single base (16). Prior studies have shown that it is possible to identify posttranslationally altered proteins using MS, as well as to identify highly abundant abnormal proteins, such as those responsible for amyloidosis (17–22). In this work, we sought to develop an MS approach that could identify and quantify somatically mutant proteins in a generally applicable fashion. We were particularly interested in working out a strategy that could be applied to complex biological samples such as those encountered clinically.
Results
Although MS-based technologies are capable of detecting attomole quantities of proteins (23), their sensitivity can be compromised by many factors, including sample preparation and the biochemical complexity of clinical specimens (24). For this reason, the work described here involved the implementation of two independent components: enrichment of the protein of interest and the targeted analysis of peptides derived from this protein.
Enrichment of Proteins for Selected Reaction Monitoring Experiments.
Among the available methods for enrichment of proteins, we chose immunoprecipitation (IP) for several reasons. First, antibodies have been generated against most proteins of interest, and selected reaction monitoring (SRM) does not require the antibodies to be absolutely specific for the antigens or specific for the mutations of interest; this specificity comes from the subsequent MS analysis. Second, IP removes the most abundant proteins, including cytoskeletal proteins, immunoglobulins, and serum albumin, from biological samples (25, 26). Third, it is scalable and can be applied readily to samples containing large volumes or high concentrations of irrelevant proteins.
We used cancer cells in culture to optimize the IP methods, with the protein encoded by KRAS as the target. The KRAS gene is commonly mutated in human colorectal and pancreatic cancers, with most mutations clustered at residues 12 or 13 of the protein. Several methods for lysing cells and capturing Ras proteins were explored to obtain the great majority of the Ras protein in a form compatible with subsequent MS analysis. We found that cell lysis in a detergent-containing buffer followed by binding to antibody-coupled magnetic beads achieved these goals (Materials and Methods and ref. 25). Covalent coupling of the antibody to magnetic beads was performed using dimethyl pimelimidate. After the antigen was bound to the immobilized antibodies, Ras was eluted and concentrated. Of the elution methods tried (various concentrations of acids, bases, glycine, detergents, and denaturants at various temperatures and times), we found that 3% (vol/vol) acetic acid most reproducibly eluted Ras proteins in a fashion that facilitated subsequent protease digestion.
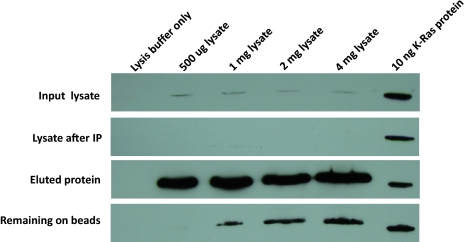
This experimental scheme for IP (Fig. 1) was applied to the human colorectal cancer cell line SW480, one of the cell lines in which KRAS mutations were identified originally (27). Analysis of the IP results by Western blotting with an antibody that reacts with K-Ras is shown in Fig. 2. There was a linear relationship between the amount of cellular protein used for IP and the amount of K-Ras protein eluted from the beads when up to 4 mg of total protein (5.6 million cells) was used as starting material. As assessed by densitometry of the Ras-specific band, >90% of the total cellular K-Ras protein was captured successfully from the lysates and eluted from the beads.
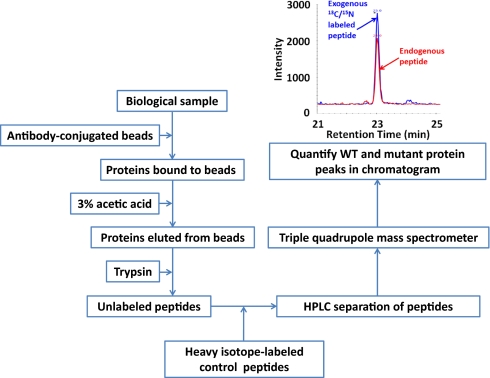
Fig. 1.
Schematic of the approach.
Fig. 2.
IP of Ras proteins. An antibody directed against a common epitope of all three forms of mutant and WT forms of Ras (K-Ras, N-Ras, and H-Ras) was used to immunoprecipitate the indicated amounts of protein in SW480 cell lysates. Western blots were performed using an HRP-conjugated monoclonal antibody to K-Ras. Ten nanograms of recombinant K-Ras protein were loaded on the right-most lane of each gel for comparison purposes. The “input lysate” and “lysate after IP” lanes contained 4% of the proteins used for IP; all the eluted protein and protein remaining on beads were loaded into the corresponding lanes.
MS Optimization.
SRM is becoming the method of choice for selective detection of specific proteins in complex samples (28). Classic liquid chromatography (LC)-MS/MS experiments scan a large mass range to characterize proteins in cellular extracts comprehensively. In contrast, SRM monitors only a small number of preselected ions, greatly increasing the sensitivity of detection.
In SRM, the output fractions from LC are directed to a triple-quadrupole instrument by electrospray. The first and third quadrupoles act as filters to monitor predefined m/z values corresponding to the peptides of interest, and the second quadrupole acts as a collision cell to fragment the parent peptide. Generally, two to four product ions are monitored in the third quadrupole for each peptide molecular ion in the first quadrupole. The simultaneous appearance of the product ions at the same LC retention time provides exquisite specificity. The approach is analogous to that used for monitoring small molecules, widely applied in pharmacokinetic and toxicologic studies (29).
Heavy isotope-labeled synthetic peptides can serve as internal controls for such experiments, increasing the confidence of identification and facilitating absolute quantification (9,30, 31). We therefore synthesized peptides labeled at their C terminus with 13C/15N-lysine or 13C/15N -arginine as internal controls. Based on MS analysis of these synthetic peptides, as well as control experiments with unlabeled synthetic peptides, the best fragments (transitions) for monitoring were chosen for further analysis. A complete list of parent and product ions that were used for SRM, together with their optimal collision energies and m/z ratios, is provided in Table S1. These peptides included those representing trypsinized normal (WT) Ras protein as well as the two most common mutants of Ras in pancreatic cancers, K-Ras G12V and G12D.
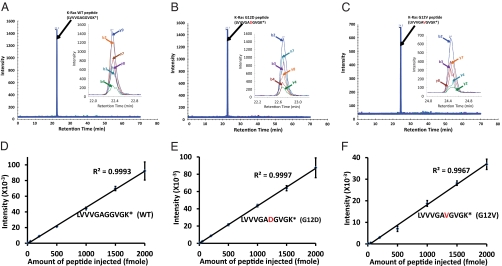
Chromatograms of the MS data obtained with synthetic peptides representing the WT and mutant Ras proteins are shown in Fig. 3 A–C. In all these experiments, chromatographic elution times of the product ions from the 13C/15N heavy isotope-labeled and unlabeled synthetic peptides were identical. The summed peak intensities for the ions corresponding to the heavy and light versions of peptides representing WT and mutant proteins showed that they were related linearly to abundance across more than two orders of magnitude (10–2,000 fmol; R2 > 0.99 for WT and mutant proteins) (Fig. 3 D–F). The variation from experiment to experiment was very small, with coefficients of variation less than 10% even for the smallest amounts of peptide used (Fig. 3 D–F).
Fig. 3.
Extracted ion chromatograms of 13C/15N-labeled synthetic peptides. The retention times of the indicated peptides are shown above the peaks in A–C, and the insets at the right of each panel represent an expanded view. Asterisks indicate the heavy isotope (13C6/15N2)-labeled lysine. D–F illustrate the relationship between the amount of peptide injected into the mass spectrometer and the integrated intensity of the transitions. The b and y peaks indicate the detected intensities of b ions and y ions (as designated in Table S1).
Analysis of Cultured Cells.
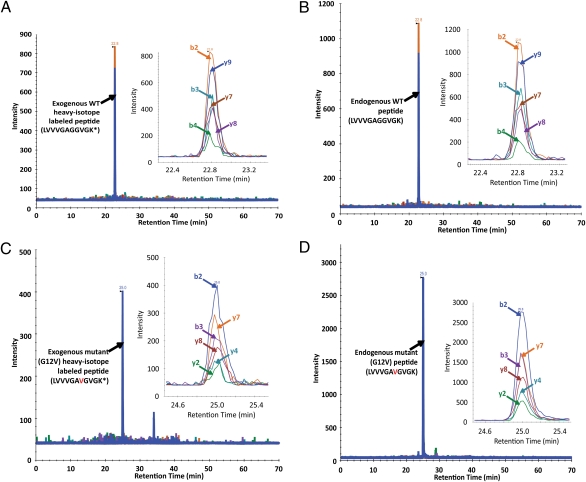
We next applied the complete procedure described in Fig. 1 to SW480 colorectal cancer cells growing in culture. Endogenous WT Ras protein was quantified by spiking a known amount (1 pmol) of heavy isotope-labeled synthetic peptide into the endogenous peptide mixture following IP. A chromatogram of selected product ions of the WT Ras synthetic peptide LVVVGAGGVGK(13C615N2) is shown in Fig. 4A. A chromatogram of the selected product ions of the corresponding unlabeled peptide from the endogenous WT Ras protein present in the cells is shown in Fig. 4B. By comparing the intensity of the MS signal of peptide from endogenous Ras protein with that of the spiked heavy isotope-labeled peptide, the amount of Ras protein was estimated to be 1.6 ± 0.22 pmol per 2 mg of cell lysate protein, corresponding to 1.5 ± 0.20 million molecules of WT Ras protein per cell.
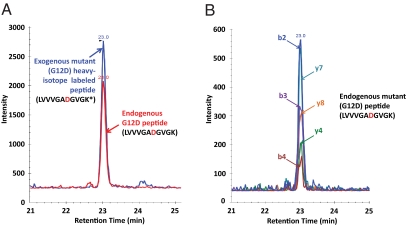
Fig. 4.
SRM of endogenous proteins from SW480 cells. (A and B) Extracted ion chromatograms of transitions from (A) exogenously added heavy isotope-labeled WT peptide and (B) corresponding endogenous WT peptide, illustrating the identical retention times. (C and D) Extracted ion chromatograms of the (C) exogenous and (D) endogenous mutant peptides. In each panel, the inset at the right represents an expanded view of the major peaks. Asterisks indicate the heavy isotope (13C6/15N2)-labeled lysine.
The SW480 cell line is known to harbor a KRAS G12V mutation (27). Chromatograms representing a known amount (1 pmol) of spiked peptide LVVVGAVGVGK(13C615N2) and unlabeled endogenous G12V-containing peptides are shown in Fig. 4 C and D, respectively. By comparison with the internal control peptides, the ratio of mutant to WT Ras protein was calculated to be 5.6, and no signals corresponding to the other tested mutations (G12D and G13D) were detectable in these cells (Table 1).
Table 1.
Levels of WT and mutant Ras proteins in cells and tissues (pmoles/2 mg cellular protein)
| WT Ras LVVVGAGGVGK | G12V Ras LVVVGAVGVGK | G12D/G13D Ras LVVVGADGVGK/ LVVVGAGDVGK | Mutant/WT Ras | |
| Cancer cell lines | ||||
| SW480 | 1.6 ± 0.28 | 9.0 ± 2.1 | 0 | 5.6 ± 0.28 |
| Pa08C | 2.6 ± 0.49 | 4.1 ± 0.57 | 0 | 1.7 ± 0.071 |
| Pa16C | 1.8 ± 0.14 | 0 | 0.88 ± 0.021 | 0.49 ± 0.028 |
| Pa02C | 2.5 ± 0.14 | 0 | 0 | NA |
| Normal tissues | ||||
| Spleen 10 | 3.5 ± 0.4 | 0 | 0 | NA |
| Spleen 12 | 4.1 ± 0.22 | 0 | 0 | NA |
| Colorectal mucosa 102 | 2.6 ± 0.21 | 0 | 0 | NA |
| Colorectal mucosa 104 | 3.2 ± 0.11 | 0 | 0 | NA |
| Tumor tissues | ||||
| CRC 2640 | 0.87 ± 0.31 | 0 | 0.60 ± 0.19 | 0.70 ± 0.025 |
| CRC 2966 | 1.3 ± 0.17 | 0 | 0.35 ± 0.040 | 0.28 ± 0.026 |
| CRC 3106 | 2.0 ± 0.25 | 0.87 ± 0.10 | 0 | 0.45 ± 0.010 |
| CRC 3107 | 1.6 ± 0.28 | 1.2 ± 0.22 | 0 | 0.79 ± 0.056 |
| CRC 3108 | 1.9 ± 0.74 | 0 | 0.47 ± 0.080 | 0.28 ± 0.090 |
| Pancreatic cyst fluids | ||||
| Cyst 3950 | 0.17 | 0.18 | 0 | 1.1 |
| Cyst 10592 | 0.15 | 0 | 0.18 | 1.2 |
| Cyst 8296 | 0.47 | 0 | 0 | NA |
NA, not applicable.
To determine whether the amounts or ratios of the WT and mutant peptides were dependent on the amount of cell lysate used in SRM, we varied the input from 0.5 mg (0.7 million cells) to 4 mg (5.6 million cells) per lysate. The amounts of both WT and mutant Ras proteins were linearly related to the input, as expected (R2 > 0.98) (Fig. S1). Importantly, the ratio of mutant to WT Ras proteins was 5.0 and was independent of the amount of input protein. This result is consistent with previous reports showing that the majority of KRAS mRNA transcripts in SW480 cells contain the G12V mutation (27).
To assess the efficiency of the combined steps involved in our approach, we added known amounts of WT K-Ras proteins to cells before performing the procedure. The WT protein was produced in vitro using a wheat germ extract. We found that 22.4 ± 1.4% of the input K-Ras protein was recovered in the MS analysis (Fig. S2). Using this correction factor, we calculated that there were an average of 1.5 and 8.6 million molecules of WT and mutant Ras proteins, respectively, per SW480 cell (Table 1).
This approach also was used to analyze three pancreatic cancer cell lines, two with KRAS mutations. The mutations known to occur in these two lines were identified correctly, and no mutant was identified in the third (Table 1). The average ratio of mutant to WT Ras proteins was 0.49 and 1.7 in the two lines with mutations (Table 1). The average amount of total Ras protein molecules (WT plus mutant) in these cells therefore varied from 1.0 to 4.0 million. DNA sequencing confirmed that the KRAS mutations were heterozygous in these lines as well as in SW480.
To confirm the presence of mutant peptides in the immunoprecipitates, we performed full MS/MS scanning on an ultra–high-definition accurate-mass quadrupole-TOF mass spectrometer interfaced with a nanoflow chip cube-based LC system. Several peptides from mutant (as well as WT) Ras proteins were identified unambiguously using a 1% false discovery rate cutoff, as shown in Fig. S3. These peptides included but were not limited to LVVVGAGGVGK, LVVVGAVGVGK, SFEDIHHYR, and SFADINLYR from SW480 cells and LVVVGAGGVGK, LVVVGADGVGK, and SFADINLYR from Pa16C cells.
Analysis of Human Tissues.
The procedure outlined in Fig. 1 then was applied to frozen pulverized tissue instead of tissue culture cells. A representative result is shown in Fig. 5 for a colorectal tumor harboring a G12D mutation of KRAS (details for this tumor and four others are provided in Table 1). The mutations identified by SRM in all five samples were identical to those previously found in these tumors (32). The relative proportion of mutant to WT Ras proteins varied from 0.28 to 0.70. Histopathologic analysis showed that tumors with ratios of mutant to WT protein <0.5 contained a relatively large proportion of nonneoplastic cells, which presumably contributed WT proteins to the lysates. As controls for the tumor tissues, we analyzed two samples each of normal colorectal mucosae and spleen; no mutant Ras proteins were identified (Table 1).
Fig. 5.
SRM of endogenous proteins from a colorectal tumor obtained at surgery. (A) Integrated intensities of the exogenously added mutant peptide and the endogenous mutant peptide from the tumor, as indicated. The integrated intensities correspond to the sum of the peak areas of the transitions described in Table S1, which are shown in B for the endogenous peptide. The asterisk indicates the heavy isotope (13C6/15N2) labeled lysine.
Analysis of Pancreatic Cyst Fluid.
Pancreatic cysts represent an increasingly common condition, often discovered incidentally during diagnostic procedures such as CT scans (33, 34). Certain types of cysts are precursors of pancreatic adenocarcinomas, a generally incurable cancer. It is notoriously difficult to distinguish cyst types from one another and to determine when surgery, which can have severe sequelae, should be performed. The identification and quantification of mutant Ras proteins in cyst fluids therefore could prove useful for diagnostic purposes.
We evaluated fluids obtained from three intraductal pancreatic mucinous neoplasms, a common cyst type that can evolve to adenocarcinoma. In these cases, we did not know which, if any, of the cysts contained KRAS mutations. Fluid from each cyst contained detectable Ras proteins, and we identified Ras protein mutations in two of the three cases (Table 1). Subsequently, we used the same cyst fluids to determine whether these mutations could be identified at the DNA level. DNA sequencing confirmed the exact mutations identified by SRM and showed that the sample without a SRM-detectable mutation did not have a RAS mutation at the analyzed positions. Notably, histopathologic analysis of the cyst walls demonstrated that these lesions had not yet become malignant.
Analysis of Relative Abundance of K-Ras, N-Ras, and H-Ras Proteins.
One of the advantages of SRM-based technologies is that multiple different proteins can be analyzed at once. Three highly conserved Ras proteins—K-Ras, neuroblastoma rat sarcoma viral oncogene homolog (N-Ras), and Harvey rat sarcoma viral oncogene homolog (H-Ras)—are expressed in human cells. Antibodies exquisitely specific to each protein have been difficult to generate. In the process of evaluating the levels of mutant and WT Ras proteins, we simultaneously measured the relative abundance of the three normal isoforms.
We first ensured that the antibody used was equivalently effective in capturing the three Ras protein types. By comparing SRM analysis of synthetic Ras proteins before and after IP, we confirmed that the efficiency was 26 ± 1.2%, 24 ± 0.17%, and 25 ± 1.9% for K-Ras, N-Ras, and H-Ras, respectively. The tryptic peptide (residues 6–16) containing the most common mutants of any of these proteins (residues 12 and 13) are identical in K-Ras, N-Ras, and H-Ras. However, trypsin produces nine-residue peptides from each protein, spanning residues 89–97, which are distinguishable by SRM (Fig. S4). After optimization of the transition parameters for these three peptides (Fig. S5 and Table S1), their levels were measured in the cell lines and tissues described above.
We found that the estimated levels of Ras proteins were similar when assessed by analysis of residues 6–16 (Table 1) and residues 89–97 (Table S2). In the 13 samples analyzed, the ratio of Ras proteins assessed by peptides containing residues 6–16 to that assessed by peptides containing residues 89–97 in the same samples were 1.02 ± 0.30 (mean ± SD). Although the total amount of Ras proteins in 2 mg of total cellular protein varied considerably, the relative levels of the three individual Ras proteins were similar: 63 ± 10% for K-Ras, 23 ± 5% for N-Ras, and 14 ± 7% for H-Ras (Table S2). Because each protein is encoded by an independent gene, and the normal tissues, tumor cell lines, and tumors represented disparate cell types, this result suggests that the relative levels of the three Ras proteins are regulated by similar mechanisms in many cell types. This regulation likely occurs at the posttranscriptional level, because the relative levels of mRNA were not highly correlated with the levels of protein (2). These analyses also permitted us to estimate the relative ratios of mutant and WT K-Ras (rather than total RAS) polypeptides in cell lines; these ratios varied from 0.8 (in Pa16C) to 8.0 (in SW480).
Discussion
The results described above show that SRM can be used to detect and quantify the levels of WT and mutant proteins in cell lines and in clinically relevant tissue samples and biologic fluids. Several advantages are apparent from the data: The technique is sensitive, allowing detection of as little as 10 fmol; the calculated levels of WT and mutant proteins are linearly related to input over a wide range (Fig. S1); the use of internal controls and the monitoring of multiple product ions ensure exquisite specificity; and the technique is relatively simple to implement, requiring only commercially available reagents, including an antibody against the normal form of the protein and a state-of-the-art mass spectrometer. In particular, SRM does not require mutant-specific antibodies, which can be difficult to develop, especially when the target protein can be mutated at many positions.
What are the limitations of this approach? One potential limitation is its sensitivity. Based on the results presented above, we estimate that SRM can be used to detect mutant proteins reliably when they are present at levels as low as 25 fmol/mg of total protein. We thus could detect mutant and WT Ras proteins in as few as 6,000 cells. However, this sensitivity may be inadequate to detect mutant proteins in some clinical samples, such as sputum, serum, or urine. Although the success of detection can be increased simply by increasing the amount of sample used for enrichment, this solution is not feasible when the amount of sample is limited (sputum) or too dilute (urine). Further improvements in mass spectrometer instrumentation can be expected to improve this sensitivity. Additionally, the various steps involved in the procedure before MS—pulverization, homogenization, IP, elution, trypsinization, and chromatography—presumably can be improved to reduce sample loss.
In sum, the approach described here fulfills a need in cancer research, permitting the determination of the relative amounts of missense mutant and WT proteins and allowing comparisons among the amounts of DNA, RNA, and polypeptides. The determination of the relative levels of mutant and WT proteins can help inform the mechanisms underlying the abnormal protein's function, e.g., through supporting the basis for dominant-negative effects or haploinsufficiency. The approach also opens up diagnostic opportunities, as illustrated by the results obtained in this study on pancreatic cysts. One advantage of this approach over DNA-based approaches is that numerous independent proteins can be assessed simultaneously, thereby preserving precious clinical samples and reducing the costs of clinical analyses. Another advantage is that no amplification is needed, thereby minimizing the contamination issues that can plague PCR-based approaches (35).
Materials and Methods
The Pa02C, Pa08C, and Pa16C pancreatic cancer cell lines were derived as described (36). Colorectal tumors and cyst fluids were obtained from surgical resection specimens at the Johns Hopkins Hospital. All samples were obtained in accordance with the Health Insurance Portability and Accountability Act and had Institutional Review Board approval. Conjugation of antibodies to beads was performed using slight modifications of methods described by Whiteaker et al. (26). After IP and trypsin digestion, peptides were separated using a reversed-phase column with a gradient that was generated in 0.1% formic acid with increasing acetonitrile concentrations. The peaks of y and b ions that were generated from peptides with 2+ and 3+ charge states were optimized by altering the collision energy for each transition. The Skyline program (37) preloaded with WT and mutant Ras peptide sequences was used to analyze the data. Detailed methods for each of the procedures described in the text are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Lustgarten Foundation for Pancreatic Cancer Research, National Cancer Institute Contract NO1 CN-43302, Department of Defense Award W81XWH-06-1-0428, the Sol Goldman Center for Pancreatic Cancer Research, the Virginia and D. K. Ludwig Fund for Cancer Research, the Michael Rolfe Foundation, the Joseph L. Rabinowitz Fund for Pancreatic Cancer Research, and National Institutes of Health Grants CA62924, RR020839, CA130938, and CA57345.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019203108/-/DCSupplemental.
References
- 1.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann M, Kelleher NL. Precision proteomics: The case for high resolution and high mass accuracy. Proc Natl Acad Sci USA. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciordia S, de Los Ríos V, Albar JP. Contributions of advanced proteomics technologies to cancer diagnosis. Clin Transl Oncol. 2006;8:566–580. doi: 10.1007/s12094-006-0062-4. [DOI] [PubMed] [Google Scholar]
- 9.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Old WM, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 12.Schnölzer M, Jedrzejewski P, Lehmann WD. Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 13.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 14.Tabb DL, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Little DP, Braun A, O'Donnell MJ, Köster H. Mass spectrometry from miniaturized arrays for full comparative DNA analysis. Nat Med. 1997;3:1413–1416. doi: 10.1038/nm1297-1413. [DOI] [PubMed] [Google Scholar]
- 16.Murray KK. DNA sequencing by mass spectrometry. J Mass Spectrom. 1996;31:1203–1215. doi: 10.1002/(SICI)1096-9888(199611)31:11<1203::AID-JMS445>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Connors LH, Yamashita T, Yazaki M, Skinner M, Benson MD. A rare transthyretin mutation (Asp18Glu) associated with cardiomyopathy. Amyloid. 2004;11:61–66. doi: 10.1080/13506120410001682569. [DOI] [PubMed] [Google Scholar]
- 18.Lim A, et al. Characterization of transthyretin variants in familial transthyretin amyloidosis by mass spectrometric peptide mapping and DNA sequence analysis. Anal Chem. 2002;74:741–751. doi: 10.1021/ac010780+. [DOI] [PubMed] [Google Scholar]
- 19.Nepomuceno AI, Mason CJ, Muddiman DC, Bergen HR, 3rd, Zeldenrust SR. Detection of genetic variants of transthyretin by liquid chromatography-dual electrospray ionization fourier-transform ion-cyclotron-resonance mass spectrometry. Clin Chem. 2004;50:1535–1543. doi: 10.1373/clinchem.2004.033274. [DOI] [PubMed] [Google Scholar]
- 20.Rosenzweig M, et al. A new transthyretin variant (Glu61Gly) associated with cardiomyopathy. Amyloid. 2007;14:65–71. doi: 10.1080/13506120601116625. [DOI] [PubMed] [Google Scholar]
- 21.Bunger MK, et al. Detection and validation of non-synonymous coding SNPs from orthogonal analysis of shotgun proteomics data. J Proteome Res. 2007;6:2331–2340. doi: 10.1021/pr0700908. [DOI] [PubMed] [Google Scholar]
- 22.Muñoz J, et al. Mass spectrometric characterization of mitochondrial complex I NDUFA10 variants. Proteomics. 2008;8:1898–1908. doi: 10.1002/pmic.200701085. [DOI] [PubMed] [Google Scholar]
- 23.Hanke S, Besir H, Oesterhelt D, Mann M. Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J Proteome Res. 2008;7:1118–1130. doi: 10.1021/pr7007175. [DOI] [PubMed] [Google Scholar]
- 24.Tu C, et al. Depletion of abundant plasma proteins and limitations of plasma proteomics. J Proteome Res. 2010;9:4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson NL, et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 26.Whiteaker JR, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capon DJ, et al. Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature. 1983;304:507–513. doi: 10.1038/304507a0. [DOI] [PubMed] [Google Scholar]
- 28.Picotti P, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 29.Kostiainen R, Kotiaho T, Kuuranne T, Auriola S. Liquid chromatography/atmospheric pressure ionization-mass spectrometry in drug metabolism studies. J Mass Spectrom. 2003;38:357–372. doi: 10.1002/jms.481. [DOI] [PubMed] [Google Scholar]
- 30.Addona TA, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopalan H, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 33.Megibow AJ. Update in imaging of cystic pancreatic masses for gastroenterologists. Clin Gastroenterol Hepatol. 2008;6:1194–1197. doi: 10.1016/j.cgh.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Anderson MA, Kwon RS, Scheiman JM. PANDA cyst-fluid analysis: Eats, shoots and leaves? Gastrointest Endosc. 2009;69:1103–1105. doi: 10.1016/j.gie.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 35.Millar BC, Xu J, Moore JE. Risk assessment models and contamination management: Implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol. 2002;40:1575–1580. doi: 10.1128/JCM.40.5.1575-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaffee EM, et al. Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. Cancer J Sci Am. 1998;4:194–203. [PubMed] [Google Scholar]
- 37.MacLean B, et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.