Abstract
A central paradigm in island biogeography has been the unidirectional “downstream” colonization of islands from continents (source to sink) based on the idea that less-diverse island communities are easier to invade than biologically more-diverse continental communities. Recently, several cases of “upstream” colonization (from islands to continents) have been documented, challenging the traditional view. However, all these cases have involved individual island species that have colonized mainland regions. Here, using molecular phylogenetic data, divergence time estimates, lineage diversity distributions, and ancestral area analyses, we reconstruct the spread of a species-rich (>700 species) passerine bird radiation (core Corvoidea) from its late Eocene/Oligocene origin in the emerging proto-Papuan archipelago north of Australia, including multiple colonizations from the archipelago to Southeast Asia. Thus, islands apparently provided the setting for the initiation of a major songbird radiation that subsequently invaded all other continents. Morphological and behavioral adaptations of the core Corvoidea as generalist feeders in open habitats, which facilitated dispersal and colonization, apparently evolved in the descendants of sedentary forest birds that invaded the proto-Papuan archipelago. The archipelago evidently provided islands of the right size, number, and proximity to continental areas to support the adaptation and diversification of vagile colonizers that went on to increase avian diversity on a global scale.
Keywords: Indo-Pacific, Passeriformes, community assembly, macroecology, range expansion
Islands and archipelagos have fascinated ecologists and biogeographers since Darwin (1) and Wallace (2, 3) demonstrated their value as model systems. Although recognizing that biodiversity on islands develops as a dynamic equilibrium between colonization and extinction, many biogeographers regard islands as biodiversity sinks for colonists from diverse mainland source biotas (4–7). Recently, however, documentation of numerous examples of colonization from islands to mainland areas (8–11) have led to a reassessment of the evolutionary and biogeographic roles of islands in a broader geographic context (12–15).
Oscine passerine birds (songbirds) comprise a globally distributed, species-rich group of birds (approximately 4,500 species or nearly half of all avian species) (16), which originated in the early Tertiary in Australia (17–21). At that time, Australia was isolated by wide oceanic expanses from the northern landmasses (22, 23) that the oscine passerines later colonized. The northern rim of the Australo-Papuan plate (present-day New Guinea) was submerged as a continental shelf, but began to rise out of the sea in the late Eocene/Oligocene as the Indo-Australian plate approached and collided with other plates (24, 25). Although our understanding of global spread of passerine birds is improving, we know little about the timing, dispersal routes, and taxonomic selectivity of colonization events of oscine passerines moving from Australia to Asia or Africa.
The relatively small, basal oscine passerine families comprising Menuridae, Climacteridae, Maluridae, and Pardalotidae are mostly restricted to present day Australia, with only a few members having secondarily colonized New Guinea. Other basal oscine families are broadly distributed in Australia and New Guinea (Ptilonorhynchidae), and two families (Meliphagidae and Acanthizidae) are more widely distributed across the Indo-Pacific. However, other oscine lineages—the core Corvoidea (crows and allies), comprising more than 700 species, and the Passerida (warblers, thrushes, finches, and other songbirds), comprising more than 3,500 species—originated in the mid-Tertiary and dispersed all over the world (17, 18, 20). Although the deep phylogenetic relationships of the Passerida are still obscured by gaps and uncertainties (26, 27), a phylogenetic hypothesis including all major lineages now exists for the core Corvoidea (Fig. 1). Our analysis focuses on the diversification of this group and the escape of some lineages across the water barrier separating Australia from the rest of the globe.
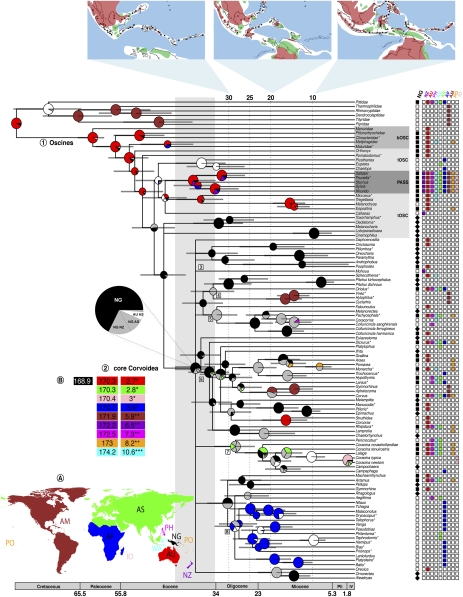
Fig. 1.
Chronogram and ancestral areas for oscine passerine birds. A summary of the Bayesian dispersal-vicariance analysis for oscine passerine birds. The tree is a chronogram based on a 50% majority-rule consensus tree of a Bayesian analysis of a combined data set of nuclear introns (ODC and Myo2), nuclear exons (RAG-1 and RAG-2), and mitochondrial (ND2) DNA sequences. The distribution for each taxon, as delimited in the map (A), is presented in the columns to the right of the taxon names (bOSC, basal oscines; tOSC, transitional oscines; PASS, Passerida). For New Guinea, a diamond indicates that the taxon is endemic. Pie charts at internal nodes represent the marginal probabilities for each alternative ancestral area derived by using dispersal-vicariance analysis (DIVA) and integrating over tree topologies using MCMC. These probabilities account for the phylogenetic uncertainty in the rest of the tree and the biogeographic uncertainty at each node, conditional on this node to occur. The colors of the pie charts reflect the probability of the area of origin according to the geographical delimitations in A. Gray and light gray colors indicate multiple areas of origin shared with New Guinea. White indicates multiple areas of origin not shared with New Guinea. All other colors indicate only one area of origin. Large pies indicate well-supported nodes (PP ≥ 0.95 and ML bootstrap ≥ 70). Nodes discussed in the text are indicated with numbers before the node. Taxa for which two different species from the same genus were used in the analyses are indicated with an asterisk after the genus name. (A) Biogeographic regions: AF, Africa; AM, America; AS, Asia; AU, Australia; IO, Indian Ocean islands; NG, New Guinea; NZ, New Zealand; PH, Philippines; WA, Wallacea; PO, Pacific Ocean islands. The inset maps above, of the transition zone between Southeast Asia and Australia (after refs. 22 and 23), show the distribution of land and see from the Oligocene to the present at 10-million y intervals: red, land; blue, deep sea; white, shallow sea; green, calcareous plateaus, which may at times have been above sea-level. Triangles represent volcanism and emergence of oceanic islands. (B) Summary of the BayesTraits analysis. The left column indicates the harmonic mean, ordered with the highest at the top. New Guinea (black) is the most likely area of origin with the highest harmonic mean. The right column indicates the Bayes factor compared against New Guinea as the favored area of origin. * Indicates positive evidence and ** indicates strong evidence for the favored hypothesis.
The early radiation of the core Corvoidea (Fig. 1) coincided with the emergence of the first archipelagos in the Papuan epicontinental seas (22–25), and many lineages that occur outside the Australo-Papuan area are nested among lineages that are endemic to Australo-Papua. In the present study, we investigate the timing and location of the origins of the core Corvoidea to assess the role of the proto-Papuan archipelago in the dispersal of several lineages out of the Australo-Papuan region. Analyzing the diversification of the core Corvoidea within an explicit spatiotemporal framework, we call attention to the potential importance of the proto-Papuan islands for the early diversification of this large songbird radiation and its spread to the rest of the world. Evidently, the proto-Papuan islands in the Late Eocene/Oligocene provided an ideal biogeographic context for the evolutionary maintenance of dispersal and colonization capabilities, which allowed several lineages of the otherwise sedentary Australian oscines to escape their ancestral home.
Results
Molecular Phylogenetics and Dating.
Mixed-models Bayesian analyses of the concatenated DNA sequence dataset and its individual partitions yielded 50% majority-rule consensus trees that were topologically congruent with the maximum likelihood trees generated in RAxML (Fig. S1). The Bayesian runs of the concatenated dataset failed to converge within 60 million generations. Lack of convergence with large datasets has been encountered in other recent studies (e.g., ref. 28). However, both Bayesian and maximum likelihood (ML) analytic approaches find support for a monophyletic core Corvoidea (Fig. 1).
Analyses in MULTIDIVTIME generated a chronogram (consistent with the “2% rule” for the rate of mitochondrial DNA sequence divergence per million years) (29) for the oscine passerine birds, of which 77 taxa in our sample represent the lineages of the core Corvoidea (Fig. 1). An additional chronogram of basal oscines comprised seventeen species (Fig. S2). The chronogram for the core Corvoidea, together with ancestral area reconstruction (Fig. 1), suggests that this group (node 2) originated in the Australo-Papuan area in the late Eocene and that initial dispersal events out of the region of origin that left modern descendants began in the Oligocene.
Biogeographic Analyses.
The Bayes-DIVA analysis (Fig. 1) provided strong support for an Australian origin of all oscine passerine birds: P = 0.93 for node 1, and P > 0.90 for the two other well-supported basal nodes. For the core Corvoidea (node 2), however, there was support for a New Guinean (or proto-Papuan) origin; P = 0.68; New Guinea/New Zealand: P = 0.16; New Guinea/Asia: P = 0.10, meaning that this node is placed in New Guinea by the present distribution of core Corvoidea lineages. The results of the BayesTraits analysis (Fig. 1B and Table S1) also supported a New Guinean origin of the core Corvoidea. Lineages of the core Corvoidea colonized the rest of the world during the mid-Tertiary, presumably through the developing proto-Papuan archipelago in the northern Australo-Papuan region. The Bayes-diva analysis suggests that at least five dispersal events connect the Papuan region to contemporary avifaunas elsewhere, in some cases at great distance: Mohoua (node 3) in New Zealand, Vireonidae (node 4) in the New World, members of Pachycephalidae (node 5) in Asia and Oceania, Pericrocotus (node 7) in Asia, and the African bush-shrikes and helmet-shrike complex (node 8) in Africa.
Lineage Diversity Maps.
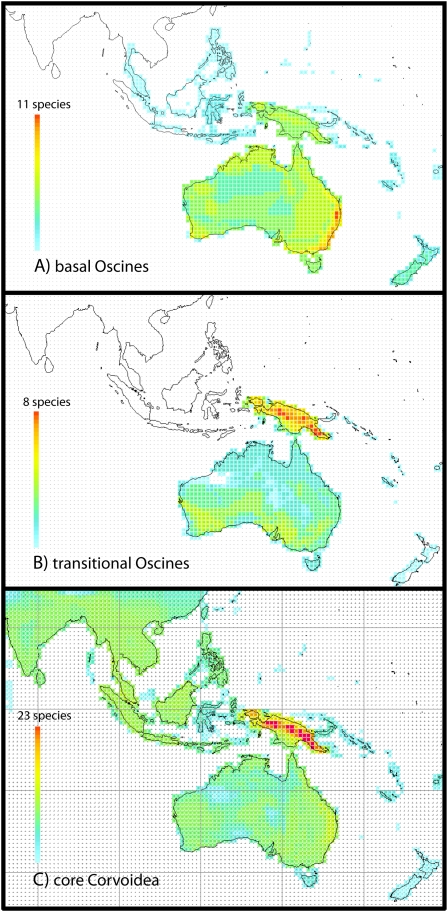
Among the core Corvoidea, 34 lineages with modern descendants existed 25 Mya (Fig. 2, Table 1, and Table S2). Of these, 26 occur in New Guinea (five endemic) and 17 in Australia (two endemic). Two additional lineages are restricted to New Zealand, and 18 lineages occur elsewhere. Of the latter, eight occur only outside Australo-Papua, five of these lineages representing one monophyletic group of mainly African taxa. Of the 20 lineages occurring entirely or partly outside the Australo-Papuan area, all have Papuan sister taxa and some are nested within a larger complex of Papuan taxa. Furthermore, many endemic Papuan genera represent small clades that date back to the mid-Tertiary, as is also the case for the “transitional oscines.”
Fig. 2.
Lineage diversity maps of oscine passerine birds. Each lineage is mapped out as the distribution of all extant members in a single layer (for details on taxa and number of lineages, see Table S2 and S3). (A) Present distribution of the 11 basal oscine lineages that existed 25 Mya. All of these lineages occur in Australia. Maximum pixel value (red) in Australia is 11. (B) Present distribution of the 12 transitional oscine lineages (Passerida excluded) that existed 25 Mya. Maximum pixel value (red) in New Guinea is 8. (C) Present distribution of the 34 lineages of core Corvoidea that existed 25 Mya. Maximum pixel value (red) in New Guinea is 23.
Table 1.
Lineage diversity at 25 Mya
| Number of lineages | Number of endemic lineages | |
| Basal oscines | 11 | |
| Australia | 11 | 6 |
| New Guinea | 4 | 0 |
| Outside Australo-Papua | 2 | 0 |
| Transitional oscines excl. Passerida | 12 | |
| Australia | 4 | 0 |
| New Guinea | 8 | 4 |
| Outside Australo-Papua | 4 | 4 |
| core Corvoidea | 34 | |
| Australia | 17 | 2 |
| New Guinea | 26 | 5 |
| Outside Australo-Papua | 20 | 8 |
The 11 lineages of basal oscines that existed 25 Mya (Fig. 2, Table 1, and Table S3) all presently occur in Australia (six of them endemic), with their peak diversity in forests of the east coast. Four lineages are represented in New Guinea, mainly by terminal subgroups, as secondary dispersal is indicated for two or three clades. Only two of the 25 My basal oscine lineages (Meliphagidae and Acanthizidae) include species that presently occur outside Australo-Papua.
The modern descendents of 12 “transitional” oscine lineages present 25 Mya (Fig. 2, Table 1, and Table S2) are concentrated in New Guinea (eight taxa, four of which are endemic). Four lineages occur in Australia (none endemic) and four lineages exist outside Australo-Papua. Passerida is not evaluated here because its early radiation occurred outside Australo-Papua (26), and the clade was not properly sampled in our study.
Morphological analysis.
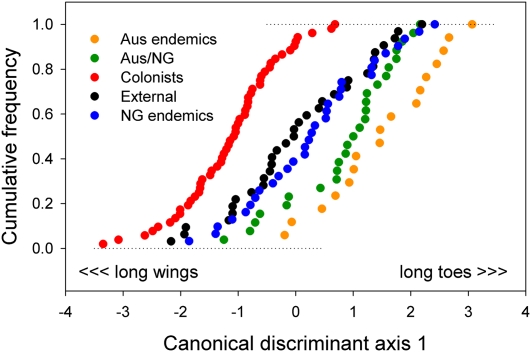
Three significant (P < 0.01) discriminant axes based on seven external measurements of 158 species (83 species of core Corvoidea, 37 species of transitional oscines, and 38 species of basal oscines) contained 63, 25, and 11% of the total variance among five distributional groups of species in the Australo-Papuan oscine radiation: (i) endemics to Australia, (ii) endemics to New Guinea, (iii) restricted to both, (iv) spanning Australo-Papua and Asia/Africa, and (v) presently distributed only outside of Australo-Papua. Posterior classification placed 88 of 158 species (57%) in the correct group. Only the sample of New Guinea endemics and lineages shared between New Guinea and Australia could not be readily separated. The group of lineages that bridges Australo-Papua to Asia/Africa was morphologically the most distinctive, with 75% of species correctly placed (generalized squared distances to the other groups ranged between 2.9 and 7.5 times the pooled within-groups variance). Variation that distinguished these colonizing lineages was concentrated on the first discriminant axis, where positive values, contrasting short wings (standardized canonical coefficient = −2.00) and long middle toes (2.56), were associated with noncolonists. Lineages that extend from Australo-Papua into southeast Asia and to Africa are clearly distinguished as having large negative values on this axis (mean = −1.14); that is, relatively long wings and short toes, whereas birds restricted to Australia proper have the opposite morphology (1.52) (Fig. 3). Species in lineages that no longer occur in Australo-Papua (0.06) group with species endemic to New Guinea (0.30); lineages present in both Australia and New Guinea (0.84) are closer to the Australian endemics.
Fig. 3.
Colonizing lineages among the oscines have distinctive morphology associated with dispersal ability. Cumulative distribution of scores on canonical discriminant axis 1 among species of basal oscines (bOSC, n = 38), transitional oscines (tOSC, n = 37), and core Corvoidea (n = 83) belonging to each of the five distribution groups: endemic to Australia (including New Zealand), endemic to Australia plus New Guinea (Aus/NG), endemic to New Guinea (NG), widely distributed across Australasia to southeast Asia or Africa (Colonists), and species presently occurring only outside of the Australo-Papuan region (External).
Discussion
According to our phylogenetic hypotheses, combined with several others based on DNA sequences (e.g., refs. 17–21), the basal oscine lineages are entirely or predominantly restricted to Australia (Figs. 1 and 2A). The oscines originated in what is now Australia, which was isolated from other land masses by a large expanse of ocean. Most of the basal oscine lineages are presently represented by forest species (30) that are unlikely to cross water (31). Some basal oscine lineages secondarily became established in New Guinea, which was occasionally connected to the Australian mainland during periods of sea-level lows in the Upper Tertiary (22, 23); only a few members of the families Acanthizidae and Meliphagidae dispersed beyond the Australo-Papuan region.
In contrast, the origin of core Corvoidea appears to be Papuan (Fig. 1). This clade, which has successfully colonized other continents and the Pacific archipelagos, began to diversify in the late Eocene/Oligocene (Fig. 1), when the northern part of the Australo-Papuan plate became subaerial (22, 23), with large calcareous island plateaus emerging above sea level several hundred kilometers from the Australian mainland. These islands provided new opportunities for species adapted to crossing oceanic stretches of water, and we argue that this archipelagic setting favored the maintenance of well-developed dispersal capabilities and eventually allowed several lineages to cross the water gap separating Australo-Papua and Asia.
The history of the area in which the core Corvoidea is currently most diverse (Fig. 1) suggests that the group diversified in a regional setting with access to emerging archipelagos in the mid-Tertiary, well before extensive terrestrial environments appeared in the northern part of the Australian plate. Dispersal of many lineages out of the Papuan area, to New Zealand (Philesturnus, Mohoua) and other parts of the world (e.g., African-Malagasy radiation, Dicruridae, Pericrocotus, Platylophus, Vireonidae, Corvidae/Lanidae), appears to have occurred over a brief period in the late Eocene/Oligocene, judging from the depth of phylogenetic splits between Australo-Papuan endemics and outside representatives of these groups. The core Corvoidean lineages also have been successful recent colonizers of the Pacific archipelagos, currently representing 44% of all passerine birds in remote Micronesia and Polynesia (32), consistent with adaptation to island life. Considering the association of basal songbird groups with forested habitats (30), the early core Corvoidea groups would have undergone a marked change in life strategy to become dispersalists. The new Papuan island environments must have favored morphology and behaviors that facilitated dispersal and establishment of new colonists (7, 13).
Although it is unlikely that selection would favor overwater dispersal per se, certain types of ecological distribution might be associated with traits that would facilitate long-distance colonization. In particular, generalized foraging in relatively open environments might be conducive to flocking and to dispersal between seasonal feeding areas. The consequential colonization of remote islands permitted by these adaptations created opportunities for geographic isolation and lineage splitting, leading to a build-up of diversity in the proto-Papuan archipelago, and eventually to the colonization of Southeast Asia across a stepping-stone chain of emerging islands on the developing Sunda shelf.
During this period of diversification and colonization, various “key innovations” might also have been important. In the case of the core Corvoidea, these might have included, in addition to a morphology compatible with long-distance dispersal: (i) a bill suitable for generalized foraging (i.e., massive and hooked) (33); (ii) increased brain size, enhancing flexibility in food selection, feeding innovation (e.g., food hoarding in crows, shrikes and petroicids) (34), and complex group structure (35, 36); and (iii) exploratory behavior, which may be associated with changes in life strategy, including social behavior, increasing the likelihood of successful long-distance dispersal. Innovative behavior and ability to adapt to new environments is associated with enlargement of the hyperstriatum ventrale of the brain, which is exceptionally developed in crow-like birds (37). Few data on relative sizes of various brain parts are available, but it might be worthwhile to compare the endemic taxa (basal oscines and endemic Papuan core corvoids) and the colonizing taxa (core corvoids that dispersed) in this respect.
The lineage diversity maps (Fig. 2) suggest that many evolutionary lines of the core Corvoidea secondarily became adapted to forest environments on the developing island of New Guinea. As the Papuan landmass began to emerge, birds invaded the developing montane forest habitats, where their powers of dispersal began to diminish as they adapted to the closed environments. This scenario, in which old endemic populations on large islands become sedentary specialists of montane forest, characterizes many taxa [e.g., birds (13, 38–42), ants (43, 44), butterflies (45)]. The core Corvoidean lineages that became restricted in this way to Papuan cloud forests include Aleadryas, Androphobus, Chaetorhynchus, Eulacestoma, Ifrita, Melampitta, Ornorectes, and Rhagologus.
Our results suggest that island colonization during the early evolution of the core Corvoidea reinforced adaptations for long-distance dispersal and led to a proliferation of species with these traits in an apparent adaptive radiation within a developing archipelago. Species “pumps,” whether associated with taxon cycles in archipelagos (39, 40) or biodiversity hotspots in continental areas (46), might be most productive at an early stage of radiation, when open ecological space places a selective premium on innovation and dispersal (47, 48).
Although we find support for the Papuan island origin hypothesis, one alternative hypothesis cannot be ruled out definitively. It is plausible that in the Oligocene/Miocene when Australia was further to the south and the climate there was wetter, the core Corvoidea had already radiated within Australia and some members had begun to disperse across to proto-Papua and mainland Asia, involving some island-hopping events. Subsequently, Australia moved further north and became hotter and drier causing many core Corvoidean lineages to go extinct there. This process would leave a print of the basal core Corvoidean lineages in New Guinea. However, because the basal oscines restricted to Australia apparently persisted there since their origin, there is little reason to believe that core Corvoidean taxa would not also have been able to survive in Australian monsoon rain forest pockets in the eastern and northern parts of the continent had they originated in Australia.
Based on the biogeographic and dating analyses presented here, we propose that the core Corvoidea originated in the proto-Papuan islands among lineages that were morphologically and behaviorally adapted in ways that facilitated overwater dispersal. This process led to a proliferation of species within the ancient archipelago and eventually to dispersal to distant continental and Oceanic regions, where core Corvoidea have since radiated further. This theory implies that an initial phase of island life, to which the core Corvoidea were eminently suited, preceded their worldwide expansion following successful “upstream” colonization from islands to mainland areas. Thus, several lineages of the core Corvoidea represent, beyond additional examples of colonization from islands to mainlands, the beginnings of global radiations from within an island archipelago. Clearly, some lineages of this diversifying clade became adapted to inner forest environments and stayed put in New Guinea. Others remained in open habitats, developing morphology and behavior suitable to overwater dispersal. Our hypothesis that islands can be centers of evolution and adaptation depends on persistence of island populations and contrasts with the widely held idea that populations on small islands are prone to extinction (5). We note, however, that highly diversified lineages have survived for millions of years in archipelagoes, such as the Hawaiian and Galapagos Islands, and that estimated ages of many individual island populations of lesser Antillean birds range into the millions of years (42).
The role of distribution within the Papuan and southeast Asian archipelagoes in selecting for overwater dispersal ability is emphasized by the fact that corvoids occurring only outside of the Australo-Papuan region have apparently reverted to a morphology more like that of the New Guinea endemics (Fig. 3), which is evidently unsuitable for overwater dispersal. Thus, the proto-Papuan archipelago and the islands presently lying between New Guinea and Southeast Asia appear to be a cauldron of interisland movement and colonization, selecting in many lineages against adaptation for life in closed forest environments—a route taken by many endemic lineages—and maintaining the potential for long-distance colonization.
Methods
Taxon Sampling and Phylogenetic Analyses.
We initially compiled a dataset including all core Corvoidean lineages dating back to the mid-Miocene or beyond for which at least one of the markers Myo2, ODC, RAG-1, RAG-2 or ND2 was available (e.g., refs. 10, 17, 18, 49–54). A few additional sequences were generated specifically for this study. Our dataset of the core Corvoidea is not complete at the genus level but it likely includes all lineages that existed 25 Mya. We subsequently pruned the dataset to include 102 oscine passerine bird taxa and 77 members of the core Corvoidea, representing all of the deeper lineages (Fig. 1 and Table S2). Some suboscine species were also included and Acanthisitta chloris was used to root the tree. An additional dataset of recombination activating protein 1 (RAG-1) was compiled for all seventeen documented deep lineages of basal oscines (Fig. S2 and Table S3).
Sequences were aligned using MegAlign (DNASTAR, Inc.) and the concatenated core Corvoidea alignment consisted of 6,676 bp and the basal oscine RAG-1 alignment included 2,872 bp. We used Bayesian inference (e.g., refs. 55 and 56), as implemented in MrBayes 3.1.2 (57, 58), to estimate phylogenetic relationships. Substitution models (Figs. S1 and S2) were determined with MrModeltest 2.0 (59). In all MrBayes analyses, four Metropolis-coupled Markov Chain Monte Carlo (MCMC) simulations, one cold and three heated, were run for 20 million iterations (individual partitions) or 60 million iterations (concatenated alignment) with trees sampled every 500 iterations. The burn-in and convergence diagnostics were graphically estimated using AWTY (Are We There Yet?) (60, 61). Partitioned ML analyses were performed using RAxML version 7.0.4 (62), by applying to each partition the same models as in the MrBayes analyses. Nodal support was evaluated with 100 nonparametric bootstrap pseudoreplications.
Dating Analyses.
We used the relaxed Bayesian molecular clock implemented in Multidivtime to estimate divergence dates (63). Parameters and priors were specified following the Multidivtime guidelines (SI Material and Methods). The posterior distributions of node times were approximated through four independent MCMC runs of 50 million generations and the “burn-in” was set at 1 million generations. Finally, convergence diagnostics were checked graphically. To calibrate the tree to estimate absolute dates, we used a combination of geological calibration points (64) and secondary calibration points (18) (SI Material and Methods). To further corroborate the absolute date estimates, we compared with the “2% rule” for the rate of mitochondrial DNA sequence divergence per million years (SI Material and Methods).
Biogeographic Analyses.
We used Bayes-diva to elucidate ancestral patterns (65) with particular emphasis on the nodes around the core Corvoidea. We sampled 20,000 trees (by thinning the chain; i.e., sampling every nth generation) from the MCMC output and ran diva (66) on all of them. The frequency of ancestral areas for clades was then recorded and plotted as marginal distributions on the majority-rule consensus tree derived from the MCMC. We assigned ten geographic areas for the Bayes-DIVA analysis (Fig. 1A), considering evidence of historical relationships of geological plates and terranes in the Indo-Pacific (22–24) but otherwise keeping to major continental areas. The analysis used maxareas = 2, but we also explored the importance of changing the maxareas (setting maxareas = 3, 4, and 5) (SI Material and Methods).
We used BayesTraits ver. 1.0 (67) to test the validity of New Guinea as the area of origin of the core Corvoidea. BayesTraits estimates the instantaneous forward and backward rates among all specified states (in this case areas of origin) to compute probabilities of a state change in a given branch (68). To contrast the New Guinean origin hypothesis for the core Corvoidea clade, we used a local approach (69) and compared state reconstructions at this specific node (Node 2: core Corvoidea). Thus, we constrained the Most Recent Common Ancestor for the core Corvoidea node at one of the ten potential states (areas of origin). MCMC was used to explore the samples and the space of rate parameter values of 20,000 trees generated in the combined MrBayes analysis. Bayes factors were then calculated to determine the support for New Guinea compared with the nine remaining alternative biogeographic states (SI Material and Methods).
To generate a visual overview of the distributions of the various clades of oscine passerine birds, we generated lineage diversity maps at the time when the proto-Papuan archipelago emerged in the Oligocene (Fig. 2) (SI Material and Methods). We generated lineage diversity maps for three time frames (35, 30, or 25 Mya) to accommodate the uncertainty associated with any date estimate. We chose these points in time for two reasons: (i) they mark the transition between initial (deeper) branching of the core Corvoidea and several strong secondary radiations in some subgroups outside Australo-Papua; and (ii) they mark the transition between the existence of isolated Papuan islands and continuous land connections with Australia (22, 23) (see maps in Fig. 1). Our assumption here is that lineage endemism and the peak of lineage diversity reflects the center of radiation in the Late Eocene/Oligocene.
Morphological Analyses.
We used discriminant analysis (DISCRIM and CANDISC procedures of the Statistical Analysis System, SAS Institute, Cary, NC) based on seven morphological traits (SI Materials and Methods) to determine whether lineages that diversified on the proto-Papuan islands and finally crossed into Southeast Asia possessed particular morphologies that might have been associated with long-distance dispersal. Species were assigned to one of five distributional groups according to the present distribution of the 25 Mya lineages to which they belong: Australia (including New Zealand; 17 species), Australia plus New Guinea (26), endemic to New Guinea (31), Australasia and Southeast Asia and/or Africa (52), and outside of Australasia (32).
Supplementary Material
Acknowledgments
We thank the number of institutions and field workers who provided samples used in the study, and Martin Irestedt and Jan Ohlson for assisting with laboratory work. World bird databases have been developed by the Center for Macroecology, Evolution and Climate, University of Copenhagen, Denmark. K.A.J., P.-H.F., and J.F. acknowledge the Danish National Research Foundation for support to the Center for Macroecology, Evolution, and Climate. R.E.R. acknowledges the generous financial support of the curators of the University of Missouri and the Alexander von Humboldt Foundation, and assistance from ornithological collection managers at the Field Museum of Natural History in Chicago, IL, The United States National Museum in Washington, DC, the American Museum of Natural History in New York, NY, and the Zoologisches Staatssamlung in Munich, Germany.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ456526, HQ456530, HQ456531, HQ612121, HQ612122, HQ612130, HQ612165, HQ612166, HQ612174). All other Genbank numbers from previously published work are listed in the tables in SI Materials and Methods.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018956108/-/DCSupplemental.
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection. London: Murray; 1859. [Google Scholar]
- 2.Wallace AR. The Geographical Distribution of Animals. New York: Harper; 1876. [Google Scholar]
- 3.Wallace AR. Island Life: Or, The Phenomena and Causes of Insular Faunas and Floras, Including a Revision and Attempted Solution of the Problem of Geological Climates. New York, NY: Harper; 1881. [Google Scholar]
- 4.MacArthur RH, Wilson EO. An equilibrium theory of insular zoogeography. Evolution. 1963;17:373–387. [Google Scholar]
- 5.MacArthur RH, Wilson EO. The Theory of Island Biogeography. Princeton, NJ: Princeton University Press; 1967. [Google Scholar]
- 6.Mayr E. Avifauna: Turnover on Islands. Science. 1965;150:1587–1588. doi: 10.1126/science.150.3703.1587. [DOI] [PubMed] [Google Scholar]
- 7.Diamond JM. Continental and insular speciation in Pacific island birds. Syst Zool. 1977;26:263–268. [Google Scholar]
- 8.Filardi CE, Moyle RG. Single origin of a pan-Pacific bird group and upstream colonization of Australasia. Nature. 2005;438:216–219. doi: 10.1038/nature04057. [DOI] [PubMed] [Google Scholar]
- 9.Balke M, et al. New Guinea highland origin of a widespread arthropod supertramp. Proc Biol Sci. 2009;276:2359–2367. doi: 10.1098/rspb.2009.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jønsson KA, et al. Historical biogeography of an Indo-Pacific passerine bird family (Pachycephalidae): Different colonization patterns in the Indonesian and Melanesian archipelagos. J Biogeogr. 2010;37:245–257. [Google Scholar]
- 11.Bellemain E, Ricklefs RE. Are islands the end of the colonization road? Trends Ecol Evol. 2008;23:461–468. doi: 10.1016/j.tree.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Schluter D. The Ecology of Adaptive Radiation. New York, NY: Oxford University Press; 2000. [Google Scholar]
- 13.Mayr E, Diamond J. The Birds of Melanesia: Speciation, Ecology and Biogeography. New York, NY: Oxford University Press; 2001. [Google Scholar]
- 14.Grant PR, Grant BR. How and Why Species Multiply: The Radiation of Darwin's Finches. Princeton, NJ: Princeton University Press; 2008. [Google Scholar]
- 15.Whittaker RJ, Fernández-Palacios JM. Island Biogeography. Ecology, Evolution, and Conservation. Oxford, United Kingdom: Oxford University Press; 2007. [Google Scholar]
- 16.Dickinson EC. The Howard and Moore Complete Checklist of the Birds of the World. 3rd Ed. Princeton, NJ: Princeton University Press; 2003. [Google Scholar]
- 17.Barker FK, Barrowclough GF, Groth JG. A phylogenetic hypothesis for passerine birds: Taxonomic and biogeographic implications of an analysis of nuclear DNA sequence data. Proc Biol Sci. 2002;269:295–308. doi: 10.1098/rspb.2001.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci USA. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beresford P, Barker FK, Ryan PG, Crowe TM. African endemics span the tree of songbirds (Passeri): molecular systematics of several evolutionary ‘enigmas’. Proc Biol Sci. 2005;272:849–858. doi: 10.1098/rspb.2004.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ericson PGP, et al. A Gondwanan origin of passerine birds supported by DNA sequences of the endemic New Zealand wrens. Proc Biol Sci. 2002;269:235–241. doi: 10.1098/rspb.2001.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards SV, Boles WE. Out of Gondwana: The origin of passerine birds. Trends Ecol Evol. 2002;17:347–349. [Google Scholar]
- 22.Hall R. In: Biogeography and Geological Evolution of SE Asia. Hall R, Holloway JD, editors. Leiden, The Netherlands: Backhuys Publishers; 1998. pp. 133–163. [Google Scholar]
- 23.Hall R. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: Computer-based reconstructions, model and animations. J Asian Earth Sci. 2002;20:353–431. [Google Scholar]
- 24.Audley-Charles MG. Tectonics of the New Guinea area. Annu Rev Earth Planet Sci. 1991;19:17–41. [Google Scholar]
- 25.Pigram CJ, Symonds PA. A review of the timing of the major tectonic events in the New Guinea Orogen. J Southeast Asian Earth Sci. 1991;6:307–318. [Google Scholar]
- 26.Johansson US, Fjeldså J, Bowie RCK. Phylogenetic relationships within Passerida (Aves: Passeriformes): A review and a new molecular phylogeny based on three nuclear intron markers. Mol Phylogenet Evol. 2008;48:858–876. doi: 10.1016/j.ympev.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Irestedt M, Ohlson JI. The division of the major songbird radiation into Passerida and “core Corvoidea” (Aves: Passeriformes)—The species tree vs. gene trees. Zool Scr. 2008;37:305–313. [Google Scholar]
- 28.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 29.Weir JT, Schluter D. Calibrating the avian molecular clock. Mol Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- 30.Pizzey G, Knight F. Field Guide to the Birds of Australia. London: Harper Collins Publishers; 1997. [Google Scholar]
- 31.Moore RP, Robinson WD, Lovette IJ, Robinson TR. Experimental evidence for extreme dispersal limitation in tropical forest birds. Ecol Lett. 2008;11:960–968. doi: 10.1111/j.1461-0248.2008.01196.x. [DOI] [PubMed] [Google Scholar]
- 32.Pratt D, Bruner PL, Berrett DG. A Field Guide to the Birds of Hawaii and the Tropical Pacific. Princeton, NJ: Princeton University Press; 1987. [Google Scholar]
- 33.Manegold A. Composition and phylogenetic affinities of vangas (Vangidae, Oscines, Passeriformes) based on morphological characters. J Zoological Syst Evol Res. 2008;46:267–277. [Google Scholar]
- 34.Burns KC. Fine-scale food hoarding decisions in New Zealand Robins (Petroica australis): Is inter-sexual competition important? J Ornithol. 2009;150:321–328. [Google Scholar]
- 35.Sol D, Timmermans S, Lefebvre L. Behavioural flexibility and invasion success in birds. Anim Behav. 2002;63:495–502. [Google Scholar]
- 36.Sol D, Duncan RP, Blackburn TM, Cassey P, Lefebvre L. Big brains, enhanced cognition, and response of birds to novel environments. Proc Natl Acad Sci USA. 2005;102:5460–5465. doi: 10.1073/pnas.0408145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmermans S, Lefebvre L, Boire D, Basu P. Relative size of the hyperstriatum ventrale is the best predictor of feeding innovation rate in birds. Brain Behav Evol. 2000;56:196–203. doi: 10.1159/000047204. [DOI] [PubMed] [Google Scholar]
- 38.Greenslade PJM. Island patterns in the Solomon Islands bird fauna. Evolution. 1968;22:751–761. doi: 10.1111/j.1558-5646.1968.tb03475.x. [DOI] [PubMed] [Google Scholar]
- 39.Ricklefs RE, Cox GW. Taxon cycles and the West Indian avifauna. Am Nat. 1972;106:195–219. [Google Scholar]
- 40.Ricklefs RE, Cox GW. Stage of taxon cycle, habitat distribution, and population density in the avifauna of the West Indies. Am Nat. 1978;112:875–895. [Google Scholar]
- 41.Diamond JM. Flightlessness and fear of flying in island species. Nature. 1981;293:507–508. [Google Scholar]
- 42.Ricklefs RE, Bermingham E. History and the species-area relationship in Lesser Antillean birds. Am Nat. 2004;163:227–239. doi: 10.1086/381002. [DOI] [PubMed] [Google Scholar]
- 43.Wilson EO. Adaptive shift and dispersal in a tropical ant fauna. Evolution. 1959;13:122–144. [Google Scholar]
- 44.Wilson EO. The nature of the taxon cycle in the Melanesian ant fauna. Am Nat. 1961;95:169–193. [Google Scholar]
- 45.Holloway JD. The Lepidoptera of Norfolk Island. The Hague: Junk; 1977. [Google Scholar]
- 46.Fjeldså J, Bowie RCK. New perspectives on Africa's ancient forest avifauna. Afr J Ecol. 2008;46:235–247. [Google Scholar]
- 47.Lovette IJ, Bermingham E. Explosive speciation in the New World Dendroica warblers? Proc Biol Sci. 1999;266:1629–1636. [Google Scholar]
- 48.Phillimore AB, Price TD. Density-dependent cladogenesis in birds. PLoS Biol. 2008;6:e71. doi: 10.1371/journal.pbio.0060071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs J, Bowie RCK, Fjeldså J, Pasquet E. Phylogenetic relationships of the African bush-shrikes and helmet-shrikes (Passeriformes: Malaconotidae) Mol Phylogenet Evol. 2004;33:428–439. doi: 10.1016/j.ympev.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Jønsson KA, et al. Explosive avian radiations and multi-directional dispersal across Wallacea: Evidence from the Campephagidae and other Crown Corvida (Aves) Mol Phylogenet Evol. 2008;47:221–236. doi: 10.1016/j.ympev.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 51.Jønsson KA, Irestedt M, Ericson PGP, Fjeldså J. A molecular phylogeny of minivets (Passeriformes: Campephagidae: Pericrocotus): Implications for biogeography and convergent plumage evolution. Zool Scr. 2010;39:1–8. [Google Scholar]
- 52.Jønsson KA, et al. Phylogeny and biogeography of Oriolidae (Aves: Passeriformes) Ecography. 2010;33:1–10. [Google Scholar]
- 53.Norman JA, Ericson PGP, Jønsson KA, Fjeldså J, Christidis L. A multi-gene phylogeny reveals novel relationships for aberrant genera of Australo-Papuan core Corvoidea and polyphyly of the Pachycephalidae and Psophodidae (Aves: Passeriformes) Mol Phylogenet Evol. 2009;52:488–497. doi: 10.1016/j.ympev.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Nyari AS, Benz BW, Jønsson KA, Fjeldså J, Moyle RG. Phylogenetic relationships of fantails (Aves: Rhipiduridae) Zool Scr. 2009;38:553–561. [Google Scholar]
- 55.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 56.Holder MT, Lewis PO. Phylogeny estimation: Traditional and Bayesian approaches. Nat Rev Genet. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- 57.Huelsenbeck JP, Ronquist F. 2003. MrBayes: A program for the Bayesian inference of phylogeny Version 3.1.2. Available from: http://mrbayes.scs.fsu.edu/index.php. Accessed January 8, 2011.
- 58.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 59.Nylander JAA. 2004. MrModeltest2. Available from: http://www.abc.se/∼nylander/. Accessed January 8, 2011.
- 60.Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. 2004. Available at http://king2.scs.fsu.edu/CEBProjects/awty/awty_start.php. Accessed January 8, 2011. [DOI] [PubMed]
- 61.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 62.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 63.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 64.Jønsson KA, et al. Biogeographical history of cuckoo-shrikes (Aves: Passeriformes): transoceanic colonization of Africa from Australo-Papua. J Biogeogr. 2010;37:1767–1781. [Google Scholar]
- 65.Nylander JAA, Olsson U, Alström P, Sanmartín I. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus) Syst Biol. 2008;57:257–268. doi: 10.1080/10635150802044003. [DOI] [PubMed] [Google Scholar]
- 66.Ronquist F. 1996. DIVA version 1.1. Computer program and manual. Available on request from Fredrik Ronquist ( ronquist@csit.fsu.edu)
- 67.Pagel M, Meade A. 2007. BayesTraits version 1.0 http://www.evolution.rdg.ac.uk/BayesTraits.html. Accessed January 8, 2011.
- 68.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 69.Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst Biol. 1999;48:612–622. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.