Abstract
To clarify the molecular pathways governing hematopoietic stem cell (HSC) development, we screened a fetal liver (FL) HSC cDNA library and identified a unique gene, hematopoietic expressed mammalian polycomb (hemp), encoding a protein with a zinc-finger domain and four malignant brain tumor (mbt) repeats. To investigate its biological role, we generated mice lacking Hemp (hemp−/−). Hemp−/− mice exhibited a variety of skeletal malformations and died soon after birth. In the FL, hemp was preferentially expressed in the HSC and early progenitor cell fractions, and analyses of fetal hematopoiesis revealed that the number of FL mononuclear cells, including HSCs, was reduced markedly in hemp−/− embryos, especially during early development. In addition, colony-forming and competitive repopulation assays demonstrated that the proliferative and reconstitution abilities of hemp−/− FL HSCs were significantly impaired. Microarray analysis revealed alterations in the expression levels of several genes implicated in hematopoietic development and differentiation in hemp−/− FL HSCs. These results demonstrate that Hemp, an mbt-containing protein, plays essential roles in HSC function and skeletal formation. It is also hypothesized that Hemp might be involved in certain congenital diseases, such as Klippel-Feil anomaly.
Keywords: fetal liver hematopoiesis, skeletal abnormality
Stem cells are characterized by their ability to balance self-renewal activity with differentiation into mature cell lineages. Among tissue-specific stem cells, hematopoietic stem cells (HSCs) are most intensively studied and well characterized (1, 2). Functional and physiological studies have identified a number of genes that regulate and maintain HSC activity (3–5); however, the overall molecular mechanisms governing HSC function are not fully understood.
To provide insights into these mechanisms, we generated and sequenced a subtracted cDNA library from mouse fetal liver (FL), in which hematopoietic cells are produced and expanded (6). Enrichment of HSC-specific gene products after subtraction was verified by the presence of Runx1, CD34, and flk2/flt3, and the absence of β-actin and other housekeeping genes (6). The experimental procedures, primary datasets, and the results of the computational analyses are contained in the Stem Cell Database (http://stemcell.mssm.edu/v2/).
Among the genes presented in the Stem Cell Database, we identified one that encodes a protein containing a C2C2 zinc finger domain and four tandem malignant brain tumor (mbt) repeats. Mbt is a protein domain originally identified in the protein product of a Drosophila polycomb group (PcG) gene and lethal(3)malignant brain tumor [d-l(3)mbt], whose recessive mutations cause the malignant transformation of larval and adult brain tissues (7). Because the mbt repeats are also found in another Drosophila PcG gene, Scm (sex comb on midleg), and are highly conserved in their human counterparts, H-l(3)mbt (8, 9), SCMH1 (sex comb on midleg homolog 1) (10), and SCML2 (11), we named the this gene hemp [hematopoietic expressed mammalian polycomb, also deposited in the database as mbtd1 (mbt domain containing 1)].
To investigate the biological roles of Hemp, we generated mice deficient in hemp and analyzed their general embryonic development and HSC functions.
Results
Structure and Characterization of Hemp, a Unique mbt-Containing Protein.
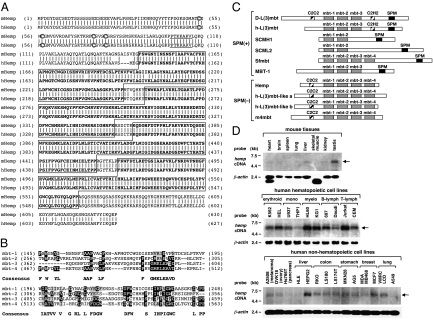
The comparison of amino acid sequences of mouse and human Hemp proteins is shown in Fig. 1A. In both mouse and human, Hemp is composed of a C2C2 zinc-finger domain at the N terminus (circled), a nuclear localization signal (underlined) and four mbt repeats (boldface and boxed). Alignment of the four mbt repeats of mouse Hemp to the consensus sequence is shown in Fig. 1B and the comparative structural characteristics of known mbt-containing proteins are shown in Fig. 1C. The mbt proteins are divided into two groups, depending on whether or not they possess an SPM (Scm, Ph, and MBT) domain, which mediates protein–protein interactions (12, 13). SPM-containing mbt proteins include d-L(3)mbt (7), H-L(3)mbt (8), SCMH1 (10), SCML2 (11), Sfmbt (Scm-related gene containing four mbt domains) (14), and MBT-1 (15) [shown as SPM(+) in Fig. 1C]; SPM-lacking mbt proteins include Hemp, h-L(3)mbt-like a (9), h-L(3)mbt-like b (9), and M4mbt (16) [shown as SPM(−) in Fig. 1C]. It is noteworthy that Hemp and other mbt-containing, SPM-lacking proteins share a structural similarity in that they contain four tandem mbt repeats preceded by a C2C2 zinc-finger domain [SPM(−) in Fig. 1C]. Therefore, these proteins may constitute a previously unexplored subfamily of mbt-containing proteins.
Fig. 1.
Structure and expression of Hemp. (A) Comparison of amino acid sequences of mouse Hemp (Upper) and human Hemp (Lower) proteins. Vertical lines indicate identical amino acids. Zinc-finger domain cysteine residues are circled, a nuclear localization signal is underlined, and each of the four mbt repeats is shown in boldface surrounded by a box. (B) Alignment of the four mbt repeats of mouse Hemp with the mbt consensus sequences. Identical and similar amino acids to the consensus sequences are indicated by black and gray backgrounds, respectively. Amino acid numbers are shown in parentheses. (C) Comparison of the primary structures of previously reported mbt-containing proteins. The proteins are divided into two groups, SPM(+) and SPM(−), depending on whether or not they possess the SPM domain (shown as a black box). The mbt domain and the C2C2/C2HC Zinc-finger domain are indicated by gray and shaded boxes, respectively. (D) Expression of hemp RNA in mouse tissues and in human hematopoietic and nonhematopoietic cell lines. The position of hemp is indicated by an arrow. Hybridization with β-actin cDNA is shown as an internal control.
Because the expression of hemp mRNA in distinct hematopoietic lineages has been reported previously (6), in this study the expression patterns of hemp were examined in mouse tissues and in human hematopoietic and nonhematopoietic cell lines. The results are shown in Fig. 1D. In adult mouse tissues, hemp showed restricted expression, with high expression in the testis (Top). In human cell lines, hemp was expressed abundantly in most of the hematopoietic cell lines (Middle), and it exhibited weak expression in all of the nonhematopoietic cell lines, except HEPG2 (Bottom). These results indicated that hemp is preferentially expressed in hematopoietic cell lines and suggested that Hemp functions largely in hematopoietic cells.
Neonatal Lethality and Skeletal Abnormalities in hemp−/− Mice.
The hemp-deficient mice were generated by replacing the mbt-coding exons with IRES-GFP-pA and the floxed Neo resistance gene (Fig. S1A). As shown in Fig. S1 B and C, the hemp gene was correctly targeted and hemp-null mutant (hemp−/−) mice were obtained.
No live homozygotes were found at 3-wk postnatal littermates [postnatal day (P) 21.5] (Table S1), indicating that hemp−/− mice die perinatally. To determine the time of lethality, embryos and neonates, obtained from timed hemp+/− intercrosses, were genotyped. At embryonic day (E) 12.5 and E17.5, hemp+/+, hemp+/−, and hemp−/− embryos were present almost at Mendelian ratios. However, at P0.5, numbers of hemp−/− pups decreased and most of them were found dead (Table S1). In addition, 1 d later, no live hemp−/− pups were obtained (Table S1), indicating that all hemp−/− neonates die by P1.5.
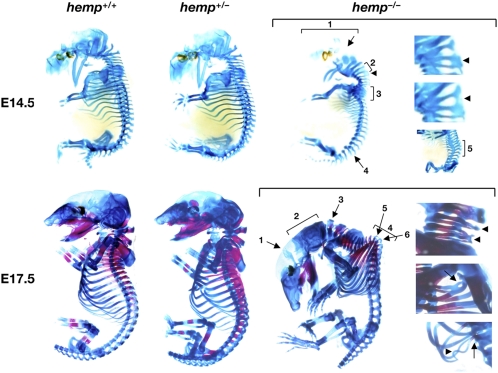
To investigate the cause of hemp−/− postnatal lethality, prenatal and neonatal mice were subjected to pathological analysis. However, despite intensive analysis, no obvious changes were detected in the internal organs of hemp−/− mice. The next step was to examine skeletal formation in hemp−/− mice, because previous studies showed that mice with deficiencies in several of the PcG proteins exhibited abnormal skeletal development (17). Representative whole-mount pictures of hemp+/+, hemp+/−, and hemp−/− embryos at E14.5 and E17.5 are shown in Fig. 2. At E14.5 (Fig. 2, Upper Right), hemp−/− embryos generally exhibited poor skeletal development, most noticeably, the cartilage primordium of the occipital bone at the base of the cranium (No. 1 and an arrow). In addition, insufficient formation of the atlas (C1) and the axis (C2) vertebrae (No. 2), a curvature of the spine (Nos. 3 and 5 in the right lower section), a misalignment of the spine (No. 4), and fusions of the cervical vertebrae [an arrowhead (C2 and C3 fusion), also shown by arrowheads in the right, upper, and center sections (C1 and C2 fusion and C1 to C3 fusion, respectively)] were found in hemp−/− embryos. At E17.5 (Fig. 2, Lower Right), the defects were more pronounced and various abnormalities were observed in hemp−/− embryos, especially in the cranium, vertebrae, and ribs. The most common and prominent feature in hemp−/− embryos was the short lambdoidal suture and flattened interparietal and occipital bones (Nos. 1 and 2). In addition, in the hemp−/− embryo presented in Fig. 2, the atlas (C1) vertebra is adjacent to the occipital bone, there is a prominent curvature of the spine at the seventh and eighth vertebrae (T7 and T8), and three ribs are attached to thoracic vertebra T7 and eight ribs to T8 (Nos. 3–6). Other skeletal malformations in hemp−/− embryos included fusions of cervical vertebrae [arrowheads in the right upper section (C3 and C4, and C5 and C6 fusions)], a fusion and a split at the first ribs (Fig. 2, Right Center, arrow), and the attachment of four ribs to the same vertebra and a deformity of the 12th rib (Fig. 2, Right Lower, arrow and arrowhead). No obvious alterations were found in other parts of the skeleton, such as the limbs. In addition, no abnormalities were found in hemp+/− embryos (Fig. 2, Center), indicating that a haplo-insufficiency of hemp does not cause skeletal abnormalities and there is no dosage effect of Hemp in skeletal development.
Fig. 2.
Representative skeletal photographs of hemp+/+ (Left), hemp+/− (Center), and hemp−/− (Right) embryos at E14.5 (Upper) and E17.5 (Lower). Abnormal skeletal features in hemp−/− embryos are indicated by arrows, arrowheads, and numbers.
Reduction of Mononuclear Cells and HSCs in hemp−/− FL During Early Development.
Because hemp was originally isolated from a highly enriched mouse FL HSC cDNA library, we investigated whether a Hemp deficiency affected embryonic hematopoiesis. Hemp expression levels were initially examined in the FL at E11.5, E14.5, and E18.5. As shown in Fig. S2A, hemp expression was highest at E11.5 and decreased thereafter. Next, the expression levels of hemp were examined at different stages of hematopoietic differentiation using the FL HSC marker, CD150 (18). Cells from E11.5, E14.5 and E18.5 FLs were separated into CD150+, lineage marker (Lin)-negative, Sca-1+, and c-Kit+ (LSK); CD150– LSK (putative HSCs); progenitor, Lin– and Lin+ cell fractions, and RNA, extracted from each fraction, was subjected to quantitative real-time PCR. As shown in Fig. S2B, at E11.5, hemp was expressed at roughly the same level in all four fractions, whereas at E14.5 and E18.5 it was predominantly expressed in the CD150+ and CD150– LSK fractions. These results indicated that hemp is preferentially expressed in primitive hematopoietic cells, including HSCs, and may play a major role in these types of cells.
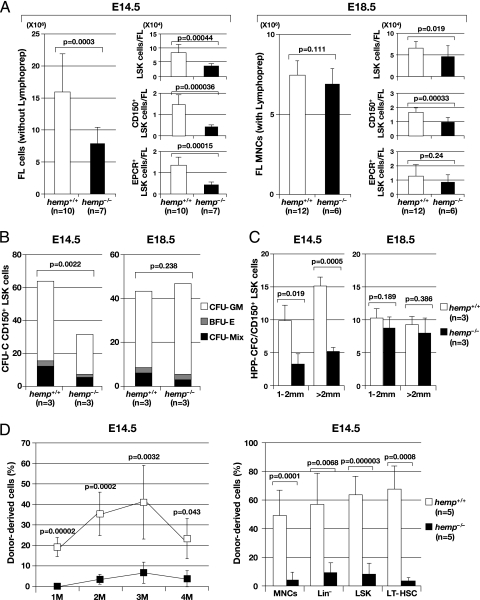
Next, cell numbers were examined in the FL of hemp+/+ and hemp−/− embryos at E14.5 and E18.5. At E14.5, the FL sizes of hemp−/− embryos were significantly smaller than those in hemp+/+ embryos. Thus, to ensure the recovery of HSCs at this time point, we collected FL cells without using lymphoprep gradients (Lymphoprep). On the other hand, at E18.5, because the FL sizes of hemp−/− and hemp+/+ embryos were almost comparable, we separated FL mononuclear cells by using Lymphoprep. Our previous study demonstrated that the use of these two different methods (preparation with or without Lymphoprep) does not affect the characteristics of the isolated cells, in terms of surface markers, functional abilities, or number of HSCs (19). As shown in the left panels of E14.5 and E18.5 in Fig. 3A, at E14.5, hemp−/− FL cell numbers were reduced approximately twofold relative to controls (E14.5 of Fig. 3A, Left), whereas at E18.5, both hemp+/+ and hemp−/− FLs contained comparable numbers of mononuclear cells (E18.5 of Fig. 3A, Left).
Fig. 3.
Phenotypic characterization and functional assays of hemp+/+ and hemp−/− FL cells. (A) Comparison of total cell numbers (Left) and absolute cell numbers for LSK (Right Top) CD150+ LSK (Right, Middle) and EPCR+ LSK (Right, Bottom) fractions between hemp+/+ and hemp−/− FLs at E14.5 and E18.5 (data ± SD). At E14.5, FL cells were collected without using lymphoprep gradients (Lymphoprep), whereas at E18.5, FL mononuclear cells (MNCs) were separated with the use of Lymphoprep. (B) Average numbers of colony forming units in culture (CFU-C) generated from hemp+/+ and hemp−/− CD150+ LSK cells at E14.5 and E18.5. One hundred and fifty cells were cultured for 8 d in a semi-solid culture with cytokines, and the colony types formed were classified into colony-forming unit-granulocyte, macrophage [(CFU-GM) open bar], burst-forming unit-erythoid [(BFU-E) gray bar], CFU-Mix (closed bar) and counted (data ± SD). (C) Average numbers of high proliferative potential-colony forming cells (HPP-CFCs) generated from hemp+/+ (open bar) and hemp−/− (closed bar) CD150+ LSK cells at E14.5 and E18.5. One hundred and fifty cells were cultured for 16 d in a semi-solid culture with cytokines, and colonies from 1–2 mm and >2 mm were classified and counted independently (data ± SD). (D) Competitive repopulation assay of hemp+/+ (open box) and hemp−/− (closed box) CD150+ LSK cells at E14.5. One hundred and fifty cells (Ly5.2) were transplanted into recipient mice (Ly5.1) together with 4×105 competitors (Ly5.1). Donor-derived chimerism in the peripheral blood (PB) at 1, 2, 3 and 4 months after transplantation and chimerism in the MNC, Lin−, LSK and LT-HSC fractions in the bone marrow (BM) at 4 mo after transplantation are shown in the left and right panels, respectively (data ± SD).
To investigate whether the reduction in cell numbers in the hemp−/− FL was caused by a differentiation block at a specific developmental stage, or by a proliferative defect of HSCs, flow cytometric analysis was performed using the lineage markers, Sca-1, c-Kit, CD150, and endothelial protein C receptor (EPCR), another marker for FL-HSCs (19). The absolute cell numbers per FL in each fraction are shown in E14.5 and E18.5 in Fig. 3A Right. The absolute LSK number in the hemp−/− FL was significantly reduced compared with that in the hemp+/+ FL at both E14.5 and E18.5 (Fig. 3A, Top Right: E14.5 and E18.5). Regarding the HSC frequencies in the LSK fractions, hemp−/− FL showed a significant decrease in absolute cell numbers of CD150+ LSK and EPCR+ LSK fractions at E14.5 (Fig. 3A, Right Middle, and Bottom: E14.5). At E18.5, although the cell number of CD150+ LSK were still low in hemp−/− FL (Fig. 3A, Right Middle: E18.5), those of EPCR+ LSK were comparable between the two types of embryos (Fig. 3A, Right Bottom: E18.5). These results indicated that a deficiency in Hemp impairs HSC development, especially at an early developmental stage.
Impaired Proliferative and Repopulating Ability of hemp−/− HSCs.
To test the functional properties of primitive hemp−/− hematopoietic cells, we first investigated the colony-forming units in culture (CFU-C) and then performed high proliferative potential colony-forming cell (HPP-CFC) assays, which reflect progenitor cell capacity, using E14.5 and E18.5 CD150+ LSK cells (Fig. 3 B and 3C). At E14.5, hemp−/− CD150+ LSK cells generated a significantly reduced number of colonies in both CFU-C and HPP-CFC assays compared with hemp+/+ CD150+ LSK cells, indicating a marked decrease in the proliferative ability of hemp−/− HSCs (Fig. 3 B and 3C, Left). In contrast, at E18.5 no apparent differences were observed between the two assays (Fig. 3 B and 3C, Right). These results indicated that a Hemp deficiency impaired the proliferative potentials of hematopoietic stem/early progenitor cells at an early developmental stage.
Next, a competitive repopulation assay was performed to investigate the stem cell capacity of hemp−/− CD150+ LSK cells at E14.5. As shown in Fig. 4C, the repopulation capacity of hemp−/− CD150+ LSK cells was significantly reduced compared with hemp+/+ CD150+ LSK cells, as indicated by a marked decrease in the percentages of donor-derived cells in peripheral blood (Fig. 3D, Left) and bone marrow (Fig. 3D, Right) in the recipient mice. These results demonstrated that a Hemp deficiency resulted in a severe functional defect of FL HSCs.
To gain mechanistic insights into HSC defects, the cell-cycle status and apoptotic ratios for CD150+ and CD150– LSK cells from E14.5 and E18.5 FLs were examined. A significant reduction in short-term (90 min) BrdU incorporation was observed in CD150+ LSK cells from hemp−/− FL at E14.5 (Fig. S3A, Left), indicating increased cell-cycle arrest in this population. In addition, a marked increase in cleaved caspase-3–positive cells was detected in CD150+ and CD150– LSK cells from hemp−/− FL at E14.5 (Fig. S3B, Left), indicating enhanced apoptosis in these cell populations. In contrast, At E18.5 these changes were attenuated and no significant differences were found (Fig. 3 A and B, Right). These results demonstrated that Hemp contributed significantly to the reconstitution ability of FL HSCs, potentially through the fine-tuning of the cell-cycle and cell-survival pathways.
Altered Gene-Expression Profiles in hemp−/− FL HSCs.
To elucidate the detailed molecular mechanism underlying the impaired reconstitution ability of hemp−/− FL HSCs, we searched for genes whose expression levels were altered in FL HSCs by the Hemp deficiency. Total RNAs extracted from the lin–/c-kithigh HSC-enriched fraction of E12.5 hemp+/+ and hemp−/− FLs were subjected to microarray analysis. Among 23,522 genes examined, 308 genes were identified that exhibited absolute expression levels of more than 50 (SI Materials and Methods) in either hemp+/+ or hemp−/− arrays, and showed alterations in expression of more than 1.5-fold between the two genotypes. Gene names are listed in Table S2 (>1.5-fold up-regulated in hemp−/− compared with hemp+/+) and Table S3 (>1.5-fold down-regulated in hemp−/− compared with hemp+/+), along with additional information concerning each gene. The marked reduction of mbtd1 (=hemp) in the hemp−/− arrays verified the quantitative accuracy of the analysis (top row of Table S3). Of the 308 genes, 210 and 229 genes were classified according to “Biological process” and “Molecular function” Gene Ontology (GO) annotation, respectively (Fig. S4A).
Among the 308 genes, 39 genes were further selected based on their greater than twofold expression differences between the hemp+/+ and hemp−/− arrays. The gene names, classified according to the GO category and expression-difference ratios are listed in Table 1 and the actual gene-expression levels in hemp+/+ and hemp−/− arrays are shown in Fig. S4B. Interestingly, these genes included those reported to play important roles in hematopoietic development and HSC reconstitution ability, as discussed below.
Table 1.
Genes with expression levels showing greater than a twofold difference between hemp+/+ and hemp−/− arrays
| Category | Gene symbol | Global normalization ratio |
| Transcription | Prdm16 | 2.3↑ |
| Sox4 | 0.5↓ | |
| Zmynd11 | 0.5↓ | |
| Stat4 | 0.5↓ | |
| Hnrnpul1 | 0.4↓ | |
| Signal transduction | Arhgap21 | 4.1↑ |
| Eltd1 | 0.4↓ | |
| Arhgef18 | 0.4↓ | |
| Olfr1461 | 0.4↓ | |
| Metabolic process | Acot2 | 0.5↓ |
| Sepp1 | 0.5↓ | |
| Aqp9 | 0.4↓ | |
| Pitpnc1 | 0.3↓ | |
| Translation | Eif2s3y | 2.3↑ |
| Aars2 | 0.4↓ | |
| Oxygen transport | Hbb-b1 | 0.5↓ |
| Hba-a1 | 0.4↓ | |
| Immune response | Ccl4 | 0.4↓ |
| Cxcl2 | 0.4↓ | |
| Chemotaxis | Ear2 | 2.2↑ |
| Protein folding | Fkbp11 | 2.1↑ |
| Apoptosis | Nfkbia | 0.5↓ |
| Others | Dcdc5 | 4.4↑ |
| Evc2 | 3.9↑ | |
| Acbd7 | 3.3↑ | |
| Adamtsl1 | 2.9↑ | |
| Ear1 | 2.2↑ | |
| Ear10 | 2.1↑ | |
| Tlcd2 | 2.0↑ | |
| Ear6 | 2.0↑ | |
| Dolpp1 | 0.5↓ | |
| EG625796 | 0.5↓ | |
| Slc6a20a | 0.4↓ | |
| Gal3st2 | 0.4↓ | |
| Erdr1 | 0.4↓ | |
| Dusp2 | 0.4↓ | |
| Xist | 0.3↓ | |
| Lelp1 | 0.3↓ | |
| Mboat2 | 0.3↓ |
Genes with a greater than twofold increase in expression in hemp−/− arrays are shown in boldface.
Discussion
In this study, we generated mice deficient in Hemp, an mbt-encoding protein, and found that a loss of Hemp induced severe abnormalities in HSC function and skeletal formation.
To investigate whether these abnormalities were caused directly by the Hemp deficiency, Hemp expression patterns during embryogenesis were examined using an anti-Hemp antibody. As shown in Fig. S5, various tissues were stained positively at E11.5 and E14.5 (Fig. S5, Left). The liver and cartilage, where phenotypical and functional abnormalities were found in hemp−/− embryos, were examined in detail. In the liver, interstitial cells, including hematopoietic cells, showed positive staining; and in the cartilage, most of the chondrocytes and some undifferentiated mesenchymal cells exhibited positive signals (Fig. S5, Right: E11.5 and E14.5). These results strongly suggested that the defects in hemp−/− embryos were primarily attributed to the Hemp deficiency. The reason for the lack of obvious defects in other tissues expressing Hemp, such as the heart (Fig. S5), is not yet clear. One possibility is that Hemp-related proteins (Fig. 1C) might compensate for Hemp function in these tissues.
The skeletal abnormalities were considered to be responsible for the postnatal lethality of the hemp−/− mice. The impaired thoracic formation would cause respiratory failure, lead to hypoxia, and eventually result in neonatal death. Skeletal malformations are one of the characteristic abnormalities in mice lacking several of the PcG genes, such as bmi-1 (20), mel-18 (21), and M33 (22). However, these mice exclusively displayed anterior/posterior homeotic transformations of vertebrae because of the ectopic or deregulated expression of Hox gene families (17), in contrast to hemp−/− mice, which exhibited very complex patterns that did not correspond to the homeotic transformations (Fig. 2). Therefore, Hemp appears to regulate skeletal development in a manner distinct from that of the PcG proteins.
Interestingly, the cervical vertebral fusion and an atlas (C1) vertebra located adjacent to the occipital bone (common skeletal abnormalities in hemp−/− mice) closely resembled the features of the Klippel-Feil anomaly in humans (23, 24). In addition, rib anomalies are frequently associated with this disease (25). The Klippel-Feil anomaly is etiologically heterogeneous, but several cases bear chromosomal aberrations and thus are considered to have a genetic component (26, 27). Of note, one case with Klippel-Feil anomaly had a translocation involving chromosome band 17q23 (26), which is close to the human hemp orthologous locus at 17q21.3. In addition, several patients with unrelated skeletal abnormalities were shown to bear chromosomal aberrations involving 17q21.3 (28–30). Therefore, it would be intriguing to investigate whether human hemp expression is affected in patients with Klippel-Feil anomaly and patients with skeletal abnormalities carrying chromosomal aberrations involving 17q21.3.
In the hematopoietic analyses, we observed a significant reduction in FL cell numbers in hemp−/− embryos at E14.5 (Fig. 3A). Flow cytometric analysis revealed that the reduction occurred at the HSC level (Fig. 3A), demonstrating a critical role for Hemp in HSC development at an early developmental phase and leading to further investigations into the proliferative and repopulating abilities of HSCs. CFU-C and HPP-CFC assays of CD150+ LSK cells revealed that hemp−/− HSCs exhibited a significantly reduced proliferative ability at E14.5 (Fig. 3 B and C). In addition, transplantation of CD150+ LSK cells at E14.5 revealed a marked impairment in the repopulating activity of hemp−/− HSCs (Fig. 3D). These results indicated that Hemp plays a pivotal role, not only in hematopoietic development, but also in HSC proliferative and repopulating abilities. The BrdU and cleaved caspase-3 assays revealed significantly decreased proliferation and significantly enhanced apoptosis of hemp−/− CD150+ LSK cells at E14.5 (Fig. S3 A and B), which would explain, at least partly, the defects in hemp−/− HSCs. The reason why the defects in hemp−/− HSCs were most prominent at E14.5 but were less remarkable at E18.5 is not clear. One possibility is that hemp expression in the FL is highest at E11.5 and is down-regulated thereafter (Fig. S2A). However, at E18.5, hemp expression in the CD150+ and CD150– LSK fractions remained dominant (Fig. S2B). Thus, the decline of the overall hemp expression in the FL at a later stage of development could be attributed to decreased hemp expression in Lin– and Lin+ fractions at E18.5, because these cell types expand massively and occupy the majority of FL cells. Another, more likely, possibility is that Hemp-related molecules (Fig. 1C) may compensate for the defects of Hemp deficiency during late gestation.
To gain insights into the molecular mechanisms underlying the impaired hematopoietic activities of hemp−/− HSCs, a microarray analysis was performed. Among 23,522 genes examined, 39 genes were identified whose expression levels differed by more than twofold in enriched hemp+/+ or hemp−/− stem and progenitor cell arrays (Table 1 and Fig. S4B), which included genes that have been shown to play pivotal roles in HSC self-renewal capacity and differentiation ability.
Among genes down-regulated in hemp−/− FL HSCs, Sox4 (0.5) is necessary for lymphocyte development (31, 32) (the figure in parenthesis refers to the ratio of hemp−/−/hemp+/+, in this case, a value of 0.5). Of the two Stat genes identified, Stat4 (0.5) is required for T-cell differentiation (33), and Stat3 (0.6) plays important functions in HSC self-renewal (34). KLF6 (0.6) is required for proper hematopoietic development (35), two of the chemokine family members, Ccl4 (0.4) and Cxcl2 (0.4), contribute to proliferation, survival and homing of hematopoietic cells (36), and Erdr1 (0.4) is capable of inducing hemoglobin synthesis (37). Among the genes up-regulated in hemp−/− FL HSC and progenitor cells, SOCS2 (1.7) negatively regulates cytokine-mediated pathways (38) and MAP4K1 (1.8) induces apoptosis by activating MAP3 proteins and suppressing Bcl2 family members (39, 40). Taken together, the altered expression patterns of various genes in mutant FL HSCs appear to coordinately and cooperatively impair the expansion and reconstitution ability of FL HSCs.
Recently, a functional screen for gene products supporting HSC activity identified 18 molecules, and the overexpression of 10 of these was shown to increase HSC repopulating activity (41). Interestingly, 3 of the 10 genes overlapped with the 39 genes identified in this study (Table 1 and Fig. S4B). Two of these were Sox4 and Erdr1, which were down-regulated in hemp−/− FL HSCs (0.5 and 0.4, respectively) (Table 1 and Fig. S4B). Therefore, it is strongly postulated that the suppressed expression of these two gene products is responsible for the impaired reconstitution ability of hemp−/− FL HSCs. The third gene, Prdm16 (also known as Mel1), was up-regulated in hemp−/− FL HSCs (2.3) (Table 1 and Fig. S4B). This finding is intriguing, because Prdm16 was originally isolated as a leukemia-associated gene (42) and is known to enhance HSC activity (41). The mechanism underlying the enhanced expression of Prdm16 remains unclear, but one possibility is that Prdm16 is up-regulated to compensate for the impaired proliferative ability of FL HSCs induced by Hemp deficiency.
In summary, we have demonstrated that Hemp, an mbt-containing protein, plays essential roles in HSC function and skeletal formation. Recent studies have shown that the mbt domain binds to methylated histone residues, for example, the mbt domain of H-L(3)MBT/L3MBTL1 binds to methylated H3K4 and H4K20 (43, 44) and also compacts nucleosomal arrays by simultaneously binding to two methylated residues, H4K20 and H1bK26 (45). In addition, Sfmbt binds to mono- and dimethylated H3K9 and H4K20 (46, 47), and its gene-silencing activity is enhanced by interacting with another mbt-containing protein, Scm (48). Therefore, it is likely that Hemp exerts its biological activity by binding to methylated histone lysine residues through the mbt repeats. Further studies will be required to clarify the lysine residues to which Hemp binds and the proteins with which Hemp interacts. In addition, an association between skeletal anomalies and impaired hematopoiesis has been reported in human diseases (49), and in particular, the Klippel-Feil anomaly can be associated with Fanconi anemia (50) and Diamond-Blackfan syndrome (51, 52). Therefore, it will be interesting to determine whether human Hemp dysfunctions are involved in the pathogenesis of these diseases.
Materials and Methods
Detailed descriptions for experimental procedures for Northern blot analysis, construction of a targeting vector and generation of hemp knockout mice, generation of an anti-Hemp antibody, immunoprecipitation and Western blot analysis, skeletal analysis, quantitative real-time PCR, immunohistochemical analysis, flow cytometric analysis, CFU-C, HPP-CFC and competitive repopulation assays, cell cycle and apoptosis assays, and DNA microarray analysis, microarray scanning and data processing are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Yuki Sakai, Kayoko Hashimoto, Yuko Tsukawaki, Tai Rika, and Tomoko Muraki for the mouse care and technical assistance, and Masaaki Miyazaki for help in preparing specimens and hematopoietic analysis, Toshikazu Ushijima for providing us with non-hematopoietic cell lines, and Tetsuo Sudo and Hideo Akiyama from Toray Co., Ltd. for microarray analysis. This work was supported by a Grant-in-Aid from the Ministry of Education, Science and Culture of Japan, a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan (13-2), and a Human Frontier Science Program Organization Long-Term Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003403108/-/DCSupplemental.
References
- 1.Lemischka IR, Raulet DH, Mulligan RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- 2.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 3.Akala OO, Clarke MF. Hematopoietic stem cell self-renewal. Curr Opin Genet Dev. 2006;16:496–501. doi: 10.1016/j.gde.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 5.Dzierzak E, Speck NA. Of lineage and legacy: The development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips RL, et al. The genetic program of hematopoietic stem cells. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- 7.Wismar J, et al. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev. 1995;53:141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- 8.Koga H, et al. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- 9.Wismar J. Molecular characterization of h-l(3)mbt-like: A new member of the human mbt family. FEBS Lett. 2001;507:119–121. doi: 10.1016/s0014-5793(01)02959-3. [DOI] [PubMed] [Google Scholar]
- 10.Berger J, et al. The human homolog of Sex comb on midleg (SCMH1) maps to chromosome 1p34. Gene. 1999;237(1):185–191. doi: 10.1016/s0378-1119(99)00285-1. [DOI] [PubMed] [Google Scholar]
- 11.Montini E, et al. Identification of SCML2, a second human gene homologous to the Drosophila sex comb on midleg (Scm): A new gene cluster on Xp22. Genomics. 1999;58:65–72. doi: 10.1006/geno.1999.5755. [DOI] [PubMed] [Google Scholar]
- 12.Bornemann D, Miller E, Simon J. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development. 1996;122:1621–1630. doi: 10.1242/dev.122.5.1621. [DOI] [PubMed] [Google Scholar]
- 13.Peterson AJ, et al. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol Cell Biol. 1997;17:6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usui H, Ichikawa T, Kobayashi K, Kumanishi T. Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the Polycomb group of genes. Gene. 2000;248(1-2):127–135. doi: 10.1016/s0378-1119(00)00131-1. [DOI] [PubMed] [Google Scholar]
- 15.Arai S, Miyazaki T. Impaired maturation of myeloid progenitors in mice lacking novel Polycomb group protein MBT-1. EMBO J. 2005;24:1863–1873. doi: 10.1038/sj.emboj.7600654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markus J, Feiková S, Sramko M, Wolff L, Bies J. Proliferation-linked expression of the novel murine gene m4mbt encoding a nuclear zinc finger protein with four mbt domains. Gene. 2003;319:117–126. doi: 10.1016/s0378-1119(03)00801-1. [DOI] [PubMed] [Google Scholar]
- 17.van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki H, Arai F, Kubota Y, Dahl M, Suda T. Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood. 2010;116:544–553. doi: 10.1182/blood-2009-08-240903. [DOI] [PubMed] [Google Scholar]
- 20.van der Lugt NM, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 21.Akasaka T, et al. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 22.Coré N, et al. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 23.Klimo P, Jr, Rao G, Brockmeyer D. Congenital anomalies of the cervical spine. Neurosurg Clin N Am. 2007;18:463–478. doi: 10.1016/j.nec.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Smoker WR, Khanna G. Imaging the craniocervical junction. Childs Nerv Syst. 2008;24:1123–1145. doi: 10.1007/s00381-008-0601-0. [DOI] [PubMed] [Google Scholar]
- 25.Baga N, Chusid EL, Miller A. Pulmonary disability in the Klippel-Feil syndrome. A study of two siblings. Clin Orthop Relat Res. 1969;67:105–110. [PubMed] [Google Scholar]
- 26.Fukushima Y, et al. De novo apparently balanced reciprocal translocation between 5q11.2 and 17q23 associated with Klippel-Feil anomaly and type A1 brachydactyly. Am J Med Genet. 1995;57:447–449. doi: 10.1002/ajmg.1320570317. [DOI] [PubMed] [Google Scholar]
- 27.Goto M, Nishimura G, Nagai T, Yamazawa K, Ogata T. Familial Klippel-Feil anomaly and t(5;8)(q35.1;p21.1) translocation. Am J Med Genet. 2006;140:1013–1015. doi: 10.1002/ajmg.a.31198. [DOI] [PubMed] [Google Scholar]
- 28.Yue Y, et al. De novo t(12;17)(p13.3;q21.3) translocation with a breakpoint near the 5′ end of the HOXB gene cluster in a patient with developmental delay and skeletal malformations. Eur J Hum Genet. 2007;15:570–577. doi: 10.1038/sj.ejhg.5201795. [DOI] [PubMed] [Google Scholar]
- 29.Rooryck C, et al. A 580 kb microdeletion in 17q21.32 associated with mental retardation, microcephaly, cleft palate, and cardiac malformation. Eur J Med Genet. 2008;51:74–80. doi: 10.1016/j.ejmg.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Zahir FR, et al. A novel de novo 1.1 Mb duplication of 17q21.33 associated with cognitive impairment and other anomalies. Am J Med Genet. 2009;149A:1257–1262. doi: 10.1002/ajmg.a.32827. [DOI] [PubMed] [Google Scholar]
- 31.Schilham MW, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 32.Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. Eur J Immunol. 1997;27:1292–1295. doi: 10.1002/eji.1830270534. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 34.Hankeys PA. Regulation of hematopoietic cell development and function by Stat3. Front Biosci. 2009;1:5273–5290. doi: 10.2741/3597. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto N, et al. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broxmeyer HE. Regulation of hematopoiesis by chemokine family members. Int J Hematol. 2001;74:9–17. doi: 10.1007/BF02982544. [DOI] [PubMed] [Google Scholar]
- 37.Dörmer P, Spitzer E, Frankenberger M, Kremmer E. Erythroid differentiation regulator (EDR), a novel, highly conserved factor I. Induction of haemoglobin synthesis in erythroleukaemic cells. Cytokine. 2004;26:231–242. doi: 10.1016/j.cyto.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Krebs DL, Hilton DJ. SOCS proteins: Negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 39.Brenner D, et al. Caspase-cleaved HPK1 induces CD95L-independent activation-induced cell death in T and B lymphocytes. Blood. 2007;110:3968–3977. doi: 10.1182/blood-2007-01-071167. [DOI] [PubMed] [Google Scholar]
- 40.Shui JW, et al. Hematopoietic progenitor kinase 1 negatively regulates T cell receptor signaling and T cell-mediated immune responses. Nat Immunol. 2007;8:84–91. doi: 10.1038/ni1416. [DOI] [PubMed] [Google Scholar]
- 41.Deneault E, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mochizuki N, et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- 43.Kim JY, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min J, et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 45.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 46.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Y, et al. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 2009;37:2204–2210. doi: 10.1093/nar/gkp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimm C, et al. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charles JE, Robert PE, Anthony WB. Inborn Errors of Development. 2nd Ed. Oxford, United Kingdom: Oxford University Press; 2008. [Google Scholar]
- 50.McGaughran J. Klippel-Feil anomaly in Fanconi anemia. Clin Dysmorphol. 2003;12(3):197. doi: 10.1097/01.mcd.0000077560.66911.1f. [DOI] [PubMed] [Google Scholar]
- 51.Greenspan A, Cohen J, Szabo RM. Klippel-Feil syndrome. An unusual association with Sprengel deformity, omovertebral bone, and other skeletal, hematologic, and respiratory disorders. A case report. Bull Hosp Jt Dis Orthop Inst. 1991;51(1):54–62. [PubMed] [Google Scholar]
- 52.Lazarus KH, McCurdy FA. Multiple congenital anomalies in a patient with Diamond-Blackfan syndrome. Clin Pediatr (Phila) 1984;23:520–521. doi: 10.1177/000992288402300918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.