Abstract
Transposable elements (TEs) are often the primary determinant of genome size differences among eukaryotes. In plants, the proliferation of TEs is countered through epigenetic silencing mechanisms that prevent mobility. Recent studies using the model plant Arabidopsis thaliana have revealed that methylated TE insertions are often associated with reduced expression of nearby genes, and these insertions may be subject to purifying selection due to this effect. Less is known about the genome-wide patterns of epigenetic silencing of TEs in other plant species. Here, we compare the 24-nt siRNA complement from A. thaliana and a closely related congener with a two- to threefold higher TE copy number, Arabidopsis lyrata. We show that TEs—particularly siRNA-targeted TEs—are associated with reduced gene expression within both species and also with gene expression differences between orthologs. In addition, A. lyrata TEs are targeted by a lower fraction of uniquely matching siRNAs, which are associated with more effective silencing of TE expression. Our results suggest that the efficacy of RNA-directed DNA methylation silencing is lower in A. lyrata, a finding that may shed light on the causes of differential TE proliferation among species.
Keywords: gene silencing, transposons
Transposable elements (TEs) constitute the largest component of higher plant genomes, and they are the major contributor to genome size differences among plant species (1). Although the evolutionary forces that govern the accumulation or removal of TEs over many generations are not fully understood, it is known that TE activity in individual plants is suppressed by epigenetic pathways (2). These pathways require 24-nucleotide (nt) small interfering RNAs (siRNAs) that target specific TE insertions via sequence identity (3). The 24-nt siRNAs combine with protein complexes and other RNA transcripts to guide methylation of target DNA (4, 5). The importance of methylation for moderating TE activity has been demonstrated with Arabidopsis thaliana mutants (2, 6). For example, met1 and ddm1 mutants have decreased levels of TE methylation, with concomitant increases in the expression and activity of some TEs (7–14). These and similar studies have established a strong correlation among siRNA targeting, DNA methylation, and transcriptional gene silencing (TGS) of TEs.
DNA methylation may affect not only TE activity, but also the expression of nearby genes. Although the mechanism of TE-triggered gene silencing is not fully understood, the phenomenon has been demonstrated for several genes (15–17). The suppression of gene expression can generate adaptive variation, an example of which is provided by the down-regulation of the A. thaliana FWA gene by methylation of an upstream TE, which in turn prevents a delay in flowering (13, 18). More generally, however, methylation of TEs near genes is likely to have deleterious effects on gene and genome function, as was demonstrated recently in a population-genomic study of A. thaliana (19). This study showed that methylated TEs near genes are under stronger purifying selection than other TEs, suggesting both that TE methylation has deleterious effects and that these effects vary as a function of the distance to genes (19). Thus, the emerging picture is that TE methylation involves an evolutionary tradeoff: The benefit is reduced TE activity, but the cost is the potential perturbation of gene expression.
Most of our knowledge about the biochemical mechanisms and patterns of plant DNA methylation comes from A. thaliana (20, 21). Unfortunately, the A. thaliana genome is depauperate of TEs compared with most angiosperms (1). It is thus unclear whether A. thaliana is typical in its pattern and extent of siRNA-based TE silencing. Is targeting of TEs by 24-nt siRNAs less or more effective in other species? Does TE silencing have an association with gene expression in other species, as it does in A. thaliana? Ultimately, do differences in siRNA targeting between species help explain variation in TE copy numbers—and thus genome sizes—among angiosperms?
To begin to answer these questions, we make use of the recently sequenced Arabidopsis lyrata genome. Although A. thaliana and A. lyrata shared an ancestor only 10 million years ago (22, 23), they differ in numerous respects. First, A. lyrata has eight chromosomes, but A. thaliana experienced a series of chromosomal fusions resulting in five chromosomes (24). Second, A. lyrata has an ≈1.5-fold larger genome. Some of the difference in genome size can be attributed to TEs, but other factors—such as intron sizes, gene number, and the loss of chromosomes—contribute as well. Finally, the two species differ in mating system. A. lyrata is a (mostly) obligate outcrosser (25), whereas A. thaliana is predominantly a selfer. The difference in mating system has potential implications for TE evolution; the efficacy of selection against TEs is expected to be different in selfers and outcrossers, but the direction of the difference depends critically on the mechanism of selection (26, 27). To date, the empirical consensus is that reduced recombination in selfers like A. thaliana leads to less effective selection against TE insertions (28, 29). Yet, despite major differences between these two species, the genomes of A. lyrata and A. thaliana are largely collinear, with ∼80% sequence identity in alignable regions (including intergenic regions). As a result, orthologs can be easily identified between species.
Here we investigate TE abundance, siRNA targeting, and their potential effects on gene expression in A. thaliana and A. lyrata. To facilitate this interspecies comparison, we have assembled datasets of TEs from both genomes, used 24-nt siRNA data from both species, and complemented these data with mRNA expression information. Our analyses focus on two specific questions. First, is there evidence that TE silencing is correlated with gene expression in A. lyrata as it is in A. thaliana—and are the associations similar with respect to the distance of TEs from genes? Second, do the species differ with regard to siRNA targeting of TEs—and what might these differences mean both for the efficacy of silencing and for the accumulation of TEs?
Results
Distributions of TEs and Genes.
To compare the TE distributions between genomes, we used the same TE discovery pipeline to assemble parallel datasets (SI Materials and Methods), resulting in 22,818 A. thaliana TE insertions (covering a total of 19.2 Mb) and 67,033 A. lyrata TE insertions (48.6 Mb; see Materials and Methods). A. lyrata had twofold to threefold higher copy numbers of every major TE family examined, including both class I retrotransposons (gypsy, copia, LINE, and SINE) and class II DNA elements (Table 1). The correlation of TE copy number in different families was high between species (r2 = 0.86), indicating few (if any) family-specific expansions or reductions since the two species shared a common ancestor.
Table 1.
Comparison of major TE families in A. thaliana and A. lyrata
| TE family | Species | Copy no. | Mean length, bp | Total length, kbp | % Only multiply mapping siRNAs |
| Gypsy | Ath | 2,734 | 2,500 | 6,835 | 13 |
| Aly | 5,800 | 2,668 | 15,474 | 24 | |
| Copia | Ath | 2,042 | 1,018 | 4,245 | 15 |
| Aly | 5,632 | 1,196 | 6,735 | 30 | |
| LINE | Ath | 2,437 | 672 | 1,637 | 6 |
| Aly | 5,922 | 846 | 5,010 | 16 | |
| SINE | Ath | 802 | 174 | 140 | 10 |
| Aly | 2,793 | 263 | 735 | 17 | |
| Helitron | Ath | 3,437 | 640 | 2,130 | 13 |
| Aly | 10,452 | 523 | 5,466 | 40 | |
| MULE | Ath | 3,096 | 1,031 | 3,192 | 9 |
| Aly | 7,384 | 538 | 3,973 | 33 | |
| hAT | Ath | 1,544 | 411 | 635 | 8 |
| Aly | 5,548 | 234 | 1,298 | 24 | |
| Mariner | Ath | 269 | 212 | 57 | 12 |
| Aly | 640 | 227 | 145 | 14 | |
| Pogo | Ath | 460 | 335 | 154 | 15 |
| Aly | 917 | 299 | 274 | 44 | |
| Tc1 | Ath | 364 | 187 | 68 | 24 |
| Aly | 2667 | 217 | 579 | 27 | |
| Stowaway | Ath | 53 | 170 | 9 | 25 |
| Aly | 470 | 202 | 95 | 16 |
Ath, A. thaliana; Aly, A. lyrata.
We calculated the density of TEs and genes in 100-kilobase pair (kbp) windows. The median TE density on A. lyrata chromosome arms was 23 kbp per 100-kbp window (0.23), which was nearly fivefold higher than in A. thaliana (0.045) and significant by a Mann–Whitney U test (MWU) at P << 10−10. Conversely, gene density was higher on A. thaliana chromosome arms (0.33 vs. 0.57; MWU, P << 10−10). Nonetheless, within each species, the densities of TEs and genes were negatively correlated (Fig. S1).
The higher density of TEs on A. lyrata chromosomal arms has the consequence that A. lyrata genes are on average closer to TEs than are A. thaliana genes (MWU, P < 3−16). Indeed, 10.5% (or 2,567) of A. lyrata genes harbor TE insertions, whereas only 6.5% (or 1,641) of A. thaliana genes do [Fisher's exact test (FET), P << 10−10]. Similarly, 24% (or 5,840) of A. lyrata genes have a TE insertion located within 500 bp 5′ or 3′ of the coding region, compared with 16% (or 4,030) in A. thaliana (FET, P << 10−10).
Targeting of TEs by 24-nt siRNAs.
One of the many factors that could influence the accumulation of TEs near genes is siRNA-guided TGS (Discussion). To investigate this possibility, we mapped 24-nt siRNAs, which are primarily associated with pretranscriptional silencing of TEs (14), to genomic locations. The siRNA datasets for the two species were produced from the same floral tissues, making direct comparisons appropriate. Moreover, the broadly similar siRNA profiles of the two species indicate that a detailed comparison will not be confounded by principal differences in siRNA pathways (30, 31).
Our 24-nt siRNA data included 3.6 million reads for A. thaliana and 5.1 million for A. lyrata. We focused on reads that had perfect matches to a TE sequence, with ∼78% and ∼70% of reads in A. thaliana and A. lyrata, respectively, fulfilling this criterion. We mapped siRNAs to genomic locations and labeled a single TE as siRNA+ if it matched at least one 24-nt siRNA; TEs with no matching siRNAs were labeled siRNA−. Overall, we detected a higher proportion of siRNA+ TEs in A. lyrata, at 86%, than in A. thaliana, at 68%, perhaps reflecting greater sampling in A. lyrata. However, for both species the density of siRNAs closely mirrored the distribution of TEs (ref. 31; Fig. S1), consistent with the role of 24-nt siRNAs in silencing (2, 5, 6, 19).
siRNA Targeting and Gene Expression.
siRNA+ TEs are more likely to be methylated (14) and thus may have an effect on the expression of nearby genes (19). To investigate this possibility, we first measured gene expression in A. lyrata floral tissues using mRNA-seq data. The data provided evidence of expression for 77% of A. lyrata genes. We then standardized the mRNA-seq data for comparison with A. thaliana tiling array data based on cDNA from floral tissue (SI Materials and Methods and Fig. S2).
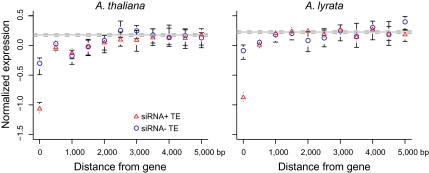
To investigate the relationship of TE proximity with gene expression, we measured the distance from a gene to its nearest neighboring TE, including both upstream and downstream TEs. (Results were qualitatively similar considering only upstream or downstream TEs separately; data not shown.) Genes were separated as to whether the nearest TE was siRNA+ or siRNA−. In both species, average gene expression increased with distance from the nearest TE (Fig. 1), but the rate of increase differed between species. In A. thaliana, maximal gene expression levels were reached when the nearest TE was ≥ 2.5 kbp away, but maximal levels in A. lyrata were reached when the nearest TE was only ∼1.0 kbp distant.
Fig. 1.
Expression levels for genes in each bin of increasing distance from the nearest TE. Whiskers indicate 95% confidence intervals. Mean and confidence intervals of expression levels of all genes without a TE within 500 bp 5′ or 3′ from the gene are indicated by solid and dashed horizontal lines, respectively. Gene expression was averaged for TE distances binned in 500-bp increments, up to a maximum of 5,000 bp. A distance of zero indicates TEs within introns or UTRs.
In both species there was a discernible difference between genes that were close to an siRNA+ compared with those close to an siRNA− TE. When the TE was located within the gene (distance = 0), then genes with siRNA+ TEs were expressed at much lower levels, on average, than genes with siRNA− TEs. However, the difference between siRNA+ or an siRNA− TEs dissipated within a distance of ∼500 bp from the gene in both species (Fig. 1). Repeating the analysis on a finer scale suggested that an siRNA+ effect was detectable up to 400 bp in A. thaliana and 200 bp in A. lyrata (Fig. S3). Overall, these analyses are consistent with an effect of TE insertion on gene expression, with stronger effects associated with siRNA targeting and, presumably, TE methylation.
TEs and Expression Divergence Between Orthologs.
A more direct approach to assess the relationship between TEs and gene expression is the comparison of expression levels between orthologous genes that differ in the presence of TEs. Orthology has been established for >20,000 A. thaliana and A. lyrata genes). Of these, 17,842 showed evidence of expression in our A. lyrata mRNA-seq dataset.
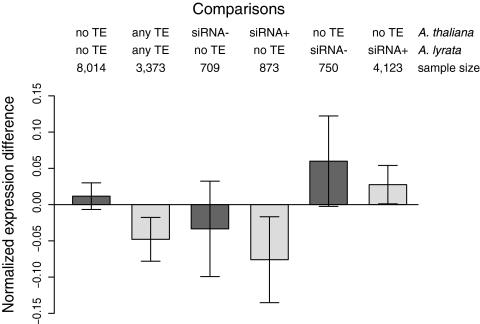
We compared expression levels of orthologs that differed in the presence/absence of any TE within 1.0 kbp either upstream or downstream of the gene. We chose 1.0 kbp based on the observed correlation of TE proximity with gene expression, which exceeds 1.0 kbp in A. thaliana but dissipates at about that distance in A. lyrata (Fig. 1). When there are no TEs within 1.0 kbp of genes in either species, then the orthologs do not differ significantly in expression (paired t test; P = 0.21). However, when both orthologs have a TE within 1.0 kbp, the A. thaliana copy is expressed at significantly lower levels than the A. lyrata copy (paired t test; P < 0.002). This observation is consistent with the fact that the correlation of TE distance with gene expression extends further in A. thaliana (Fig. 1).
We then contrasted orthologs that differed with respect to proximal TEs, and further delineated whether those TEs were siRNA+ or siRNA− (Fig. 2). When the A. thaliana gene was flanked by a TE within 1.0 kbp, but the A. lyrata ortholog was not, the expression level of the A. thaliana ortholog was fourfold lower, on average. However, the expression difference was only significant for siRNA+ TEs (P < 0.02), and not for siRNA− TEs (P = 0.32). The converse was true as well: If the A. lyrata gene had a TE within 1 kbp but the A. thaliana gene did not, then gene expression was twofold lower in A. lyrata, with the difference being only significant for genes with flanking siRNA+ TEs (P < 0.05) and only marginally significant for siRNA− TEs (P = 0.06). In some cases the siRNA− comparisons may lack statistical power due to low sample sizes (Fig. 2), but the overall pattern is clear: Proximal TEs are associated with lower expression, and reduced expression is better supported statistically when the TE is targeted by siRNAs.
Fig. 2.
Comparison of gene expression between A. thaliana and A. lyrata orthologs as a function of TE presence. Positive values of the log-transformed normalized expression difference indicate higher expression in A. thaliana.
siRNA Targeting and TE Expression.
While mapping 24-nt siRNAs to genomic locations, we noticed an unanticipated difference between species. In A. thaliana, more 24-nt siRNAs mapped to a single unique location in the genome (1,901,624), rather than to multiple locations (568,393). In A. lyrata, we detected slightly fewer uniquely mapping (1,820,527) than multiply mapping (1,821,741) siRNAs. Thus, the ratio of uniquely to multiply mapping siRNAs differed substantially between A. thaliana (3.3:1) and A. lyrata (1:1).
This apparent difference could be an artifact of sampling different numbers of siRNAs in the two species, but we do not believe this to be the case based on the following reasoning. Approximately 1.5-fold more 24-nt siRNAs were sequenced in A. lyrata, but when we considered TEs that have at least one matching siRNA, the density of siRNAs mapping to TEs was similar (median 0.81 reads per bp in A. thaliana vs. 0.80 in A. lyrata). However, the median density of uniquely mapping siRNAs in A. thaliana was approximately twofold higher (0.11 vs. 0.06). Thus, the overall coverage of individual siRNA+ TEs was similar for each species, and the major difference was in the density of uniquely mapping siRNAs.
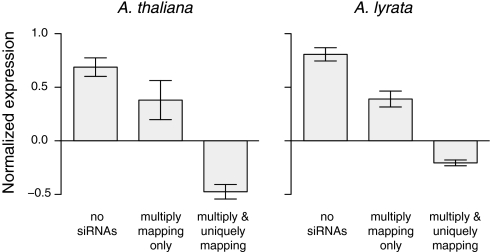
This difference may be biologically relevant, because uniquely mapping siRNAs more consistently correlate with DNA methylation than multiply mapping siRNAs (9). To test whether uniquely mapping siRNAs silence TEs more effectively, we examined expression of TE-encoded proteins. Published mRNA-seq data from A. thaliana floral tissues (9) indicated that 3% (690) of the TEs in our dataset were expressed. We then measured TE gene expression as a function of siRNA hits. TEs without any 24-nt siRNAs were expressed at the highest level, followed first by TEs that matched only multiply mapping 24-nt siRNAs (MWU, P < 0.006), and finally by TEs that were targeted by uniquely mapping siRNAs (P < 2e−11) (Fig. 3). The same pattern held for A. lyrata: 8% (5,932) of TEs were expressed in our mRNAseq data, with a clear gradient of expression from nontargeted TEs to TEs targeted by multiply mapping siRNAs, to TEs targeted by unique siRNAs (Fig. 3).
Fig. 3.
Expression levels of TEs without matching 24-nt siRNAs, with multiply mapping 24-nt siRNAs only, or with multiply and uniquely mapping siRNAs.
Could the difference in the ratio of uniquely to multiply mapping siRNAs result in genome-wide differences in silencing between species? Overall, many more A. thaliana siRNA+ TEs were targeted by uniquely mapping siRNAs (90%) than A. lyrata TEs (75%). Moreover, the direction of this difference was consistent for every major TE family except for Stowaway DNA elements (Table 1). Given differences in unique vs. multiply mapping siRNAs and their correlation with TE expression (Fig. 3), it is plausible that genome-wide siRNA-guided TE silencing is less effective in A. lyrata.
siRNA Targeting and TE Age.
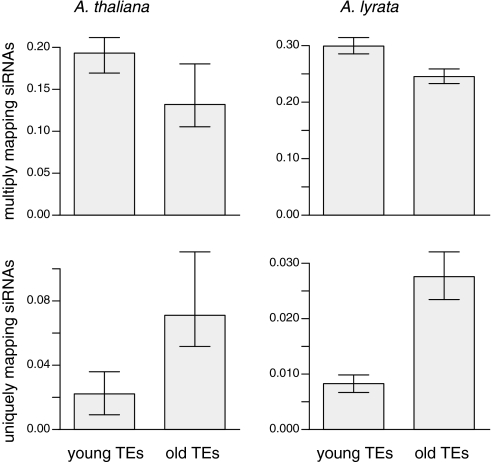
The difference between the two species in the ratio of unique to multiply mapping siRNAs could reflect an effect of TE age on silencing. If TEs proliferate rapidly, they immediately produce multiple targets for any single 24-nt siRNA. Over time, these TE targets diverge in sequence, providing potential templates for the production of uniquely mapping siRNA. We thus predicted that the ratio of uniquely to multiply mapping siRNAs correlates positively with TE age. To assess such a correlation, we inferred the ages of long terminal repeat (LTR) retrotransposons based on divergence between the two LTRs, which are exact duplicates immediately after insertion (32). We binned intact LTR retrotransposons into “old” and “young” groups, based on whether a retrotransposon was older or younger than the median age within a species, which was 1.1 MY for A. lyrata and 3.1 MY for A. thaliana.
As predicted, the age of an element and the density of uniquely mapping 24-nt siRNAs correlated positively in both species (Spearman's rho = 0.23 for A. thaliana, P < 0.001; 0.25 for A. lyrata, P < 0.0002), and there was a negative correlation between TE age and multiply mapping 24-nt siRNAs (Spearman's rho = −0.15 for A. thaliana, P < 0.03; −0.1 for A. lyrata, P < 0.00002) (Fig. 4). In summary, our data suggest that (i) multiply mapping siRNAs are, on average, less effective at silencing TE expression than uniquely mapping siRNAs, and that (ii) this effect particularly pertains to more recent TE insertions, because they are more often targeted by multiply mapping siRNAs.
Fig. 4.
Comparison of young and old TEs with multiply or uniquely mapping 24-nt siRNAs. These analyses are based on 914 copia and 897 gypsy retrotransposons in A. lyrata and 111 copia and 98 gypsy elements in A. thaliana.
Discussion
The two closely related species A. thaliana and A. lyrata differ greatly in TE copy number, with every major TE family having more copies in A. lyrata (Table 1). This global difference could be explained by two nonexclusive factors. The first is that all TEs have been more active in A. lyrata since the divergence from A. thaliana, a view supported by the fact that LTR retrotransposon insertions are younger in A. lyrata.
The second is that selection differs between the two species, such that TEs are removed less efficiently from the A. lyrata genome. Several variables could contribute to differences in selection against TE insertions (1), including demographic history and mating system (26, 27, 33). Although we cannot reject the possibility that either is the primary cause of the differences in copy number between A. lyrata and A. thaliana, we note that the lower TE copy number in A. thaliana is contrary to population genetic studies that suggest the efficacy of selection against TEs is lower in selfers (28, 29).
Interestingly, TE methylation can affect both TE activity and selection against TEs: It not only moderates TE activity, but it also has the potential to increase selection against individual TE insertions through perturbation of gene expression (19). Previous work has shown that the complement of 24-nt siRNAs, which are associated with TE silencing, is relatively similar between the two species (31) but produced from primarily nonsyntenic genomic locations (30). Here we extend these observations, revealing two interwoven themes. First, gene expression is correlated with both TE proximity and siRNA targeting. Second, TE expression (and presumably activity) is a function of both siRNA targeting and TE age. Critically, both differ between A. lyrata and A. thaliana.
TEs, 24-nt siRNAs, and Expression of Adjacent Genes.
The distributions of TEs and 24-nt siRNAs are positively correlated in both A. thaliana and A. lyrata (Fig. S1; ref. 31), consistent with 24-nt siRNAs being crucial for the initiation and maintenance of DNA methylation at TEs (2, 5, 6). Perhaps less expected is the pervasive relationship between the proximity of TEs, the presence of 24-nt siRNAs, and expression levels of adjacent genes. In both species, gene expression increases as a function of the distance to the nearest TE, and this relationship is stronger when the closest TE to a gene is siRNA+ and thus more likely to be methylated (Fig. 1). The findings from within-species analyses were corroborated by a between-species comparison of orthologous genes that differed in the presence of nearby TEs (Fig. 2). It is tempting to propose that reduced gene expression in Arabidopsis is a direct consequence of TE insertion. However, new insertions of the mping TE in rice actually enhance gene expression (34), suggesting that effects may vary across taxa, TE families, and individual TEs. Nonetheless, for all of the families in Table 1, the effects were consistent (although not always significantly so) with the general pattern—i.e., TEs and particularly siRNA+ TEs were associated with reduced gene expression (data not shown).
The potential of TEs to regulate gene function was first described by McClintock (35), who identified the Dissociation (Ds) element by its effect on genes in the anthocyanin biosynthesis pathway. Since then, there have been numerous other examples of single genes whose expression varies either with the presence of a TE (36) or with the methylation of a TE (13). Yet, despite much discussion of the potential regulatory effects, there have been few genome-wide studies of these effects of TEs in plants. In animals, genome-wide studies have produced evidence that TEs contribute to divergence in gene expression between rodent species (37) but not between primates (38).
Surprisingly, the relationship between TE proximity and gene expression level varies between A. lyrata and A. thaliana; in all of our analyses, gene expression in A. thaliana appeared more sensitive to the proximity of TEs. It is not clear whether this difference reflects, for example, greater robustness in A. lyrata gene expression, or perhaps reduced efficacy of TE silencing in A. lyrata compared with A. thaliana.
TE Copy Number and the Efficacy of Silencing.
A second major theme of our results is the interplay among copy number, the age of TEs, and the strength of silencing. Our analysis suggests that a higher proportion of TEs in A. lyrata is targeted by siRNAs (31), but confirms that the density of siRNA targeting, with regard to uniquely mapping reads, is higher in A. thaliana. As a result, a higher proportion of siRNA+ TEs lack uniquely mapping reads in A. lyrata (25%) than in A. thaliana (10%). This difference is particularly apparent among high-copy-number TE families. For example, 40% of siRNA+ Helitrons lack uniquely mapping reads in A. lyrata, whereas only 13% lack them in A. thaliana (Table 1). Altogether, these observations suggest that TEs in A. lyrata are less often targeted by unique 24-nt siRNAs.
In agreement with previous work (9), we find TEs targeted by multiply mapping 24-nt siRNAs to be more highly expressed (Fig. 1). We suggest that this is probably due to the effects of multiply mapping siRNAs being diluted across many targets. If this conjecture is accurate, then the expression level of uniquely mapping siRNAs should be higher, on average, than multiple mapping siRNA, after expression is corrected for the number of targets. This is indeed the case. The mean expression level for unique mapping reads was 4.40 and 2.74 reads per million for A. thaliana and A. lyrata, respectively. After correction for the number of mapping locations, the mean expression level for multiple-mapping reads was 0.26 and 0.03 reads per million for A. thaliana and A. lyrata, respectively. We thus see evidence of dilution of multiply mapping siRNAs with respect to the number of potential target sequences in both species, with a stronger effect in A. lyrata.
Based on these results, we speculate that there is a relationship between TE copy number, the rate of production of new copies through transposition, and the efficacy of siRNA-directed TE silencing. If new TE copies are produced at a high rate and are not quickly lost by purifying selection, then these copies will have a high degree of sequence similarity, because molecular divergence between copies is essentially a function of time (e.g., ref. 39). Therefore, siRNAs that are generated from a recently transposed copy are more likely to match multiple other copies in the genome (40). This conjecture is supported by the relationship between the age of LTRs and targeting by unique vs. multiply mapping siRNAs (Fig. 4).
In effect, then, a given 24-nt siRNA can potentially be recruited for silencing any of the TE copies that are identical (or highly similar) to its locus of origin. Assuming that the nuclear concentration of enzymes associated with pretranscriptional silencing is limited, it seems plausible that above a certain number of highly similar TEs, the efficacy of silencing mechanisms is insufficient to curb transposition on a genome-wide scale, perhaps leading to rapid increases in copy number among active TEs (41, 42). In other words, an initial burst of TE activity could lower the efficiency of silencing, causing a feedback loop that allows TE copy number to increase rapidly. Unfortunately, we do not know the triggers of TE activity, but they may include hybridization events, polyploid events, and other biotic and abiotic stresses (1, 2).
Two Genomes Going in Opposite Directions?
There are at least two attributes that could make A. lyrata less effective at purging TEs than A. thaliana. The first is that gene expression in A. lyrata seems to be less perturbed, on average, by the presence of TEs, perhaps leading to less efficacious selection against TE insertions near genes (19). The second is that TE silencing seems to be less efficient, such that A. lyrata may be in the midst of a feedback loop that favors TE proliferation. Both of these ideas—and their possible interdependence—need to be tested further with more epigenomic data from A. lyrata, additional information about patterns of TE expression, and population genetic data to infer the strength of selection against TE insertions.
In contrast, A. thaliana appears to be better at controlling TE activity. Most TEs within the genome present unique siRNA targets, so that silencing should be not only highly effective but also carry a potentially higher cost with respect to gene expression, leading to strong selection against TEs near genes (19). Although we observe differences in silencing and gene expression between species, it cannot be accentuated too strongly that the dynamics of TE prevalence within genomes is a complex function of population history (33), mating system (26), invasion dynamics (43), and other factors (reviewed in ref. 1). Nonetheless, differences in 24-nt siRNA targeting and its effects on gene expression could contribute to the observed twofold to threefold differences in TE copy numbers between the two Arabidopsis species.
Materials and Methods
Details of the data and the analyses can be found in the SI Materials and Methods. Briefly, datasets of TE insertions were assembled with RepeatMasker and applied to the A. thaliana (TAIR 8) genome and the A. lyrata final 8× assembly (http://genome.jgi-psf.org/Araly1). The siRNA data for A. lyrata have been described (31). The A. thaliana (Col-0) siRNA dataset came from small RNAs extracted from stage 1-to-14 flowers, as for A. lyrata, and sequenced on the Illumina platform. The 24-nt siRNAs were mapped to the A. thaliana and A. lyrata reference genomes by using the SHORE pipeline (44), without mismatches.
We used two sources of data for gene expression. For A. thaliana, RNA was extracted from whole inflorescences, up to stage 14, in three biological replicates. Labeled RNA was applied to a tiling array and analyzed by using published methods (45, 46). For A. lyrata, RNA was extracted from floral tissue up to stage 14 as well. A strand-specific mRNA dataset was generated by sequencing two technical replicates each of two biological replicates. mRNA-seq data were mapped to the A. lyrata genome by using SHORE (44). To quantify expression, we weighted multiple mapping mRNA reads by the reciprocal of their number of mapping locations. To compare the two expression datasets, we standardized the distribution of expression values.
Supplementary Material
Acknowledgments
We thank J. Wendel and T. Bureau for comments on the manuscript, T. T. Hu and P. Pattyn for sharing unpublished results, and S. Henz for generating summary values for gene expression from tiling array data. L.M.S. was supported by European Community FP7 Marie Curie Fellowship PIEF-GA-2008-221553 and an EMBO Long-Term Fellowship. Small RNA studies in the D.W. laboratory were supported by European Community FP6 IP SIROCCO Contract LSHG-CT-2006-037900, FP7 Collaborative Project AENEAS Contract KBBE-2009-226477, and the Max Planck Society. Work in the B.S.G. laboratory was supported by National Science Foundation Grant DEB-0723860 (to B.S.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE24571 and GSE24569).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018222108/-/DCSupplemental.
References
- 1.Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15:471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:43–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- 3.Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. doi: 10.1016/j.tcb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X. The epigenetic landscape of plants. Science. 2008;320:489–492. doi: 10.1126/science.1153996. [DOI] [PubMed] [Google Scholar]
- 5.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 7.Rangwala SH, et al. Meiotically stable natural epialleles of Sadhu, a novel Arabidopsis retroposon. PLoS Genet. 2006;2:e36. doi: 10.1371/journal.pgen.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 9.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia Y, et al. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5:e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lisch D, Carey CC, Dorweiler JE, Chandler VL. A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc Natl Acad Sci USA. 2002;99:6130–6135. doi: 10.1073/pnas.052152199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukahara S, et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 13.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, He Y, Amasino R, Chen X. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 2004;18:2873–2878. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 19.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 22.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Mable BK, Robertson AV, Dart S, Di Berardo C, Witham L. Breakdown of self-incompatibility in the perennial Arabidopsis lyrata (Brassicaceae) and its genetic consequences. Evolution. 2005;59:1437–1448. [PubMed] [Google Scholar]
- 26.Wright SI, Schoen DJ. Transposon dynamics and the breeding system. Genetica. 1999;107:139–148. [PubMed] [Google Scholar]
- 27.Morgan MT. Transposable element number in mixed mating populations. Genet Res. 2001;77:261–275. doi: 10.1017/s0016672301005067. [DOI] [PubMed] [Google Scholar]
- 28.Wright SI, Le QH, Schoen DJ, Bureau TE. Population dynamics of an Ac-like transposable element in self- and cross-pollinating Arabidopsis. Genetics. 2001;158:1279–1288. doi: 10.1093/genetics/158.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockton S, Gaut BS. The evolution of transposable elements in natural populations of self-fertilizing Arabidopsis thaliana and its outcrossing relative Arabidopsis lyrata. BMC Evol Biol. 2010;10:10. doi: 10.1186/1471-2148-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z, Coruh C, Axtell MJ. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell. 2010;22:1090–1103. doi: 10.1105/tpc.110.073882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahlgren N, et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 33.Lockton S, Ross-Ibarra J, Gaut BS. Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata. Proc Natl Acad Sci USA. 2008;105:13965–13970. doi: 10.1073/pnas.0804671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 35.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Coen ES, Carpenter R, Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986;47:285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- 37.Pereira V, Enard D, Eyre-Walker A. The effect of transposable element insertions on gene expression evolution in rodents. PLoS ONE. 2009;4:e4321. doi: 10.1371/journal.pone.0004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warnefors M, Pereira V, Eyre-Walker A. Transposable elements: insertion pattern and impact on gene expression evolution in hominids. Mol Biol Evol. 2010;27:1955–1962. doi: 10.1093/molbev/msq084. [DOI] [PubMed] [Google Scholar]
- 39.Hollister JD, Gaut BS. Population and evolutionary dynamics of Helitron transposable elements in Arabidopsis thaliana. Mol Biol Evol. 2007;24:2515–2524. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- 40.Slotkin RK, Freeling M, Lisch D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet. 2005;37:641–644. doi: 10.1038/ng1576. [DOI] [PubMed] [Google Scholar]
- 41.Piegu B, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Rouzic A, Boutin TS, Capy P. Long-term evolution of transposable elements. Proc Natl Acad Sci USA. 2007;104:19375–19380. doi: 10.1073/pnas.0705238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naouar N, et al. Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 2009;57:184–194. doi: 10.1111/j.1365-313X.2008.03662.x. [DOI] [PubMed] [Google Scholar]
- 46.Laubinger S, et al. At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 2008;9:R112. doi: 10.1186/gb-2008-9-7-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.