Summary
Objective
There is debate whether hyaluronan (HA) can be enzymatically degraded within the extracellular matrix of cartilage and other tissues or whether its catabolism occurs strictly within the lysosomal compartment of chondrocytes and other cell types. Previous studies have suggested that one of the lysosomal hyaluronidases (hyaluronidase-2) can be expressed as a functionally-active glycosyl phosphatidylinositol-linked protein at the surface of mammalian cells. If this form of hyaluronidase expression occurs in chondrocytes, this could represent a possible mechanism for extracellular HA cleavage. Thus, which hyaluronidases are expressed and where was the objective of this study.
Methods
mRNA for hyaluronidases was quantified by reverse transcription-polymerase chain reaction (RT-PCR) and enzymatic activity by HA zymograms. Recombinant forms of hyaluronidase-2 were generated and expressed in model cell lines. A peptide-specific polyclonal antiserum was prepared to localize endogenous human hyaluronidase-2 in human articular chondrocytes.
Results
Hyaluronidase-2 is the principal mRNA transcript expressed by primary human articular chondrocytes as well as various model cell lines. Recombinant hyaluronidase-2, containing N-terminal or C-terminal epitope tags, was strictly localized intracellularly and not released by treatment with a phosphatidylinositol-specific phospholipase. Endogenous hyaluronidase-2 expressed by human chondrocytes as well as HeLa cells could only be detected following detergent permeabilization of the plasma membranes.
Conclusions
These data suggest that on chondrocytes and other cell types examined, hyaluronidase-2 is not present or functional at the external plasma membrane. Thus, local turnover of HA is dependent on receptor-mediated endocytosis and delivery to low pH intracellular organelles for its complete degradation.
Keywords: Hyaluronidase, Hyaluronan, CD44, HYAL-2, Chondrocytes
Introduction
Hyaluronan (HA) is a high molecular mass non-sulfated polymer composed of repeating disaccharide units of N-acetylglucosamine and glucuronic acid1. It is a major component of the extracellular matrix and plays an important role in matrix assembly, wound healing and embryonic development2. In addition to potential free radical-mediated degradation3,4, HA catabolism depends on the enzymatic activity of hyaluronidases, a family of degradative endoglucosaminidase enzymes with an exclusive specificity for the β-1,4-glycosidic bonds between glucuronic acid and N-acetylglucosamine5.
Six human hyaluronidase-like genes have been identified and occur in clusters of three at two chromosomal locations6. HYAL-1, HYAL-2 and HYAL-3 are located on chromosome 3p21.3 while SPAM-1 (also known as PH-20), HYAL-4 and HYALP1 are located on chromosome 7q31.3. In addition, meningioma-expressed antigen 5 (MGEA5) located on chromosome 10 was shown to be a hyaluronidase as well as an N-acetylglucosaminidase7,8. All the hyaluronidases with the possible exception of HYAL-4 and HYALP1 are capable of degrading HA6.
HYAL-1 is present in many tissues but is predominantly found in the plasma and urine6,9,10, has an optimum pH of 3.8 and cleaves HA to small molecular mass fragments less than 20 kDa11. Transcripts for HYAL-3 have been detected in brain, liver, bone marrow and cartilage but little is known about HYAL-3 protein or enzymatic activity9,10,12. PH-20 was identified as a glycosyl phophatidylinositol (GPI)-linked sperm acrosomal enzyme with a neutral pH optimum that cleaves HA into fragments of less than 20 kDa13,14. PH-20 mRNA has been shown to be expressed by chondrocytes, synoviocytes and fibroblasts but its functional role has not been established15. MGEA5, found in meningioma patients, is also a neutral pH optimum hyaluronidase7.
Another prominent hyaluronidase expressed in many tissues including cartilage is HYAL-29,12,16. It has a unique substrate specificity in that it hydrolyzes high molecular mass HA into intermediate-sized fragments of about 20 kDa, has an optimum pH of 4.0, and is localized predominately in lysosomes17. However, recent reports have also indicated that HYAL-2 may be anchored by a GPI-anchor to the plasma membrane18,19, exhibit a broad pH activity spectrum and thus have the capacity to affect the extracellular degradation of HA20. In Xenopus laevis, a HYAL-2-type hyaluronidase exists both intracellularly as well as being attached to the cell surface through a GPI-anchor21. The Xenopus enzyme degrades HA not only at acidic pH but also under physiological conditions, albeit at a slower rate.
In cartilage, HA turnover occurs locally at the cellular level22. Chondrocytes internalize HA by way of a CD44-mediated mechanism, delivering the HA to lysosomes for its complete degradation23. As shown in the present study and others12,16, of the three hyaluronidases species, HYAL-2 is the predominant mRNA transcript expressed by chondrocytes. Recent evidence suggests that hyaluronidase is up-regulated in both osteoarthritis and rheumatoid arthritis and may partly account for the reduction in the concentration and size of HA observed in these disease conditions24. It has been suggested that HYAL-1 and HYAL-2 may work in concert to degrade HA9. For example, it has been suggested that there may be an initial cleavage of HA at the cell surface or within unique acidic endocytic vesicles by a GPI-linked HYAL-225. These fragments would then be transported to the lysosomes for further digestion by HYAL-126. Since chondrocytes express HYAL-1 and HYAL-2, our interest was to determine whether these cells utilize a similar two-step pathway for the degradation of HA and to determine whether this pathway is applicable to other cell types. In this paper, we investigated the expression and localization of both endogenous and recombinant HYAL-2 in chondrocytes and a number of other model cell types.
Materials and methods
CELL CULTURE
Chick embryo chondrocytes were isolated from 12-day-old tibiae by sequential enzymatic digestion of trypsin (Sigma, St. Louis, MO) followed by collagenase (Roche, Mannheim, Germany)27. Human articular chondrocytes were isolated from the normal talocrural ankle joint of human donors (obtained from the Gift of Hope Organ and Tissue Donor Network with consent and IRB Institutional approval). Full thickness articular cartilage slices were dissected under aseptic conditions and then subjected to sequential pronase/collagenase (Calbiochem, San Diego, CA and Boehringer Manheim, Indianapolis, IN, respectively) digestion to liberate chondrocytes as described previously28. Human and chick primary chondrocytes as well as HeLa, COS-7 and human embryonic kidney (HEK-293) cell lines (ATCC) were all cultured in Dulbecco’s modified Eagles medium (DMEM, Gibco-BRL, Grand Island, NY) with 10% fetal bovine serum (FBS, Summit Biotechnology, Ft. Collins, CO). The immortalized human chondrocytes cell line (C-28/I2) was maintained in a 1:1 mixture of DMEM/Ham’s F12 medium containing 10% FBS. All cells were maintained in a 5% CO2 humidified incubator at 37°C.
GENERATION OF THE HYAL-2 CONSTRUCTS
The full length coding region of human HYAL-2 was obtained by reverse transcription-polymerase chain reaction (RT-PCR) amplification of human chondrocyte total RNA, using Platinum Pfx DNA polymerase. The amplification primers were engineered to include EcoRI and NotI restriction sites for cloning into expression plasmids. Human HYAL-2 cDNA was cloned into a green fluorescence protein (GFP) containing pTracer™-EF/V5-His vector (Invitrogen, Carlsbad, CA) in the sense orientation and into pcDNA 3.1 (−) (Invitrogen) in the reverse sense orientation. To introduce an myc tag at the N-terminus, the human HYAL-2 cDNA was cloned into a pSecTag2 vector (Invitrogen), using primers engineered with BamHI and EcoRI restriction sites.
TRANSFECTION OF CELLS
To optimize transfection efficiency, cells were released with trypsin (0.25% trypsin, Gibco-BRL) and re-plated one day prior to transfection. Chick embryonic chondrocytes were transfected with HYAL-2 containing plasmid in the presence FuGene 6 (Roche), HeLa cells transfected using HeLa MONSTER® (Mirus, Madison, WI), and COS-7 cells using Lipofectamine 2000 (Invitrogen), all according to manufacturers’ directions. In some experiments, 24 h post-transfection the cells were subsequently transfected a second time with morpholino antisense oligonucleotides (5′-GTGTCACCTGCCTGGCACCAGCTCA-3′, Gene Tools, Philomath, OR) using Endo-Porter reagent (Gene Tools) according to manufacturer’s directions to knockdown gene expression. In other experiments, transfected cells were subsequently treated with a phosphatidylinositol-specific phospholipase C (PI-PLC); 1 unit/ml of PI-PLC in 10 mM Tris, pH 7.4, containing 144 mM NaCl and 0.05% bovine serum albumin (BSA). At various time points, the enzyme-containing cell supernatants were collected and the remaining cell layers extracted for Western blot analysis or RT-PCR as described below. As a positive control, chondrocytes were treated under similar conditions and probed with an anti-MT4-MMP antibody (Sigma, St. Louis, MO). MT4-MMP is a known GPI-linked protein that is expressed by chondrocyte29.
RT-PCR
Total cytoplasmic RNA was extracted from monolayer cultures of primary human chondrocytes, C-28/I2 cells, HeLa cells, COS-7 cells and HEK-293 cells using Trizol reagent, purified and then subjected to RT-PCR analysis as described previously30. The amplified products were analyzed by electrophoresis on 1.5% agarose gels followed by staining with ethidium bromide. The stained products were scanned and quantified using a Fluor-S MultiImager fluor-imaging system with Quantity One 4.1.1 software (Bio-Rad, Hercules, CA). The primer sequences were HYAL-1: sense 5′-CCAGAATGCCAGCCTGA TGCC-3′, antisense 5′-GTCATTTTGGGCACGGATGCC-3′; HYAL-2: sense 5′-GGCACAATATGAGTT TGAGTTCGC-3′, antisense 5′-TTGAGGTACTGGCAGGTCTCCG-3′; and HYAL-3: sense 5′-CATTTTCTACAAGAACCAACTCGGCC-3′, antisense 5′-CCAATGCAGTTGAGTGT TGCGG-3′. These same primer pairs were also used for quantitative real time RT-PCR. Thermal cycling and fluorescence detection was performed in the Smart Cycler system (Cepheid, Sunnyvale, CA) and PCR products were detected by SYBR® Green I Nucleic Acid Gel Stain (Molecular Probes, Inc., Eugene, OR).
POLYCLONAL RABBIT ANTI-HUMAN HYAL-2 ANTISERA
For the preparation of polyclonal rabbit anti-human HYAL-2 antisera, a specific synthetic peptide (11 amino acids unique for the human HYAL-2 sequence, GDAGYTT-STET) was prepared by NeoMPS (San Diego, CA). The purified peptide was coupled through the terminal cysteine thiol to Keyhole Limpet Hemocyanin using the heterofunctional cross-linking agent maleimidobenzoyl-N-hydroxysuc-cinimide ester. The antigen was suspended in phosphate buffered saline (PBS), emulsified by mixing with an equal volume of Complete Freund’s Adjuvant and injected into the subcutaneous dorsal sites for primary immunization. Subsequent immunizations were performed using Incomplete Freund’s Adjuvant. The animals were bleed, the crude serum immunoaffinity purified and the anti-peptide antisera titer determined by enzyme-linked immunosorbent assay. Aliquots of antisera from each rabbit were then tested by Western blotting of lysates derived from human articular chondrocytes.
WESTERN BLOT ANALYSIS
At indicated times, conditioned media were isolated and concentrated by lyophilization while the cell layers were extracted for 1 h on ice with 1% Triton X-100 containing 100 mM Tris, pH 7.4, 0.15 M NaCl and 1× protease inhibitor cocktail (Sigma). The lysates were then cleared by centrifugation at 10,000 × g for 10 min and the supernatants were recovered. The cell lysates and conditioned media were separated by a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)31 and electroblotted onto nitrocellulose membranes. The nitrocellulose membranes were blocked with 5% nonfat dry milk in PBS containing 0.05% Tween 20 for 1 h, followed by 1 h incubation with the appropriate primary mouse monoclonal antibody (anti-V5 antibody at 1:5000, anti-myc antibody at 1:5000) or 3 h with rabbit anti-human HYAL-2 at 1:120. The blots were then washed extensively before detection using the Vecta-stain Elite ABC peroxidase kit (Vector Laboratories, Burlin-game, CA) and developed using the enzyme substrate diaminobenzidine tetrahydrochloride. As controls, blots were probed with the appropriate secondary antibody alone followed by color development. The bands were imaged using the Fluor-S fluorimaging systems.
IMMUNOFLUORESCENCE MICROSCOPY
Cells were fixed in 1% paraformaldehyde in PBS, pH 7.0, for 5 min, washed and then permeabilized with 0.2% Triton in PBS for 5 min. The cells were washed again and then blocked in 1% BSA in PBS for 1 h. In some instances, the permeabilization step was omitted to selectively detect extracellular HYAL-2 expression. Following blocking, the cells were incubated with either anti-V5 or anti-myc antibody for 1 h, washed and incubated with fluorescent secondary anti-mouse antibody, Cy3-conjugated Affipure goat anti-mouse IgG (1:3000, Jackson ImmunoResearch, West Grove, PA), for 40 min. With the rabbit anti-human HYAL-2 antisera, the cells were incubated overnight with the primary antibody, washed and incubated with goat anti-rabbit biotinylated secondary antibody for 1 h in PBS containing 3% goat serum. Following extensive washing, the cells were incubated with 1 μg/ml neutravidin Texas Red (Molecular Probes, Inc., Eugene, OR) in PBS with 1% BSA for 30 min. The cells were then washed, mounted in mounting media containing the nuclear stain, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Molecular Probes), and visualized using a Nikon Eclipse E600 microscope equipped with Y-Fl Epi-fluorescence (Melville, NY), a 60× 1.4 n.a. oil-immersion objective and Rhodamine Red (red), FITC (green) and DAPI (blue) filters. Images were captured digitally in real time using a Spot-RT camera (Diagnostic Instruments, Sterling Heights, MI) and processed using MetaView imaging software (Universal Imaging, West Chester, PA).
HA ZYMOGRAPHY
Hyaluronidase activity in cell lysates and conditioned media from embryonic chondrocytes were assayed by HA zymography as previously described32. Briefly, the samples were separated on a 10% SDS polyacrylamide gel containing 0.17 mg/ml HA (Genzyme, Framingham, MA) followed by a 1 h incubation in 0.3% Triton X-100 and then overnight incubation at 37°C in 0.1 M sodium formate, pH 3.7, containing 0.15 M NaCl. After the overnight incubation, the gel was treated for 1 h with 1 mg/ml pronase in 20 mM Tris HCl, pH 8.0 and then stained with 0.5% Alcian Blue in 3% acetic acid for 30 min. The gel was next destained with 40% ethanol in 3% acetic acid until the background was sufficiently reduced to visualize the bands. The bands were imaged using the Fluor-S MultiImaging system.
Results
ENDOGENOUS EXPRESSION OF HYALURONIDASES IN CULTURED CELLS
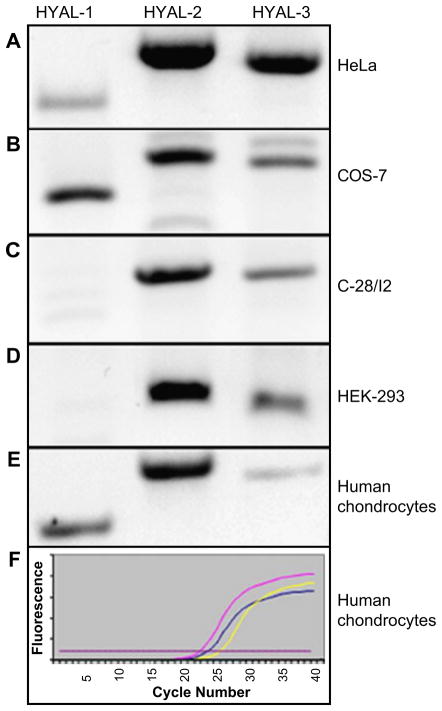
Semi-quantitative RT-PCR was used as an initial screen to determine the relative expression of HYAL-1, HYAL-2 and HYAL-3 mRNA in chondrocytes as well as a variety of cultured cells that we use as model systems. As shown in Fig. 1, HeLa cells, COS-7 cells and human articular chondrocytes each expressed mRNA for all three hyaluronidases. The immortalized human chondrocytes cell line (C-28/I2) and human embryonic kidney cells (HEK-293) both expressed HYAL-2 and HYAL-3 but no detectable HYAL-1. In all of the cultured cells tested, HYAL-2 was the predominant transcript. The real time RT-PCR results shown in Fig. 1(F) indicate that differences in the relative expression of HYAL-1, HYAL-2 and HYAL-3 by human articular chondrocytes are also observed at low cycle number. These results supported our continued focus of attention on the expression and cellular localization of HYAL-2.
Fig. 1.
Several cell types express HYAL-2 mRNA as the principal endogenous lysosomal hyaluronidase transcript. Replicate aliquots of total RNA isolated from (A) HeLa; (B) COS-7; (C) C-28/I2; (D) HEK-293 cells; or (E and F) primary human articular chondrocytes were subjected to RT-PCR using primers specific for human HYAL-1, HYAL-2 and HYAL-3. Panel F depicts a real time RT-PCR profile (SYBR-green fluorescence vs cycle number); red line trace = HYAL-2; blue line trace = HYAL-1; yellow line trace = HYAL-3.
EXPRESSION OF RECOMBINANT HUMAN HYAL-2 IN CULTURED CELLS
Embryonic chick tibial chondrocytes were used as a chondrocyte model system because of their high transfection efficiency. These cells were transfected with a full-length, human HYAL-2 construct containing a V5-epitope tag at the C-terminus. Examination of cells for GFP fluorescence demonstrated that greater than 50% of the cells were successfully transfected (data not shown). RT-PCR analysis documented the expression of recombinant human HYAL-2 mRNA [Fig. 2(A), lane 3]. No detectable expression of endogenous chick hyaluronidase was observed in untreated control cells or empty vector-transfected cells using this primer pair [Fig. 2(A), lanes 1 and 2, respectively]. Expression of the recombinant HYAL-2 was also confirmed in transfected HeLa and COS-7 [Fig. 2(B and C), lane 3, respectively]. In these instances, expression of endogenous HYAL-2 was observed in control cells [Fig. 2(B and C), lanes 1 and 2] but at substantially lower levels as compared to the transfected cells. Next, the HeLa and COS-7 cell cultures were co-transfected with the full-length HYAL-2 together with a full-length HYAL-2 cloned in the reverse antisense orientation [Fig. 2(B and C), lane 4]. In both cell types, HYAL-2 mRNA expression was reduced by approximately 50%.
Fig. 2.
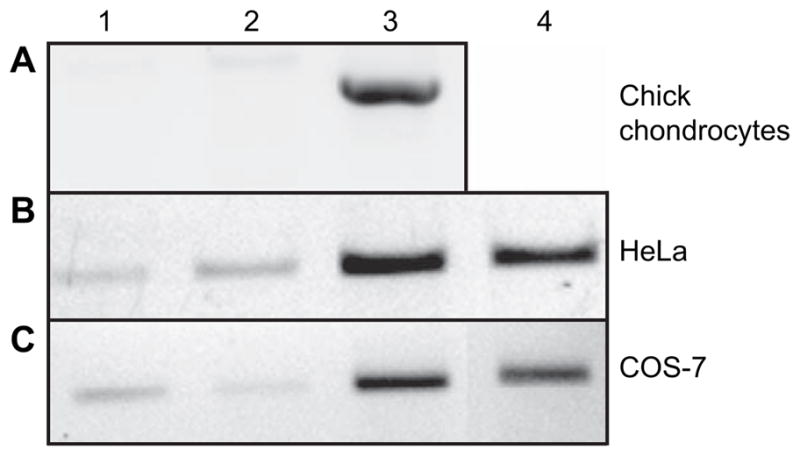
Recombinant human HYAL-2 mRNA is expressed in cultured cells following transient transfection. Replicate aliquots of total RNA were isolated from (A) embryonic chick chondrocytes; (B) HeLa; and (C) COS-7 cells and subjected to RT-PCR to detect the expression of HYAL-2 mRNA. Lane 1: control, untreated cells; lane 2: cells transfected with empty pTracer-V5His vector; lane 3: cultured cells V5-HYAL-2 plasmid alone; or lane 4: co-transfected together with a reverse-orientation (antisense) full-length HYAL-2 containing plasmid.
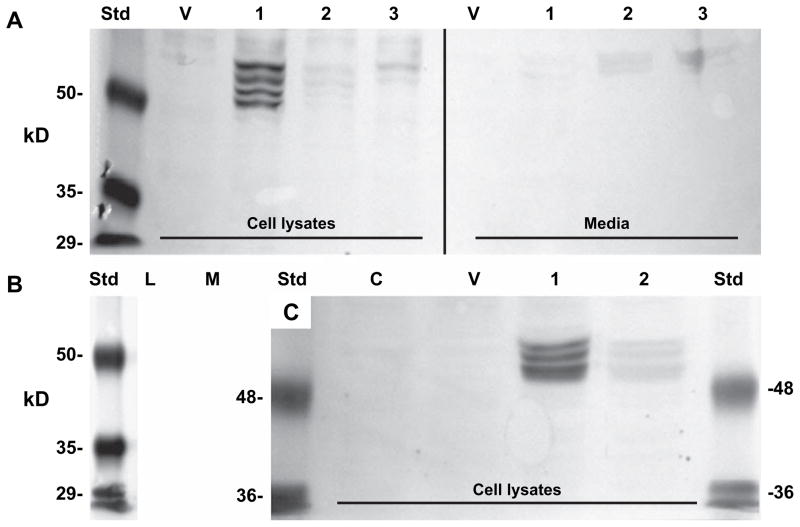
To assess the expression of recombinant HYAL-2 at the protein level, cell lysate and medium fractions were probed with an antibody directed against the V5-epitope tag. Western blot analysis of cell lysates from chick chondrocytes transfected with the human HYAL-2 construct revealed four closely spaced bands in the expected molecular mass range of HYAL-2 between 50 and 60 kDa [Fig. 3(A), lane 1]. The multiple bands could be due to differential glycosylation of the protein or the processing of signal peptides. No bands were detected in the cell lysate of control cells transfected with the empty vector alone [Fig. 3(A), lane V]. The recombinant HYAL-2 appears to be primarily retained in the cell as only faint bands were discernable in the concentrated media fractions [Fig. 3(A), Media lane 1]. This is not due to a fault in detection as we readily observe V5-HYAL-1 in medium fractions of HEK-293 cells (data not shown). No nonspecific protein bands were detected using the secondary antibody alone [Fig. 3(B)]. The identity of the recombinant HYAL-2 protein was further supported by a subsequent transfection of chick chondrocytes with full-length HYAL-2 construct together with a HYAL-2 specific morpholino antisense oligonucleotide or, co-transfection with the full-length HYAL-2 construct cloned in the reverse, antisense orientation. Both of these treatments resulted in the inhibition of the four HYAL-2 protein bands present in the chick cell lysates [Fig. 3(A), lanes 2 and 3] and medium fractions [Fig. 3(A), Media lanes 2 and 3]. Transiently transfected HeLa cells also exhibited three to four positive bands for human HYAL-2 containing the V5-epitope tag in the range of 50–60 kDa [Fig. 3(C), lane 1]. Like the embryonic chick chondrocytes, no nonspecific background bands were observed in negative control cells [Fig. 3(C), lanes C and V] and HYAL-2 protein expression was substantially inhibited by subsequent transfection of the HeLa cells with HYAL-2 morpholino antisense oligonucleotides [Fig. 3(C), lane 2].
Fig. 3.
Recombinant human HYAL-2 is expressed as protein in embryonic chick chondrocytes and HeLa cells. Cell lysates and media fractions were obtained from (A, B) embryonic chick chondrocytes and (C) HeLa cells, following transfection of these cells with a plasmid containing human HYAL-2 with a C-terminal V5-epitope tag generating a fusion protein. In panel A, the first four lanes represent cell lysates and second four lanes correspond to concentrated conditioned media. Secondary antibody alone control blots are shown in panel B using lysate (L) or medium (M) from embryonic chick chondrocytes transfected with V5-HYAL-2 plasmid. Controls also include untreated control cells (lane labeled C) and cells transfected with empty pTracer-V5His vector (lanes labeled V). Experimental conditions include cells transfected with V5-HYAL-2 plasmid alone (lanes labeled 1), cells co-transfected together with a reverse-orientation (antisense) full-length HYAL-2 containing plasmid (lanes labeled 2) and cells subsequently transfected with HYAL-2 antisense morpholino oligonucleotides (lanes labeled 3). Std, molecular mass standard proteins.
HYALURONIDASE ACTIVITY
To determine that the expressed recombinant HYAL-2 protein was functional, HA zymography was performed on embryonic chick chondrocyte lysates. As shown in Fig. 4, a small but detectable level of endogenous chick hyaluronidase activity was detected (lane 1). However, upon transient transfection of cells with the full-length human HYAL-2 construct, the hyaluronidase band intensity was substantially increased (lane 2). This increased enzymatic activity was reduced to control levels following transfection of the cells with the HYAL-2 morpholino antisense oligonucleotide (lane 3) or the antisense HYAL-2 construct (lane 4).
Fig. 4.
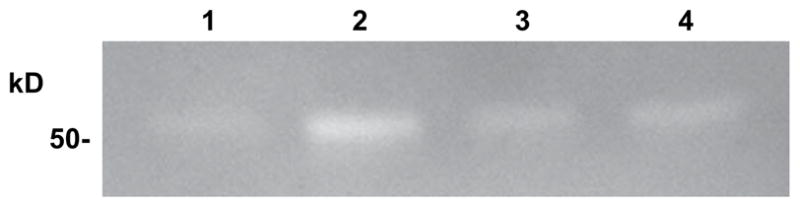
Recombinant human HYAL-2 expressed in embryonic chick chondrocytes is enzymatically active. Following transfection of embryonic chick chondrocytes with V5-HYAL-2 plasmid, cell lysates were isolated and analyzed by HA zymography. Lane 1: cells transfected with empty pTracer-V5His vector alone; lane 2: cells transfected with V5-HYAL-2 plasmid alone; lane 3: cells co-transfected with V5-HYAL-2 together with a reverse-orientation (antisense) full-length HYAL-2 containing plasmid; lane 4: cells transfected with V5-HYAL-2 followed by subsequent transfection with HYAL-2 antisense morpholino oligonucleotides. Clear bands on a darkened background depict areas of hyaluronidase activity in which Alcian blue stainable HA has been degraded and released from the gel.
The expression of recombinant human HYAL-2 protein was also confirmed by immunofluorescence staining of transfected chick chondrocytes. As shown in Fig. 5, the recombinant HYAL-2 protein appeared localized within intracellular vesicles (indicated by red fluorescence). As shown in the three-color overlay image [Fig. 5(B)], V5-tagged HYAL-2 expression was only observed in GFP-positive cells and was predominately perinuclear (nuclei depicted by blue fluorescence DAPI staining). Given that the C-terminal V5-epitope tag remains associated with the full-length recombinant HYAL-2 as a fusion protein (Fig. 3), that is enzymatically functional (Fig. 4) and, localized within vesicular-like organelles (Fig. 5), these results would suggest that HYAL-2 is not being transferred to a GPI-linkage. If transfer to a GPI-linkage had occurred, the V5-epitope tag would have been cleaved off of the protein. Nonetheless, to decipher whether GPI transfer was occurring, the human HYAL-2 sequence was next subcloned into a plasmid expression vector containing an N-terminal myc-epitope tag.
Fig. 5.
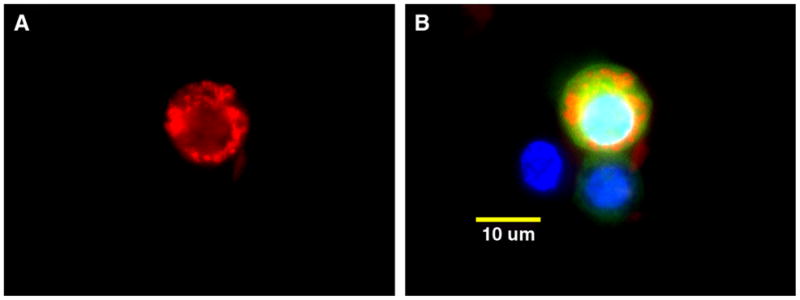
Transfected embryonic chick chondrocytes accumulate recombinant human HYAL-2 protein intracellularly. Following transient transfection of embryonic chick chondrocytes with V5-HYAL-2 expression plasmid, the cells were immunostained using a monoclonal antibody directed against the V5-epitope tag followed by a Cy3-conjugated goat anti-mouse secondary antibody (red fluorescence, panel A). Panel B depicts a three-color digital overlay of the image shown in panel A with DAPI detection of nuclei (blue fluorescence), GFP expression indicating successfully transfected cells (green fluorescence) and Cy3-positive staining for V5-epitope tag expression (red fluorescence). Magnification bar, 10 μm.
EXPRESSION OF RECOMBINANT myc-HYAL-2
As shown in Fig. 6(A and B), both HeLa and COS-7 cells transfected with the myc-tagged HYAL-2 construct exhibited intense cell-associated red fluorescence for myc-HYAL-2 but only when the cells were first detergent permeabilized prior to staining. In the absence of permeabilization, conditions that highlight extracellular cell surface staining, no myc-HYAL-2 was detected [Fig. 6(C and D), respectively]. Further, permeabilization alone is not due to a nonspecific fluorescence signal [Fig. 6(E and F), respectively]. These results indicate that the myc-HYAL-2, like the V5-tagged HYAL-2, is not present on the cell surface but rather, is localized intracellularly. Thus, it also appears unlikely that the myc-HYAL-2 is retained in these cells via a GPI-linkage.
Fig. 6.
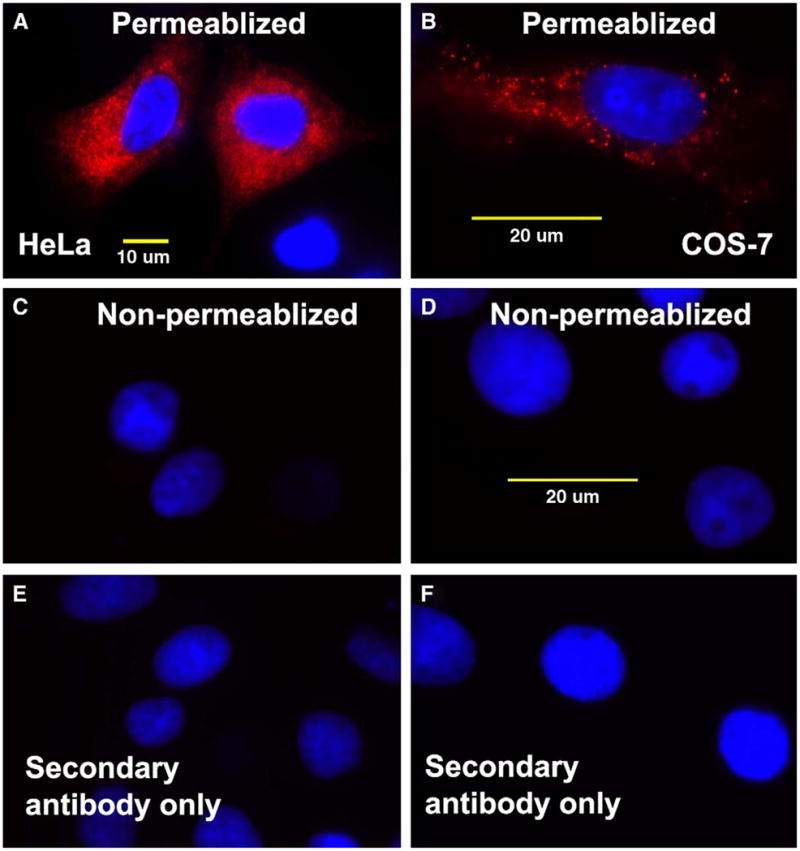
Transfected HeLa and COS-7 cells accumulate c-myc-HYAL-2 protein intracellularly as staining cannot be detected without membrane permeabilization. HeLa cells (panels A, C and E) and COS-7 cells (panels B, D and F) were transiently transfected with c-myc-HYAL-2 expression plasmid and then fixed and detergent permeabilized (panels A, B, E and F) or not permeabilized (panels C and D). The cells were then immunostained using a monoclonal antibody directed against the N-terminal c-myc-epitope tag followed by a Cy3-conjugated goat anti-mouse secondary antibody. Panel E represents control HeLa cells and panel F, the control COS-7 cells in which the permeabilized cells were incubated with the secondary Cy3-antibody alone. All images are digital two-color overlays of Cy3-expression (red fluorescence) and DAPI detection of nuclei (blue fluorescence). Magnification bars, 10 μm or 20 μm as indicated.
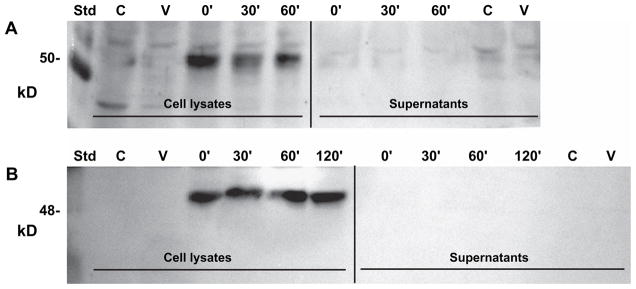
As another approach to address this question, myc-HYAL-2 transfected HeLa cells as well as transfected COS-7 cells were treated for various times with a PI-PLC. PI-PLC is used to cleave GPI-linkages and release proteins that are retained at the cell membrane via these lipid anchors18. As shown in Fig. 7, HeLa and COS-7 cells both exhibited bands for myc-HYAL-2 of ~55 kDa in total cell lysates of transfected cells [Fig. 7(A and B), lane 0′, respectively]. Only faint nonspecific bands were observed in untreated control cells [Fig. 7(A and B), lane C] or cells transfected with empty pSecTag2 vector [Fig. 7(A and B), lane V]. A similar negative reaction staining was observed in secondary antibody-only control blots (data not shown). Following 30, 60 or 120 min of treatment of the cells with PI-PLC, no major reduction in the myc-HYAL-2, associated with the cell lysates, was observed [Fig. 7(A and B), lanes 30′, 60′, 120′]. This suggests that the myc-HYAL-2, associated with either HeLa or COS-7 cells, is not susceptible to PI-PLC cleavage. Further, no myc-HYAL-2 was detected in the concentrated supernatant fractions collected following PI-PLC treatment of the cells [Fig. 7(A and B), lanes 30′, 60′, 120′]. Thus, for example, if the small diminution in HYAL-2 after 30 min of PI-PLC treatment of HeLa cells [Fig. 7(A)] were due to cleavage at the cell surface, the released enzyme would have been recovered and detected in the supernatant fraction [Fig. 7(A), lane Supernatant 30′]. Similar results were also observed with myc-HYAL-2 transfected HEK-293 cells (data not shown). As a positive control, MT4-MMP was released into the supernatant fraction when treated with PI-PLC under the same conditions (data not shown). MT4-MMP is a known GPI-linked protein that is expressed on chondrocytes29.
Fig. 7.
Recombinant c-myc-HYAL-2 protein expressed by HeLa and COS-7 cells is not depleted by PI-PLC treatment of the cells. Replicate cultures of HeLa cells (panel A) and COS-7 cells (panel B) transiently transfected with c-myc-HYAL-2 plasmid were then treated with PI-PLC for varying times and the supernatant media collected (Supernatants). The remaining cells were then washed and extracted with lysis buffer (Cell Lysates). Controls included nontransfected cells (lane C) and cells transfected with empty pSecTag2 vector (lane V); lanes 0′, 30′, 60′, 120′, c-myc-HYAL-2 transfected cells incubated PI-PLC for 0, 30, 60 or 120 min. Std, molecular mass standard proteins.
GENERATION OF PEPTIDE-SPECIFIC HUMAN HYAL-2 POLYCLONAL ANTISERA AND ITS USE IN DETECTION OF ENDOGENOUS HYAL-2 PROTEIN
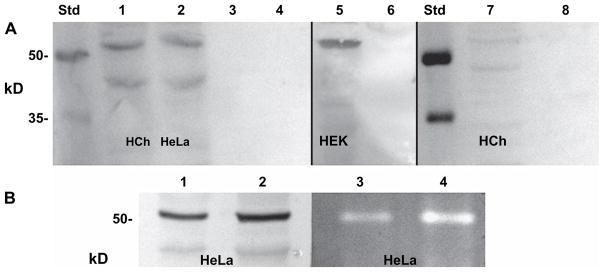
To document the expression and cellular localization of endogenous HYAL-2 protein, polyclonal antisera were generated using a peptide sequence specific to human HYAL-2. More importantly, the peptide chosen represented an amino acid sequence not present in any of the other hyaluronidases such as HYAL-1 or HYAL-3. Western blot analysis of cell lysates from human articular chondrocytes and HeLa cells, using the peptide-specific HYAL-2 antisera, revealed a band at ~55 kDa [Fig. 8(A), lanes 1 and 2, respectively]. These bands are similar to those observed with the recombinant V5-HYAL-2 (Fig. 3) or myc-HYAL-2 (Fig. 7). The concentrated conditioned media removed from these cultures also failed to express detectable levels of endogenous HYAL-2 [Fig. 8(A), lanes 3 and 4]. This observation was also supported by Western blot analysis using endogenous HYAL-2 expressed by HEK-293 cells [Fig. 8(A), lane 5]. Again, a positive band for HYAL-2 was only observed in the cell lysate fraction and not in the concentrated conditioned medium [Fig. 8(A), lane 6]. Only non-specific protein bands were observed in secondary antibody alone control blots [Fig. 8(A), lanes 7 and 8]. The identity of the bands detected by Western blotting was also confirmed by HA zymography. Fig. 8(B) depicts the analysis of 20 and 40 μg aliquots of lysates from HeLa cells on Western blots (lanes 1 and 2, respectively) as compared to the same samples analyzed by HA zymography (lanes 3 and 4).
Fig. 8.
Endogenous HYAL-2 protein, expressed by primary human chondrocytes, HeLa and HEK-293 cells, is cell-associated and not released into the culture media. Panel A: Cell lysates and concentrated media fractions from cultures of primary human chondrocytes, HeLa and HEK-293 cells were analyzed by Western blot analysis using a rabbit peptide-specific antisera directed against human HYAL-2. Lanes 1 and 3: lysates and media derived from primary human chondrocytes, respectively; lanes 2 and 4: lysates and media derived HeLa cells; and lanes 5 and 6: lysates and media derived from HEK-293 cells. In lanes 7 and 8, lysates and media derived human chondrocytes were incubated with secondary antibody reagents alone as a control. Panel B: Cell lysates from HeLa cells (20 and 40 μg aliquots) were analyzed by both Western blotting (lanes 1 and 2, respectively) and HA zymography (lanes 3 and 4, respectively). Std, molecular mass standard proteins; HCh, human chondrocytes; HEK, HEK-293 cells.
Figure 9 depicts the cellular localization of endogenous HYAL-2 in primary human articular chondrocytes [Fig. 9(A) and 9A inset] as well as HeLa cells [Fig. 9(C)]. However again, this intense immunofluorescence staining was only detected following detergent permeabilization of the cells following fixation. In the absence of membrane permeabilization, no HYAL-2 was detected in either the human chondrocytes [Fig. 9(B)] or HeLa cells (data not shown). No immunofluorescent staining was seen with the secondary antibody alone [Fig. 9(D)]. These data suggest again that the endogenous HYAL-2 is not localized on the extracellular face of the plasma membrane of these cell types.
Fig. 9.
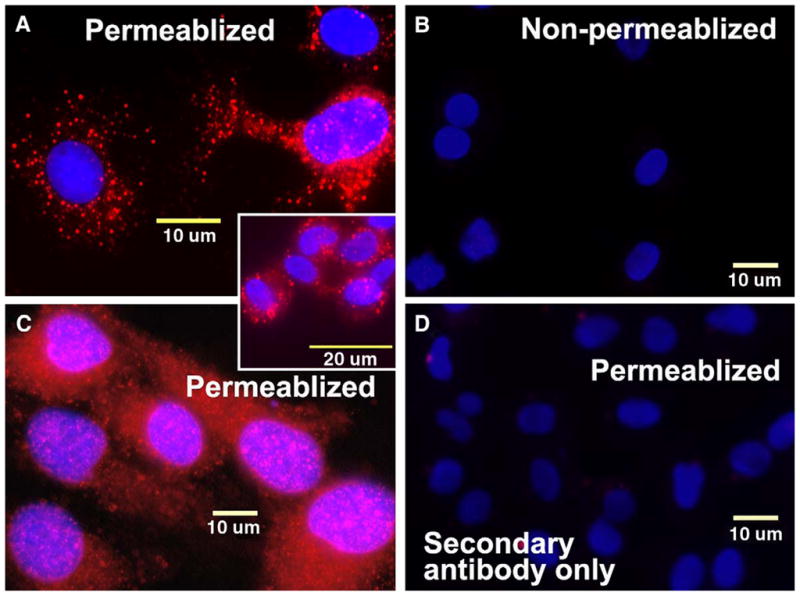
Endogenous HYAL-2 protein, expressed by primary human chondrocytes or HeLa cells, is localized intracellularly and cannot be detected without membrane permeabilization. Primary human chondrocytes (panel A and inset) and HeLa cells (panel C) were fixed and detergent permeabilized or not permeabilized (panel B). The cells were then immunostained using a rabbit peptide-specific antisera directed against human HYAL-2, followed by goat anti-rabbit biotinylated secondary antibody and neutravidin Texas Red (red fluorescence). Panel D represents control cells in which the permeabilized primary human chondrocytes were incubated with the secondary antibody and biotin detection reagents alone. All images are digital two-color overlays of neutravidin Texas Red expression (red fluorescence) and DAPI detection of nuclei (blue fluorescence). Magnification bars, 10 μm or 20 μm as indicated.
Discussion
In human, six hyaluronidase-like genes have been identified with HYAL-1 and HYAL-2 being the most predominantly expressed gene products in somatic tissues. HYAL-2 was originally thought to be a lysosomal enzyme because of its acidic pH optimum17. However, recent studies have also indicated that HYAL-2 can be found on the cell surface as a GPI-linked protein as well as a soluble form18,19,20,33. Thus, it has been proposed that HA degradation occurs in a stepwise manner whereby HA is first cleaved by cell surface bound HYAL-2 into 20 kDa fragments which are then delivered into lysosomes-like structures for further degradation by HYAL-1 and two lysosomal β-exoglycosidases, β-glucuronidase and β-N-acetylglucosaminidase26.
In this study, we investigated the expression and localization of both endogenous and recombinant HYAL-2 to determine whether chondrocytes and a number of other model cell types utilize a similar two-step pathway for the degradation of HA. Our findings indicated that both recombinant C-terminus and N-terminus tagged HYAL-2 were found localized within intracellular vesicles. Such a similar localization of epitope tags would not be expected if C-terminal cleavage had occurred during the transfer of HYAL-2 to a GPI-anchor. In addition, endogenous HYAL-2 showed a similar intracellular localization pattern (Fig. 9) which is in agreement with the recombinant expression data. Further support for an intracellular localization of the protein came from (1) the need for the cells to be permeabilized with detergent before immunoreactivity could be detected (Figs. 6 and 9) and (2) treatment with PI-PLC did not result in the reduction in cell-associated HYAL-2 protein (Fig. 7). Nonetheless, it could be possible that our analytical approaches were not sensitive enough or appropriate to detect membrane-bound HYAL-2.
These results also confirm our previous observations of HA turnover in chondrocytes. Addition of purified 3H-labeled HA to bovine or rat chondrocytes resulted in the internalization of ~5% of the labeled HA probe that was bound at the extracellular cell surface, within 24 h of incubation23. Analyses of the intracellular pool revealed only two size classes of label, one that eluted in the void volume of a Sepharose CL-2B column (i.e., >1 × 106 Da), and one that eluted in the total volume of the column (i.e., degradation products of <50 kDa). Further, the generation of small fragments was inhibited in the presence of the lysosomotropic agent chloroquine. The presence of these two pools (i.e., no intermediate sizes) is consistent with a pool of internalized HA present in endosomes and another, within lysosomes. In rat keratinocytes, the size of intracellular HA was predominately <90 kDa25. The generation of these small, extensively degraded products was also inhibited by the presence of the lysosomotropic agents chloroquine, NH4Cl or the hyaluronidase inhibitor apigenin. Therefore, the intracellular degradation of HA occurs within a low pH environment, such as that of the lysosome. Nonetheless, these data cannot rule out the possibility that some internalized HA in the degraded fraction could be present in an acidified endosome that includes a hyaluronidase activity such as HYAL-2.
Many GPI-linked proteins have been determined for the amino acid sequences of their signal peptides, by comparing the matured proteins sequences with that of the nascent proteins as predicted from their cDNA sequences 34,35. The signal for GPI-anchor is characterized by a C-terminal hydrophobic domain that is preceded by a short hydrophilic spacer linked to the GPI-anchor attachment site36 which is replaced during the transfer to the GPI moiety. Amino acid sequence analysis of HYAL-2 indicates the presence of such a signal18. However, our study seems to suggest that the presence of a putative GPI-linkage in the predicted cDNA sequence does not necessarily translate into a GPI-anchored protein. This is exemplified by LFA-3 whereby cDNA-predicted sequence of the precursor form of GPI-linked protein is the same as the transmembrane form of the protein suggesting that the fate of a GPI-anchored protein is not necessarily encoded in the primary RNA transcript but is a posttranscriptional event34 and both forms can be coexpressed in many cell types37. Depending on the cell type and/or development stage, alternate RNA processing could also result in other forms of the protein despite the presence of a putative signal in the cDNA sequence as seen with neural cell adhesion molecule38. Thus, the cellular localization of HYAL-2 is a complex issue and it could exist in various forms that might be dependent on the cell type and/or its developmental state. As such, HYAL-2 expression extracellularly might occur in some cell types as a highly transient event or in some way enhanced during certain pathological conditions.
Acknowledgments
The authors thank the donor families and the Gift of Hope Organ and Tissue Donor Network. The authors also thank Dr Mary Goldring of Harvard Institutes of Medicine for the C-28/I2 cells. Supported in part by NIH grants RO1-AR43384, RO1-AR39507, T32-AR07590 and P50-AR39239 (SCOR).
References
- 1.Knudson W, Knudson CB. An update on hyaluronan and CD44 in cartilage. Curr Opin Orthop. 2004;15:369–75. [Google Scholar]
- 2.Toole BP. Hyaluronan: from extracellular glue to peri-cellular cue. Nat Rev Cancer. 2004;4:528–39. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 3.Al-Assaf S, Navaratnam S, Parsons BJ, Phillips GO. Chain scission of hyaluronan by peroxynitrite. Arch Biochem Biophys. 2003;411:73–82. doi: 10.1016/s0003-9861(02)00724-5. [DOI] [PubMed] [Google Scholar]
- 4.Baker MS, Green SP, Lowther DA. Changes in the viscosity of hyaluronic acid after exposure to a myeloperoxidase-derived oxidant. Arthritis Rheum. 1989;32:461–7. doi: 10.1002/anr.1780320416. [DOI] [PubMed] [Google Scholar]
- 5.Meyer K. Hyaluronidases. In: Boyer PD, editor. The Enzymes. 3. New York: Academic Press; 1971. pp. 307–320. [Google Scholar]
- 6.Csoka AB, Frost GI, Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. doi: 10.1016/s0945-053x(01)00172-x. [DOI] [PubMed] [Google Scholar]
- 7.Heckel D, Comtesse N, Brass N, Blin N, Zang KD, Meese E. Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum Mol Genet. 1998;7:1859–72. doi: 10.1093/hmg/7.12.1859. [DOI] [PubMed] [Google Scholar]
- 8.Comtesse N, Maldener E, Meese E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem Biophys Res Commun. 2001;283:634–40. doi: 10.1006/bbrc.2001.4815. [DOI] [PubMed] [Google Scholar]
- 9.Csoka AB, Scherer SW, Stern R. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics. 1999;60:356–61. doi: 10.1006/geno.1999.5876. [DOI] [PubMed] [Google Scholar]
- 10.Triggs-Raine B, Salo TJ, Zhang H, Wicklow BA, Natowicz MR. Mutations in HYAL1, a member of a tan-demly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc Natl Acad Sci U S A. 1999;96:6296–300. doi: 10.1073/pnas.96.11.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost GI, Csoka AB, Wong T, Stern R. Purification, cloning, and expression of human plasma hyaluronidase. Biochem Biophys Res Commun. 1997;236:10–5. doi: 10.1006/bbrc.1997.6773. [DOI] [PubMed] [Google Scholar]
- 12.Flannery CR, Little CB, Hughes CE, Caterson B. Expression and activity of articular cartilage hyaluronidases. Biochem Biophys Res Commun. 1998;251:824–9. doi: 10.1006/bbrc.1998.9561. [DOI] [PubMed] [Google Scholar]
- 13.Cherr GN, Yudin AI, Overstreet JW. The dual functions of GPI-anchored PH-20: hyaluronidase and intracellular signaling. Matrix Biol. 2001;20:515–25. doi: 10.1016/s0945-053x(01)00171-8. [DOI] [PubMed] [Google Scholar]
- 14.Cherr GN, Meyers SA, Yudin AI, VandeVoort CA, Myles DG, Primakoff P, et al. The PH-20 protein in cynomolgus macaque spermatozoa: identification of two different forms exhibiting hyaluronidase activity. Dev Biol. 1996;175:142–53. doi: 10.1006/dbio.1996.0102. [DOI] [PubMed] [Google Scholar]
- 15.El Hajjaji H, Cole AA, Manicourt DH. Chondrocytes, synoviocytes and dermal fibroblasts all express PH-20, a hyaluronidase active at neutral pH. Arthritis Res Ther. 2005;7:R756–68. doi: 10.1186/ar1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow G, Knudson W. Characterization of promoter elements of the HYAL-2 gene. J Biol Chem. 2005;280:26904–12. doi: 10.1074/jbc.M413845200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepperdinger G, Strobl B, Kreil G. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J Biol Chem. 1998;273:22466–70. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 18.Rai SK, Duh FM, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, Miller AD. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci U S A. 2001;98:4443–8. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourguignon LY, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+–H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem. 2004;279:26991–7007. doi: 10.1074/jbc.M311838200. [DOI] [PubMed] [Google Scholar]
- 20.Vigdorovich V, Strong RK, Miller AD. Expression and characterization of a soluble, active form of the jaag-siekte sheep retrovirus receptor, Hyal2. J Virol. 2005;79:79–86. doi: 10.1128/JVI.79.1.79-86.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullegger J, Lepperdinger G. Degradation of hyaluronan by a Hyal2-type hyaluronidase affects pattern formation of vitelline vessels during embryogenesis of Xenopus laevis. Mech Dev. 2002;111:25–35. doi: 10.1016/s0925-4773(01)00593-7. [DOI] [PubMed] [Google Scholar]
- 22.Knudson W, Chow G, Knudson CB. CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 2002;21:15–23. doi: 10.1016/s0945-053x(01)00186-x. [DOI] [PubMed] [Google Scholar]
- 23.Hua Q, Knudson CB, Knudson W. Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J Cell Sci. 1993;106:365–75. doi: 10.1242/jcs.106.1.365. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida M, Sai S, Marumo K, Tanaka T, Itano N, Kimata K, et al. Expression analysis of three isoforms of hyaluronan synthase and hyaluronidase in the synovium of knees in osteoarthritis and rheumatoid arthritis by quantitative real-time reverse transcriptase polymerase chain reaction. Arthritis Res Ther. 2004;6:514–20. doi: 10.1186/ar1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tammi R, Rilla K, Pienimaki J-P, MacCallum DK, Hogg M, Luukkonen M, et al. Hyaluronan enters keratinocytes by a novel endocytic route for catabolism. J Biol Chem. 2001;276:35111–22. doi: 10.1074/jbc.M103481200. [DOI] [PubMed] [Google Scholar]
- 26.Stern R. Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology. 2003;13:105–15. doi: 10.1093/glycob/cwg112. [DOI] [PubMed] [Google Scholar]
- 27.Rousche KT, Knudson CB. Temporal expression of CD44 during embryonic chick limb development and modulation of its expression with retinoic acid. Matrix Biol. 2002;21:53–62. doi: 10.1016/s0945-053x(01)00189-5. [DOI] [PubMed] [Google Scholar]
- 28.Chow G, Knudson CB, Homandberg G, Knudson W. Increased expression of CD44 in bovine articular chondrocytes by catabolic cellular mediators. J Biol Chem. 1995;270:27734–41. doi: 10.1074/jbc.270.46.27734. [DOI] [PubMed] [Google Scholar]
- 29.Patwari P, Gao G, Lee JH, Grodzinsky AJ, Sandy JD. Analysis of ADAMTS4 and MT4-MMP indicates that both are involved in aggrecanolysis in interleukin-1-treated bovine cartilage. Osteoarthritis Cartilage. 2005;13:269–77. doi: 10.1016/j.joca.2004.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Peterson RS, Wang W, Bartnik E, Knudson CB, Knudson W. A requirement for the CD44 cytoplasmic domain for hyaluronan binding, pericellular matrix assembly and receptor mediated endocytosis in COS-7 cells. J Biol Chem. 2002;277:10531–8. doi: 10.1074/jbc.M108654200. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli UK. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Miura RO, Yamagata S, Miura Y, Harada T, Yamagata T. Analysis of glycosaminoglycan-degrading enzymes by substrate gel electrophoresis (zymography) Anal Biochem. 1995;225:333–40. doi: 10.1006/abio.1995.1163. [DOI] [PubMed] [Google Scholar]
- 33.Lepperdinger G, Mullegger J, Kreil G. Hyal2-less active, but more versatile? Matrix Biol. 2001;20:509–14. doi: 10.1016/s0945-053x(01)00170-6. [DOI] [PubMed] [Google Scholar]
- 34.Ferguson MA, Williams AF. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu Rev Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 35.Moran P, Raab H, Kohr WJ, Caras IW. Glycophospholipid membrane anchor attachment. Molecular analysis of the cleavage/attachment site. J Biol Chem. 1991;266:1250–7. [PubMed] [Google Scholar]
- 36.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–91. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 37.Dustin ML, Selvaraj P, Mattaliano RJ, Springer TA. Anchoring mechanisms for LFA-3 cell adhesion glycoprotein at membrane surface. Nature. 1987;329:846–8. doi: 10.1038/329846a0. [DOI] [PubMed] [Google Scholar]
- 38.Low MG. Biochemistry of the glycosyl-phosphatidylinositol membrane protein anchors. Biochem J. 1987;244:1–13. doi: 10.1042/bj2440001. [DOI] [PMC free article] [PubMed] [Google Scholar]