Abstract
Because centrosome amplification generates aneuploidy and since centrosome amplification is ubiquitous in human tumors, a strong case is made for centrosome amplification being a major force in tumor biogenesis. Various evidence showing that oncogenes and altered tumor suppressors lead to centrosome amplification and aneuploidy suggests that oncogenes and altered tumor suppressors are a major source of genomic instability in tumors, and that they generate those abnormal processes to initiate and sustain tumorigenesis. We discuss how altered tumor suppressors and oncogenes utilize the cell cycle regulatory machinery to signal centrosome amplification and aneuploidy.
The centrosome and cancer
It has well been established that centrosome amplification is a distinct feature of most cancer cells. With this observation came the hypothesis that this phenotype can drive genomic instability and subsequent tumorigenesis. Abnormal centrosome biology, including centrosome amplification and structural abnormalities frequently occurs in most types of solid tumors, as well some leukemias and lymphomas. Specifically, those cancer types include testicular germ cell, liposarcoma, adrenocortical, bronchial, bladder, cerebral primitive neuroectodermal, cervical, prostate, breast, squamous cell carcinomas of the head and neck, myeloma, and T-cell leukemia [1-13]. Work done in haematopoietic malignancies demonstrates that centrosome amplification in myelomas correlates with a specific gene expression signature, and can serve as a prognostic factor in patients [14].
One of the tumor types in which the relationship between centrosome amplification and cancer is better understood are breast cancers. The vast majority (80-100%) of breast tumors display centrosome amplification [15]. Breast adenocarcinoma cells have a much higher frequency of centrosome defects, including amplification of number [15,16], increased volume and supernumerary centrioles, when compared to normal breast tissue [16]. Similar phenotypes can also be found in pre-invasive in situ ductal carcinoma, and in pre-malignant breast lesions, suggesting that these aberrations occur early in breast carcinogenesis [4,15,17]. In support of this data, molecular analyses have found that the centrosome pathway is highly enriched for SNPs that are associated with breast cancer risk [18]. In addition to being involved in initiation, having extensive areas of centrosome amplification in breast tumors correlates with axillary lymph node involvement, suggesting that centrosome amplification also contributes to the most malignant characteristics of breast cancer cells [19]. Various rodent models have also given support to the idea that centrosome amplification is involved in mammary tumor initiation. For example, treatment of female Wistar-Furth rats with MNU leads to mammary tumorigenesis. MNU-induced preneoplastic lesions exhibited DNA damage, chromosomal instability, and supernumerary centrosomes [20]. Additionally, expression of Pin1 in the mammary epithelial cells of transgenic mice leads to hyperplastic lesions harboring centrosome amplification [21]. Also, our laboratory has recently shown that inducible expression of K-RasG12D results in mammary hyperplasias that harbor centrosome amplification, thus demonstrating that centrosome amplification precedes mammary tumorigenesis [22].
Therefore, there are many similar correlative studies that link centrosomal abnormalities and cancer, and there are even more studies working to discover the causal link and mechanism behind this well established correlation. Indeed, the most direct evidence showing that centrosome amplification is involved in tumorigenesis was obtained in Drosophila. In a study that specifically addressed the relationship between abnormal centrosome biology and tumorigenesis, Basto et al. assayed the long term consequences of an organism having supernumerary centrosomes. Allotransplantation of Plk4/SAK over-expressing Drosophila neuronal stem cells is sufficient to induce tumors in flies [23]. Also, transplanted cells expressing aur-a, plk, asl and dsas4 resulted in tumors with varying efficiency [24]. Aurora A, one of the first oncogenes shown to induce centrosome amplification in mammalian cells [25], proved to be the most efficient at inducing tumors [24]. These important experiments and observations are the first step in defining the link between centrosome amplification and tumors. This review will address how the G1 phase Cdks normally regulate the centrosome cycle, and how oncogenes and tumor supressors deregulate those Cdks to signal centrosome amplification.
The coordinated activities of G1 phase Cdks, centrosomal kinases and phosphatases regulate the centrosome cycle
The centrosome duplication cycle
It can be argued that faithful segregation of chromosomes into daughter cells during mitosis is essential to maintain genetic stability in most if not all organisms. The interplay between centrosomes and the mitotic microtubules results in the accurate segregation of chromosomes into daughter cells. Following cytokinesis each daughter cell receives only one centrosome; this centrosome, like DNA, must duplicate only once prior to the next mitosis. Centrosome duplication must be tightly regulated, because the generation of more than one procentriole per mother centriole results in centrosome amplification [26,27] and contributes to tumorigenesis [23,24]. The different phases of the centrosome cycle were originally assigned based on the morphology of the centriole pair throughout the cell cycle, as established by electron microscopy [28]. More recently, establishment of centriole duplication assays in Xenopus egg extracts [29] and cultured mammalian cells [30,31] remarkably improved the dissection of the centrosome cycle. Additionally, the development of centrin-2-GFP constructs has allowed following the centrosome duplication cycle relative to the different cell cycle phases in real-time [32], and allows the assessment of unregulated centrosome cycles [33].
Laser centrosomal ablation and mutants of Chlamydomonas that are defective in centriole segregation showed two pathways for centriole assembly, namely a template pathway that requires preexisting centrioles to nucleate new centriole assembly, and a de novo assembly pathway that is normally turned off when centrioles are present [34,35]. The templated pathway occurs as follows [36,37]: Throughout early G1 phase, normal cells have one mature centrosome. During late G1 and S phase, the structure of the mother and daughter centrioles differs, the mother centriole contains appendages, whereas the daughter centriole grows throughout these phases. At the beginning of S phase, centriole duplication starts with the appearance of short daughter centrioles, or procentrioles, at right angles to the two original centrioles [36,38]. Procentrioles are observed approximately 4 hours after the beginning of S phase [39]. This process culminates in the acquisition of appendages by the daughter centriole in G2 [37] and the recruitment of PCM [36,38]. By late G2, two mature centrosomes are generated. The de novo assembly pathway is first detected by the appearance of small centrin aggregates at S phase [40]. Formation of new centrosomes subsequently occurs in two steps. First, approximately 5-8 hours after centrosome ablation, clouds of pericentriolar material (PCM) containing γ-tubulin and pericentrin appear in the cell [41]. By 24 hours centrioles have formed inside of the already well-developed PCM clouds.
Recent studies identifying several centrosome-associated proteins, protein kinases and phosphatases have provided new insights into the regulation of centrosome structure and function, including their ability to control centriole duplication. Because unregulated expression of proteins controlling the synthesis of daughter centrioles can cause centriole reduplication and centrosome amplification, these proteins are potential targets of oncogenes and altered tumor suppressors, and will be thoroughly discussed in the following sections.
The G1 phase Cdks coordinate the cell and centrosome cycles
The centrosome duplication cycle must occur in coordination with the cell cycle; otherwise, unregulated centrosome duplication may culminate in centrosome amplification. Because DNA and centrosomes undergo semi-conservative duplication once every cell cycle, mammalian cells are equipped with a mechanism that coordinates these two events, so that they are duplicated only once [26]. This coordination is in part accomplished because cell cycle regulatory proteins also regulate the centrosome duplication cycle. The cell cycle is regulated as follows: The temporal overexpression of cyclins D, E, and A sequentially activates the G1 phase Cdks, Cdk4/Cdk6 and Cdk2, to trigger entry and progression through S phase [42-51]. The G1 phase Cdks trigger the initiation of DNA duplication in part through the phosphorylation of the retinoblastoma (Rb) protein and the activation of the E2F transcriptional program [49,52-73]. The Rb/E2F transcription program is essential for the correct expression and regulation of copious genes involved in DNA replication, DNA repair, mitosis and centrosome duplication [74-76].
Other studies have shown a close relationship between cell cycle regulatory molecules and the regulation of centrosome duplication. For example, ectopic expression of the cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1 blocked centrosome duplication in Xenopus dividing embryos at the blastomere stage [77]. In support of those studies, inhibition of cyclin E/Cdk2 in Xenopus egg extracts caused arrest in S phase and thus prevented centriole re-duplication; re-introduction of cyclin E/Cdk2 restored that reduplication [29]. It was then suggested, using the same system, that inhibition of Cdk2 activity prevents multiple rounds of centriole duplication, but it does not prevent the initial round of duplication [78]. However, there is other more recent evidence suggesting that Cdk2 is also involved in the initial round of centriole duplication. In Xenopus egg extracts, separase causes disengagement of centrioles during anaphase, and cyclin E/Cdk2 activity is required for the synthesis of a daughter centriole following disengagement [79].
Although various data obtained in Xenopus provided a strong correlation between Cdk2 activity and centrosome duplication, gene knockout experiments done in mammalian cells uncovered a much different scenario. Previous studies demonstrating that Cdk2-deficient mice develop rather normally [80,81], raised the question of the requirement of Cdk2 in other processes such as its ability to regulate DNA and centrosome duplication [80-82]. A surprising result was that cells derived from these mice can proliferate and undergo centrosome duplication with moderate defects [80-82], indicating that the function of Cdk2 for proliferation and initiation of the centrosome duplication can be readily and functionally replaced by other Cdks or other centrosome regulatory proteins. Likewise, ablation of the Cdk2 activating partners cyclin E1 and E2 in mouse embryonic fibroblasts was not associated with any centrosomal defects [83]. In support of studies done in mammalian cells, various combinatorial knockdowns of two mitotic cyclins (CycA, CycB, and CycB3), and reduction of the dosage of the remaining cyclins in Drosophila embryonic syncytial divisions allows centrosomes to duplicate, while cells do not enter mitosis [84].
Recent experiments have revealed both redundancy, as well as specificity, in regards to the G1 phase Cdks regulating centrosome duplication in eukaryotes. For example, chicken DT40 mutants were generated in which an analog-sensitive mutant cdk1 replaced the endogenous Cdk1. In those cells, Cdk1 could be inactivated using bulky ATP analogs [85]. In DT40 cells that also lack Cdk2, Cdk1 activity is essential for DNA replication initiation and for centrosome duplication. Also, the relative contributions of the G1-Cdks (Cdk2 and Cdk4) to regulate normal centrosome duplication were explored [86]. During these studies, experiments used to measure the centrosome cycle at various time points throughout the cell cycle in Cdk2-/- and Cdk4-/- MEFs, as well as transient down-regulation of Cdk2 and Cdk4 using RNA-mediated interference, uncovered distinct centrosome cycle defects, suggesting that Cdk2 and Cdk4 do not have redundant functions. For example, while Cdk2 deficiency allowed the separation and duplication of centrosomes, absence of Cdk4 favored the accumulation of cells with centrosomes that were slow to separate and duplicate.
Targets of the G1 phase Cdks
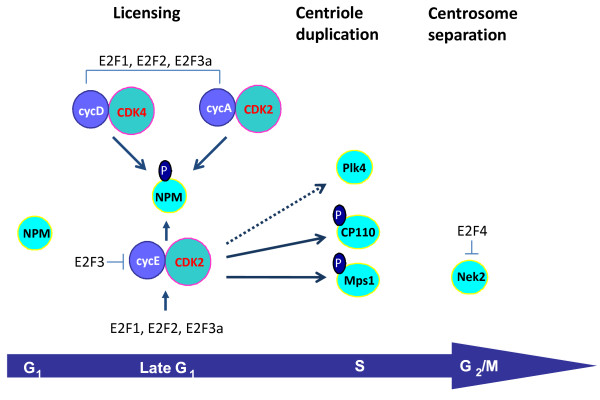
There are many structural proteins, kinases and phosphatases that regulate centrosome duplication both dependent on and independently of the G1 phase Cdk/Rb pathway [87,88]. However, those regulatory molecules acting independently of the G1 Cdks will not be covered in the scope of this review. One mode of regulation of centrosome duplication carried out by the G1 phase cyclins/Cdks is the phosphorylation of Rb family members, thus triggering de-repression and activation of E2F-responsive genes [33,74-76]. E2F-dependent centrosome regulatory targets target genes including cyclin D1 [89], cyclin E [74,90], cyclin A [76,91], Cdk2 [74], Nek2 [76], and RanBPM [76]. However, this mode of regulation remains poorly understood. A summary of known E2F targets that are known to be involved in the regulation of the centrosome cycle is presented in Figure 1.
Figure 1.
The G1 phase Cdks and the E2Fs regulate various steps in the centrosome duplication cycle. Various evidence suggests that the G1 phase Cdks directly phosphorylate NPM, CP110 and Mps1 to regulate centrosome licensing and duplication. The dotted line reflects the fact that even though Plk4 is not a direct target of Cdk2, introduction of a dominant-negative Cdk2 construct renders it ineffective in triggering centriole reduplication. The figure reflects how the E2F activators E2F1, E2F2 and E2F3 influence the centrosome duplication cycle by controlling the transcriptional levels of cyclins E, A, D, and Cdk2. The figure also reflects how E2F3 and E2F4 repress cyclin E and Nek2 to influence the centrosome cycle.
A mode of regulation that is more clearly understood is the ability of the G1 phase Cdks to phosphorylate centrosome regulatory targets modulating centrosome duplication. For example, nucleophosmin (NPM), also known as B23 [92], numatrin [93], or NO38 [94], was originally identified as a nucleolar phosphoprotein found at high levels in the granular regions of the nucleolus. NPM is a negative suppressor of licensing the centrosome cycle, and a suppressor of centrosome amplification. This was demonstrated using a genetic approach; haploinsufficiency of NPM results in unregulated centrosome duplication and centrosome amplification [95]. Conversely, microinjecting an antibody against NPM results in the suppression of centrosome duplication [96]. Licensing is modulated by G1 phase Cdks through phosphorylation and inactivation of NPM, as expression of NPM/B23 mutants whose phosphorylation sites were either deleted (NPM∆186-239) or replaced with a non-phosphorylatable residue (NPM T199A) resulted in suppression of centrosome duplication. NPM is a primary target of Cdk2/cyclin E during the initiation of centrosome duplication (Figure 1) [96]. Cdk2/cyclin A is also known to phosphorylate NPM/B23 specifically on Thr199 in vitro at a similar efficiency with Cdk2/cyclin E [97]. In addition, Cdk4/cyclinD also phosphorylates NPM on Thr 199 at mid/late G1 phase of the cell cycle [86]. NPM associates specifically with unduplicated centrosomes and dissociates from centrosomes upon Thr199 phosphorylation by Cdk2/cyclin E at the late G1 phase [96]. It is believed that the continual presence of active Cdk2/cyclin A may be responsible for preventing re-association of any cytoplasmic NPM/B23 to centrosomes during S and G2 phases. During mitosis, NPM/B23 re-associates with the centrosomes and the spindle poles [96,98]; the phosphorylation of NPM/B23 by Cdk1/cyclin B on Thr 234 and/or Thr 237 sites may play a role in re-association of NPM/B23 with centrosomes during mitosis [97]. More recently, it has been shown that NPM is also downstream of other signaling pathways, as phosphorylation of NPM by Plk2 is critical to centrosome duplication [99]. Also, NPM prevents centrosome amplification by forming a complex with BRCA2 and ROCK2 [100].
Some of the first evidence showing that centrosomal kinases are responsible for various steps in the centrosome duplication cycle was obtained from studies on the spindle pole body (SPB), the centrosome-like organelle in yeast. Like the centrosome in other organisms, the SPB duplicates only once per cell cycle commencing in G1, an event necessary for the formation of a normal bipolar spindle [101]. The Mps1 (mono polar spindle 1) family was first described in budding yeast based on its mutant phenotype, the formation of a monopolar spindle as a consequence of the failure to duplicate the SPB [102]. Localized to SPBs, Mps1 acts to control their assembly [103]. In mammalian cells, a homologous protein Mps-1 is also involved in centriole duplication. Normally, NIH3T3 cells arrested in S phase undergo only a single round of centrosome duplication [104]. In contrast, overexpression of mMps1p in these cells induced centrosome reduplication, and transfection of mMps1-KD (kinase dead) in these and other cell types (CHO, U20S) blocked centrosome duplication. The turnover of Mps1 kinases through protein degradation may be an important mechanism for their control. For example, stabilization of mMps1p within centrosomes is thought to be achieved by direct phosphorylation of mMps1p by Cdk2 (Figure 1) [104], as overexpression of cyclin A or brief proteasome inhibition increases the centrosomal levels of Mps1, whereas depletion of Cdk2 leads to the proteasome-dependent loss of Mps1 from centrosomes [105]. Also, when a Cdk2 phosphorylation site within Mps1 (T468) is mutated to alanine, Mps1 cannot accumulate at centrosomes or participate in centrosome duplication. In contrast, phosphomimetic mutations at T468 or deletion of the region surrounding T468 prevent the proteasome-dependent removal of Mps1 from centrosomes in the absence of Cdk2 activity. Moreover, cyclin A-dependent centrosome reduplication requires Mps1. Although Mps1 was reported to be involved in centrosome duplication with Cdk2 as the downstream regulator [104], another report concluded that human Mps1 does not localize to centrosomes and is not required for the ability of human U2OS cells to undergo centrosome reduplication [106]. Interestingly, it was recently shown that human Mps1 (hMps1) localizes to centrosomes after the staining of a variety of human cell types with an antibody specific to hMps1 [107]. These studies also demonstrated that overexpression of kinase dead hMps1 blocked centrosome duplication in NIH3T3, HeLa, RPE1and U2OS, and that transfection of hMps1 in U2OS cells accelerated centrosome reduplication. They also showed that siRNA silencing of hMps1 in HeLa cells induced failures in both centrosome duplication and normal progression of mitosis.
Cdk2 is responsible for regulating other proteins involved in centrosome duplication, although it is still not clear how Cdk2 controls their activity. For example, in mammalian cells, Plk4 cooperates with Cdk2, CP110 and Hs-SAS6 to induce centriole duplication [108]. Although Plk4 has not been reported to be a direct Cdk2 phosphorylation substrate, Plk4's centriole duplication activity is inefficient in the presence of a Cdk2 dominant-negative construct (Figure 1). Also, a screen for various substrates of Cdk2 revealed that CP110 is a target of Cyclin E/Cdk2, Cyclin A/Cdk2 and of Cyclin B/Cdc2 (Figure 1) [109]. CP110 is regulated by the cell cycle, as it is induced at G1/S phase, and its mRNA levels are suppressed after S phase. Down-regulation of CP110 with siRNA suppressed centriole reduplication in HU-treated U2OS cells; also, cells expressing CP110 lacking Cdk phosphorylation sites, or down-modulated CP110 also displayed centrosome separation. However, even though these studies revealed that CP110 is involved in centriole duplication and centrosome separation, the individual contribution of Cdk2 and Cdc2 sites in regulating those processes remains to be addressed.
Deregulated G1 Cdks, centrosome amplification and cancer
Oncogene-dependent centrosome amplification correlates with hyperactive Cdk2 and Cdk4
Because the centrosome cycle is regulated in part by cell cycle machinery, when the cell cycle becomes deregulated by oncogenes and altered tumor suppressors, the centrosome can also be susceptible to deregulation. This can ultimately lead to centrosome amplification, aneuploidy, and unregulated cell cycling [110,111]. Mounting evidence is showing that uncontrolled G1 phase cyclin/Cdk complexes affect two major steps in the centrosome cycle: licensing and centriole duplication.
Alterations to the centrosome duplication machinery can lead to centriole reduplication, defined as the generation of multiple procentrioles from one mother centriole; this often results in centrosome amplification. Deregulated centriole duplication and centrosome amplification was addressed using laser microsurgery to show that physical removal of all over-duplicated daughter centrioles induces reduplication of the mother in S-phase-arrested cells CHO cells [112]. In a subset of mammalian cells lacking checkpoint controls, including Chinese hamster ovary (CHO) cells [30], or p53-/- mouse embryonic fibroblasts [86], hydroxyurea (HU) treatment arrests the cells in S phase while centrosome duplication continues and results in centriole reduplication. In contrast, in CHO cells treated with mimosine, both the cell and centrosome cycles are arrested. Using that system, experiments showed that Cdk2 activity was higher in HU-treated cells than in mimosine-treated cells, suggesting a strong correlation between increased Cdk2 activity and excessive centriole duplication [30]. Also, more recent studies have shown that CHO cells arrested in G1 with mimosine can also assemble more than four centrioles, but the extent of centrosome amplification is decreased compared to cells that enter S-phase and activate the Cdk2-cyclin complex [113]. In mammalian somatic cells, centrosome reduplication is attributed to the Cdk2/cyclin A complex, since overexpression of cyclin A in cells arrested in S phase (by the expression of p16, non-phosphorylatable Rb, or in cells treated with HU), triggers centriole reduplication, while a Cdk2 dominant negative blocks reduplication [31]. Also, ectopic expression of E2F2 or E2F3 can relieve that block, suggesting that centriole re-duplication is in part mediated downstream of Cdk2 and Rb.
The first altered tumor suppressor shown to be directly associated with centrosome amplification was p53, as its genetic deletion in mouse embryonic fibroblasts promoted that abnormal process [114]. Similarly, alterations that affected p53 function resulted in centrosome amplification. For example, MDM2, an E3 ubiquitin ligase that promotes degradation of p53 [115], associates with centrosome amplification in squamous cell carcinomas of the head and neck (SCCHN) [5]. Also, the E6 viral protein from the HPV16 virus, which inactivates p53, causes centrosome amplification [116]. One of the most important functions of the p53 pathway is to trigger cell cycle arrest to allow repair of DNA damage, or cell death if the damage is unrepaired [117]. p53 exerts some of its cell cycle regulatory functions through promoting the transcription of p21Waf1/CIP1, a CKI that negatively regulates both Cdk2 and Cdk4 activities [118,119]. p53 prevents centrosome amplification through direct binding to the centrosome, and also in part through its ability to regulate p21Waf1/CIP1 [120]. Several groups have presented data supporting a role of p21Waf1/CIP1 in centrosome biology. For example, introduction of p21Waf1/CIP1 into p53-/- cells harboring centrosome amplification restored normal centrosome duplication and abrogated centrosome amplification [121]. Moreover, knock-down of p21Waf1/CIP1in murine myeloblasts stimulates excessive centriole numbers in the presence of only one mature centriole [122] and p21Waf1/CIP1 null human hematopoietic cells display elevated frequencies of centrosome amplification [123].
Consequent to the discovery that centrosome amplification in p53-null cells correlated with deregulated Cdk2 activity, many other studies began showing similar correlations. For example, when E2F3a/b, transcription factors critical to S phase entry, are ablated, elevated cyclin E-dependent Cdk2 activity correlates with constitutive centriole separation, duplication, and centrosome amplification (Figure 1) [33]. It is to note that this function is specific to E2F3-null cells, as MEFs lacking E2F1, E2F2, E2F4 or E2F5 do not display centrosome amplification. Also, the expression of the centrosome-targeting region of CG-NAP (a centrosome and Golgi-localized protein), causes centrosome amplification by anchoring excess amount of cyclin E-cdk2 to centrosomes [124]. In another correlative study disruption of Skp2, a substrate recognition component of an Skp1-Cullin-F-box protein (SCF) ubiquitin ligase, results in increased cyclin E, p27, and centrosome amplification [125]. Another example is ECRG2, a novel tumor suppressor gene which localizes to centrosomes; its depletion destabilizes p53, leading to down-regulated p21, increased cyclin E/Cdk2 activity, and centrosome amplification [126]. On the other hand, there are proteins that prevent excessive centriole duplication triggered by de-regulated G1 phase cyclins. For example, the Orc1 protein, a subunit of the origin recognition complex (ORC) that is a key component of the DNA replication licensing machinery, controls centriole and centrosome copy number in human cells [127]. Cyclin A promotes Orc1 localization to centrosomes, where Orc1 prevents Cyclin E-dependent reduplication of both centrioles and centrosomes.
Following the discovery that tumor suppressors maintained normal centrosome numbers, various laboratories showed that certain protooncogenes displayed the same activity. Some of the first observations that protooncogenes, including tyrosine kinase receptors, controlled the centrosome cycle were made in CHO cells cultured in the presence of hydroxyurea (HU) or aphidicolin. Addition of dialyzed serum to these cells stopped centriole reduplication, while addition of EGF re-initiated the process [128]. Additionally, when PTEN-/- neural precursor cells were infected with retrovirus encoding constitutively active EGFRvIII, centrosome amplification, genomic instability and glial tumors developed [129]. Furthermore, it has been shown that other EGFR family members may play a role in this story. Her2/neu (ErbB2) was first described as an oncogene when isolated from neuroglioblastomas that developed in rats treated with ethylnitrosourea (ENU) [130]. Her2 mutations are relatively rare in human cancers; however wild type ErbB2 is amplified at the genomic level or overexpressed at the protein level [131] in approximately 30% of invasive ductal breast cancers [132]. It has been shown that overexpression of this protein correlates with tumor size, spread to lymph nodes, high grade, increased percentage of S phase cells, and aneuploidy [132]. A study of mice expressing activated Her2/neu in the mammary epithelium demonstrated its ability to induce chromosomal aberrations as well as centrosome amplification in cell lines derived from primary tumors [133]. Also, analysis of fine-needle aspirations of the breast found a significant correlation between the percentage of cells with centrosome amplification, over-expression of HER2/neu and negative ER status [15]. The molecules downstream of Her2 can also become deregulated upon over-expression. Her2 induces cyclin D1 through the Ras/Rac/Rho pathway in which the ERK, JNK and p38MAPK cascades are distal mediators.
Another oncogene that has been associated with centrosome amplification is Ras. A Pubmed search for "Ras and Cancer" returns almost twenty thousand hits for articles and reviews, most discussing the oncogenic potential of Ras and the many cellular phenotypes that it affects. Probably one of the most thoroughly studied of the many Ras-mediated pathways is the MAP kinase cascade, a critical signaling cascade regulating cell proliferation by exerting control over the cell cycle. It has been shown that constitutive activation of MAPK induces defects in the normal mitotic processes of the cell [134]. For example, transduction of v-ras or v-mos into NIH 3T3 cells induced centrosome amplification and inhibition of this phenotype was possible with the introduction of MAPK inhibitors [134]. A study focusing on genomic instability in thyroid PCCL3 cells harboring wt p53, examined the effects of H-RASV12 and activated MEK1 and found that both induced centrosome amplification and chromosome misalignment [135]. Likewise, expression of the H-RasG12V or the H-RasG12V & c-Myc oncogenes in non-transformed MCF10A human mammary epithelial cells results in elevated frequencies of centrosome amplification [22]. Activation of this pathway is relevant in vivo, as ectopic expression of the K-RasG12D oncogene in mouse mammary epithelial cells resulted in centrosome amplification that greatly preceded tumorigenesis [22].
The extracellular regulated kinase (ERK) cascade, a major component of the MAPK pathway, is a critical signaling cascade, regulating cell proliferation by exerting control over the cell cycle. MEK1 and MEK2, two kinases upstream of ERK, have been shown to regulate cell cycle progression in two distinct ways [136]. Loss of MEK2 results in a mitotic delay, perhaps due to a reduction in ERK phosphorylation. When MEK2 is knocked down using siRNA in HCT116 colon cancer cells, cyclin D1 levels increase, leading to hyperactive Cdk4/6 and hyperphosphorylation of nucleophosmin (NPM); this hyperphosphorylation was independent of Cdk2. Hyperphosphorylation of NPM at T199 was accompanied by centrosome amplification and the appearance of multipolar spindles [136], making a case for Cdk4 mediation of NPM phosphorylation. In another study associating Ras/MAPK to centrosome amplification, the Hepatitis B virus (HBv) was shown to activate various signaling pathways, one of which is the Ras-Raf-MAPK [137]. The hepatitis B virus X oncoprotein HBx, is a small oncoprotein that is required for viral replication and has been associated with HBV-mediated hepatocellular carcinoma. Yun et al. discovered that the Ras-MAPK pathway is the downstream effector of HBx protein involved in abnormal amplification of centrosomes [137]. Suppression of the ERK pathway with inhibitors, and the introduction of dominant negative mutants of Ras and Mek reduce the frequency of supernumerary centrosomes in HBx expressing human Chang liver cells, thus further clarifying the role of Ras and the MAPK pathway in the HBx mediated induction of centrosome amplification [137].
Transcription of the cyclin D1 gene and subsequent interaction with its kinetically active partner, Cdk4, depends on receptor mediated Ras signaling. Various upstream and downstream effectors of the MAPK pathway up-regulate the transcription of cyclin D1 so that when it is bound to Cdk4 it is able to sequester p27Kip1 and thus activate cyclin E-Cdk2 complex [138]. Upon this activation, both cyclin-Cdk complexes are free to phosphorylate RB family proteins and cells may progress from G1 to S phase of the cell cycle [138]. In normal cells mitogenic growth factors are responsible for inducing cyclin D1; however, over-expression of cyclin D1, independent of growth factor signaling, is a common feature of many tumors [138]. For example, a great majority of small cell lung cancers, breast cancers, glioblastomas and mantle cell lymphomas have over-expression of cyclin D1 or its catalytic partner, Cdk4. In fact aberrant over-expression of cyclin D1 occurs in 70-100% of breast tumor cell lines and most breast cancers and seems to be required for neu and Ras-induced mammary epithelial transformation [89]. Along the same line, cyclin D and Cdk4 are required for neu and ras induced mammary tumorigenesis [139,140], demonstrating that the cyclin D1/Cdk4 complex is needed for mammary transformation. Unregulated expression of cyclin D1 is associated with chromosomal abnormalities and it has been documented that transient expression of cyclin D1 in hepatocytes and human mammary epithelial cells induces centrosome amplification [141]. A striking feature of this study demonstrated that centrosome abnormalities persist in a small percentage of the cells for four months after cyclin D1 is no longer expressed [141]. Interestingly, hepatocytes from Cdk2-/- mice are refractive to cyclin D1-dependent centrosome amplification, suggesting that in some contexts, either cyclin D1 uses Cdk2 to trigger centrosome amplification, or that Cdk2 is a downstream target of cyclin D/Cdk4 [142].
In support of the studies linking cyclin D1/Cdk4 with centrosome amplification, one of the primary events associated with initiation of mammary tumorigenesis is the loss of the Cdk4/Cdk6-specific inhibitor p16INK4A through hypermethylation of its promoter, which de-regulates the centrosome cycle and lead to a moderate increase in frequencies of centrosome amplification [143-145]. Concomitantly, the γ-tubulin gene is amplified [146]. Likewise, silencing the histone H3 lysine 9 methyltransferase G9a leads to centrosome amplification, reportedly by down-modulation of gene expression, including that of p16INK4A [147]. Thus, it has been postulated that loss of p16 expression coupled with increased γ-tubulin contributes to centrosome amplification and breast cancer progression.
Direct evidence demonstrating involvement of the G1 phase Cdks in centrosome amplification
Although the evidence associating hyperactive G1 phase cyclin/Cdks and centrosome amplification is convincing, it is nevertheless correlative. This is due to the fact that some of the protooncogenes, tumor suppressors, and transcription factors that control G1 phase Cdk activities, such as Her2, Ras, E2f3 and p53, also regulate a plethora of other gene products [74,76,148,149]. Table 1 lists a subset of oncogenes and altered tumor suppressors, and the G1 phase Cdk they may hyperactiate to signal centrosome amplification. How do G1 phase-CDKs signal oncogene-dependent centrosome amplification? Research showing that inhibition of specific Cdks blocks centriole reduplication was the first direct evidence of a relationship between Cdks and centrosome amplification. In HU-arrested cells, cells treated with butyrolactone I or roscovitine -inhibitors of Cdk2, Cdc2 and Cdk5 activity- [150,151], and cells treated with the Cdk2/Cdk4 inhibitor p21Waf1/Cip1 centriole reduplication was blocked [30]. Following these initial experiments, combinatorial cyclin E/A/p53 gene knockout analyses demonstrated that the G1 phase cyclins and Cdks play pivotal roles in signaling centrosome amplification. For example, in p53-/- cells arrested in early S phase, cyclin E, but not cyclin A, is important in centriole reduplication and centrosome amplification, but in the absence of cyclin E, cyclin A can drive the abnormal phenotype [152]. In p53-/- cells, Cdk2 mediated HU-induced centriole reduplication [153]. In another study, centriole reduplication triggered by the peptide vinyl sulfone proteasome inhibitor Z-L(3)VS is dependent on cyclin E/Cdk2, as well as Polo-like kinase 4 [154]. Furthermore, inhibitors of Cdk2, dominant negative mutants of Cdk2 and DP1, siRNA-mediated silencing of Cdk2, or genetic deletion of Cdk2 abrogate centrosome amplification triggered by ectopic expression of E7 [82]. These studies provided direct support to the role played by E2Fs and Cdk2 in centrosome amplification associated with the inactivation of Rb by its conditional loss [155], the acute loss of pRb by adenovirus carrying shRNA against Rb [156], or through the expression of the E7 viral protein from the HPV16 virus [116].
Table 1.
Oncogenes and inactive tumor suppressors and the G1 phase Cdk they may deregulate to signal centrosome amplification.
| Genetic alteration | Deregulated Cdk | Reference |
|---|---|---|
| Oncogenes | ||
| Cyclin D1 | Cdk2, Cdk4 | [141,142] |
| ErbB2 | Cdk4 | [139] |
| Ras | Cdk4 | [22,140] |
| Tumor Suppressors | ||
| E2F3a/b | Cdk2 | [33] |
| MEK2 | Cdk4, Cdk6 | [136] |
| p16INK4A | Cdk4, Cdk6 | [143,145] |
| p21Waf1/CIP1 | Cdk2, Cdk4 | [118,119,121,122] |
| p53 | Cdk2, Cdk4 | [86,120,121] |
| Skp2 | Cdk2 | [125] |
| Rb | Cdk2 | [82] |
Even though most evidence demonstrated that Cdk2 was the central mediator of oncogene-induced centrosome amplification, our group demonstrated that Cdk4 is also an important mediator. For example, genetic ablation of Cdk2 and Cdk4 abrogated centrosome amplification in p53-null cells [86] by restricting NPM-dependent excessive licensing of the centrosome cycle, as well as by restricting centriole reduplication in p53-null mouse embryonic fibroblasts treated with HU. Also, we showed that siRNA-mediated silencing of cyclin D1 or Cdk4 suppressed H-Ras-G12V or H-RasG12V/c-Myc-dependent centrosome amplification in MCF10A human mammary epithelial cells, while inhibition of cyclin E or cyclin B did not prevent centrosome amplification [22].
An important molecule downstream of Cdk2 that restricts centrosome separation and duplication is NPM phosphorylated at residue T199 [96,97,157]. Reasoning that this mode of deregulation was an important intermediate to centrosome amplification, our group showed that when E2F3a/b is ablated, cyclin E/Cdk2 activity is elevated, leading to the hyperphosphorylation of NPMT199 [33]. Hyperphosphorylation of NPMT199 by Cdk2 strongly correlated with constitutive centrosome duplication cycle and centrosome amplification. The role of NPM as a negative regulator of centrosome duplication was confirmed genetically through a gene knockout approach, as cells heterozygous for NPM displayed centrosome amplification [95]. Silencing of NPM in p53-/-p19Arf-/-Mdm2-/- MEFs also resulted in centrosome amplification [158]. In the same system, ectopic expression of NPMT198A could not rescue the centrosome amplification phenotype in p53-/-p19Arf-/-Mdm2-/- MEFs. In contrast, our group used a similar mutant of NPM, NPMT199A (which cannot be phosphorylated by Cdk2 or Cdk4) to demonstrate that this mutant prevented centrosome amplification in p53-null cells to the same extent as ablated Cdk2 or Cdk4 [86]. These experiments demonstrated that the G1 phase Cdks signal centrosome amplification in p53-null cells through NPM. In terms of other mechanisms linking the G1 phase Cdks and centrosome amplification, the Fry group demonstrated that nuclear export is required for centriolar satellite formation and centrosome overduplication in p53-null cells, with export inhibitors causing a Cdk2-dependent accumulation of nuclear centrin granules [153]. This group proposed an interesting model of regulation of centriole reduplication: Centrosome precursors arise in the nucleus, providing a novel mechanistic explanation for how nuclear Cdk2 can promote centrosome overduplication in the cytoplasm.
Other than the hyperphosphorylation and inactivation of NPM and the nuclear accumulation of centrin intermediates, processes that are dependent on Cdk2, the centrosomal targets controlled by oncogenes and altered tumor suppressors directly responsible for centrosome amplification are largely unknown. The sole exception is Nek2; it has been observed that silencing Nek2 abrogated centrosome amplification in human mammary epithelial cells expressing H-RasG12D and H-RasG12D/c-Myc [22]. Speculatively, we can propose the following model: Oncogene-activated G1 phase Cdks signal centrosome amplification through the stabilization of centrosome duplication kinases such as Plk4 or Mps1, or through E2F-dependent transcriptional deregulation of those centriole duplication kinases (Figure 1).
Conclusions and future directions
Because centrosome amplification is present in the vast majority of human tumors, and since supernumerary centrosomes may generate aneuploidy and genomic instability suggests that centrosome dysfunction is a potentially important contributor to cancer biogenesis. However, we are far from demonstrating a causal relationship between centrosome amplification and mammalian tumorigenesis. The observations that various pre-malignant lesions harbor centrosome amplification first mapped centrosome amplification to tumor initiation. Recent evidence demonstrating that low level aneuploidy caused by interference with spindle assembly components causes various tumors in mouse models [159,160], together with observations that merotelic attachments cause that same kind of aneuploidy [161,162] helped to bridge the gap between the correlation of centrosome amplification, aneuploidy and tumor initiation. Furthermore, two recent manuscripts showed that ectopic expression of centrosome regulatory proteins leads to benign tumors in transplanted Drosophila brain stem cells, suggesting for the first time a direct relationship between centrosome amplification and tumorigenesis [23,24]. However, unlike mammalian cancers, which are grossly aneuploid, the benign tumors in Drosophila harboring centrosome amplification displayed neither aneuploidy nor detectable gross chromosomal aberrations [24]. The classic Weinberg experiments may help shed some light on the kind of genomic changes that may be needed to transform a human epithelial cell. For example, they showed that transformation of a primary human mammary epithelial cell required ectopic expression of telomerase to protect from senescence induced by telomere shortening [163]. Ectopic expression of Ras and c-Myc as well as inactivation of p53 and Rb (via the SV40 large T antigen) was also required for transformation, suggesting that some cooperation is necessary to transform primary cells. It is to note that most of the genes that were required to transform those mammary epithelial cells affect centrosome amplification, or allow the generation of chromosome breaks and recombination [22,134,135,155,164-168]. This suggests that the centrosome amplification and genomic instability triggered by those oncogenes, combined with their ability to affect proliferation provide those cells selective advantages to initiate mammary tumors. Future experiments are needed to understand how centrosome amplification transforms cells, and whether it eventually causes ectopic proliferation and decreases apoptosis, or whether it contributes to tumorigenesis by altering other processes, such as the orientation of cells within a tissue, a concept postulated by the Gonzalez group in their Drosophila model [24]. Another pressing issue is to establish, using proteomics and transcriptomics, the centrosomal targets that are deregulated by various oncogenic and altered tumor suppressive pathways. This will allow for the ectopic expression or inactivation of various centrosome regulatory proteins in primary cell lines to more directly assess the role of centrosome amplification in transformation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MKH participated in the design, research, writing and editing of this review. AA participated in the research and writing of this review. HS conceived the review and participated in the design, research, writing, and editing of this review. All authors read and approved the final manuscript.
Contributor Information
Mary K Harrison, Email: mkharr2@emory.edu.
Arsene M Adon, Email: arsenemadon@gmail.com.
Harold I Saavedra, Email: hsaaved@emory.edu.
References
- Lothschutz D. et al. Polyploidization and centrosome hyperamplification in inflammatory bronchi. Inflamm Res. 2002;51(8):416–22. doi: 10.1007/PL00000323. [DOI] [PubMed] [Google Scholar]
- Zyss D, Gergely F. Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 2009. pp. 334–46. [DOI] [PubMed]
- Pihan GA. et al. Centrosome defects and genetic instability in malignant tumors. Cancer Research. 1998;58(17):3974–85. [PubMed] [Google Scholar]
- Pihan GA. et al. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63(6):1398–404. [PubMed] [Google Scholar]
- Carroll PE. et al. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18(11):1935–44. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- Duensing S, Munger K. Centrosomes, genomic instability, and cervical carcinogenesis. Crit Rev Eukaryot Gene Expr. 2003;13(1):9–23. doi: 10.1615/CritRevEukaryotGeneExpr.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- Chng WJ. et al. Clinical implication of centrosome amplification in plasma cell neoplasm. Blood. 2006;107(9):3669–75. doi: 10.1182/blood-2005-09-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T. et al. Centrosome amplification in adult T-cell leukemia and human T-cell leukemia virus type 1 Tax-induced human T cells. Cancer Sci. 2006;97(9):836–41. doi: 10.1111/j.1349-7006.2006.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y. et al. Centrosome hyperamplification predicts progression and tumor recurrence in bladder cancer. Clin Cancer Res. 2004;10(19):6449–55. doi: 10.1158/1078-0432.CCR-04-0773. [DOI] [PubMed] [Google Scholar]
- Weber RG. et al. Centrosome amplification as a possible mechanism for numerical chromosome aberrations in cerebral primitive neuroectodermal tumors with TP53 mutations. Cytogenet Cell Genet. 1998;83(3-4):266–9. doi: 10.1159/000015168. [DOI] [PubMed] [Google Scholar]
- Roshani L. et al. Aberrations of centrosomes in adrenocortical tumors. Int J Oncol. 2002;20(6):1161–5. [PubMed] [Google Scholar]
- Perucca-Lostanlen D. et al. Distinct MDM2 and P14ARF expression and centrosome amplification in well-differentiated liposarcomas. Genes Chromosomes Cancer. 2004;39(2):99–109. doi: 10.1002/gcc.10303. [DOI] [PubMed] [Google Scholar]
- Mayer F. et al. Aneuploidy of human testicular germ cell tumors is associated with amplification of centrosomes. Oncogene. 2003;22(25):3859–66. doi: 10.1038/sj.onc.1206469. [DOI] [PubMed] [Google Scholar]
- Chng WJ. et al. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111(3):1603–9. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- Guo HQ. et al. Analysis of the cellular centrosome in fine-needle aspirations of the breast. Breast Cancer Res. 2007;9(4):R48. doi: 10.1186/bcr1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL. et al. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95(6):2950–5. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL. et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99(4):1978–83. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JE. et al. Centrosome-related genes, genetic variation, and risk of breast cancer. Breast Cancer Res Treat. 2011;125(1):221–8. doi: 10.1007/s10549-010-0950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss A. et al. Centrosomal aberrations in primary invasive breast cancer are associated with nodal status and hormone receptor expression. Int J Cancer. 2003;107(3):346–52. doi: 10.1002/ijc.11408. [DOI] [PubMed] [Google Scholar]
- Goepfert TM. et al. Loss of chromosomal integrity drives rat mammary tumorigenesis. Int J Cancer. 2007;120(5):985–94. doi: 10.1002/ijc.22420. [DOI] [PubMed] [Google Scholar]
- Suizu F. et al. Pin1 regulates centrosome duplication, and its overexpression induces centrosome amplification, chromosome instability, and oncogenesis. Mol Cell Biol. 2006;26(4):1463–79. doi: 10.1128/MCB.26.4.1463-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X. et al. The Ras oncogene signals centrosome amplification in mammary epithelial cells through cyclin D1/Cdk4 and Nek2. Oncogene. 2010;9;29(36):5103–12. doi: 10.1038/onc.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R. et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133(6):1032–42. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18(16):1209–14. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Zhou H. et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230(1):6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J. et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Chretien D. et al. Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol. 1997;120(2):117–33. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH. et al. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. [see comments.] Science. 1999;283(5403):851–4. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Current Biology. 1999;9(8):429–32. doi: 10.1016/S0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- Meraldi P. et al. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2- cyclin A. Nat Cell Biol. 1999;1(2):88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- White RA, Pan Z, Salisbury JL. GFP-centrin as a marker for centriole dynamics in living cells. Microscopy Research & Technique. 2000;49(5):451–7. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Saavedra HI. et al. Inactivation of E2F3 results in centrosome amplification. Cancer Cell. 2003;3(4):333–46. doi: 10.1016/S1535-6108(03)00083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11(5):308–17. doi: 10.1016/S0960-9822(01)00094-X. [DOI] [PubMed] [Google Scholar]
- Khodjakov A. et al. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10(2):59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91(3 Pt 1):814–21. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93(3):938–49. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM. et al. Centriole duplication and maturation in animal cells. Curr Top Dev Biol. 2000;49:235–49. doi: 10.1016/s0070-2153(99)49011-8. full_text. [DOI] [PubMed] [Google Scholar]
- Alvey PL. An investigation of the centriole cycle using 3T3 and CHO cells. J Cell Sci. 1985;78:147–62. doi: 10.1242/jcs.78.1.147. [DOI] [PubMed] [Google Scholar]
- La Terra S. et al. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168(5):713–22. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A. et al. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158(7):1171–81. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M. et al. Cyclin A is required at two points in the human cell cycle. Embo J. 1992;11(3):961–71. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J, Hunter T. Cyclins A and B1 in the human cell cycle. Ciba Found Symp. 1992;170:187–96. [PubMed] [Google Scholar]
- Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257(5078):1958–61. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- Reed SI. et al. G1 control in yeast and animal cells. Ciba Found Symp. 1992;170:7–15. doi: 10.1002/9780470514320.ch2. discussion 15-9. [DOI] [PubMed] [Google Scholar]
- Koff A. et al. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257(5077):1689–94. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71(3):505–14. doi: 10.1016/0092-8674(92)90518-H. [DOI] [PubMed] [Google Scholar]
- Baldin V. et al. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes & Development. 1993;7(5):812–21. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Hall FL. et al. Two potentially oncogenic cyclins, cyclin A and cyclin D1, share common properties of subunit configuration, tyrosine phosphorylation and physical association with the Rb protein. Oncogene. 1993;8(5):1377–84. [PubMed] [Google Scholar]
- Peeper DS. et al. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12(5):1947–54. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7(8):1572–83. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- Chellappan SP. et al. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65(6):1053–61. doi: 10.1016/0092-8674(91)90557-F. [DOI] [PubMed] [Google Scholar]
- Pagano M. et al. Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene. 1992;7(9):1681–6. [PubMed] [Google Scholar]
- Shirodkar S. et al. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68(1):157–66. doi: 10.1016/0092-8674(92)90214-W. [DOI] [PubMed] [Google Scholar]
- Devoto SH. et al. A cyclin A-protein kinase complex possesses sequence-specific DNA binding activity: p33cdk2 is a component of the E2F-cyclin A complex. Cell. 1992;68(1):167–76. doi: 10.1016/0092-8674(92)90215-X. [DOI] [PubMed] [Google Scholar]
- Cao L. et al. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355(6356):176–9. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- Cobrinik D. et al. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7(12A):2392–404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- Fattaey AR, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13(12):7267–77. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara LR. et al. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. Embo J. 1993;12(11):4317–24. doi: 10.1002/j.1460-2075.1993.tb06116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen ME. et al. Functional interactions of the retinoblastoma protein with mammalian D- type cyclins. Cell. 1993;73(3):487–97. doi: 10.1016/0092-8674(93)90136-E. [DOI] [PubMed] [Google Scholar]
- Kato J. et al. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–42. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Dowdy SF. et al. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73(3):499–511. doi: 10.1016/0092-8674(93)90137-F. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD. et al. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8(15):1772–86. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- Krek W. et al. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78(1):161–72. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. et al. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8(15):1759–71. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- Mittnacht S. et al. Distinct sub-populations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 1994;13(1):118–27. doi: 10.1002/j.1460-2075.1994.tb06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeyesekere MN, Herbert JR, Zimmerman SO. A model of the G1 phase of the cell cycle incorporating cyclin E/cdk2 complex and retinoblastoma protein. Oncogene. 1995;11(6):1199–205. [PubMed] [Google Scholar]
- Beijersbergen RL. et al. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 1995;9(11):1340–53. doi: 10.1101/gad.9.11.1340. [DOI] [PubMed] [Google Scholar]
- Bremner R. et al. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15(6):3256–65. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnane J, Shao Z, Robbins PD. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270(15):8837–43. doi: 10.1074/jbc.270.15.8837. [DOI] [PubMed] [Google Scholar]
- Chen PL, Riley DJ, Lee WH. The retinoblastoma protein as a fundamental mediator of growth and differentiation signals. Crit Rev Eukaryot Gene Expr. 1995;5(1):79–95. [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol. 1996;8(6):805–14. doi: 10.1016/S0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- Mittnacht S, Weinberg RA. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65(3):381–93. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- Ishida S. et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21(14):4684–99. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Development. 2001;15(3):267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B. et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16(2):245–56. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(6):2817–22. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Maller JL. Calcium, calmodulin, and CaMKII requirement for initiation of centrosome duplication in Xenopus egg extracts. Science. 2002;295(5554):499–502. doi: 10.1126/science.1065693. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442(7105):947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Ortega S. et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35(1):25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- Berthet C. et al. Cdk2 knockout mice are viable. Curr Biol. 2003;13(20):1775–85. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Duensing A. et al. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 2006;25(20):2943–9. doi: 10.1038/sj.onc.1209310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y. et al. Cyclin E ablation in the mouse. Cell. 2003;114(4):431–43. doi: 10.1016/S0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- McCleland ML, Farrell JA, O'Farrell PH. Influence of cyclin type and dose on mitotic entry and progression in the early Drosophila embryo. J Cell Biol. 2009;184(5):639–46. doi: 10.1083/jcb.200810012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H. et al. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178(2):257–68. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adon AM. et al. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol Cell Biol. 2010;30(3):694–710. doi: 10.1128/MCB.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora I, Marshall WF. A mutation in the centriole-associated protein centrin causes genomic instability via increased chromosome loss in Chlamydomonas reinhardtii. BMC Biol. 2005;3:15. doi: 10.1186/1741-7007-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Lee RJ. et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20(2):672–83. doi: 10.1128/MCB.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botz J. et al. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16(7):3401–9. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S. et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Molecular & Cellular Biology. 2001;21(14):4684–99. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung BY. et al. Effects of actinomycin D analogs on nucleolar phosphoprotein B23 (37,000 daltons/pI 5.1) Biochem Pharmacol. 1985;34(22):4059–63. doi: 10.1016/0006-2952(85)90387-9. [DOI] [PubMed] [Google Scholar]
- Feuerstein N, Randazzo PA. In vivo and in vitro phosphorylation studies of numatrin, a cell cycle regulated nuclear protein, in insulin-stimulated NIH 3T3 HIR cells. Exp Cell Res. 1991;194(2):289–96. doi: 10.1016/0014-4827(91)90367-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Hugle-Dorr B, Franke WW. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 1987;6(7):1881–90. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S. et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437(7055):147–53. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- Okuda M. et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103(1):127–40. doi: 10.1016/S0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Tokuyama Y. et al. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. Journal of Biological Chemistry. 2001;276(24):1529–37. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Zatsepina OV. et al. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J Cell Sci. 1999;112(Pt 4):455–66. doi: 10.1242/jcs.112.4.455. [DOI] [PubMed] [Google Scholar]
- Krause A, Hoffmann I. Polo-like kinase 2-dependent phosphorylation of NPM/B23 on serine 4 triggers centriole duplication. PLoS One. 2010;5(3):e9849. doi: 10.1371/journal.pone.0009849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, BRCA2 and nucleophosmin co-regulate centrosome amplification and form a complex with Rho effector kinase ROCK2. Cancer Res. 2010. [DOI] [PubMed]
- Adams MR. et al. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol Cell Biol. 2000;20(10):3633–9. doi: 10.1128/MCB.20.10.3633-3639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M. et al. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114(4):745–54. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AR. et al. The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J Cell Biol. 2002;156(3):453–65. doi: 10.1083/jcb.200111025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106(1):95–104. doi: 10.1016/S0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Kasbek C. et al. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18(11):4457–69. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM. et al. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. Embo J. 2002;21(7):1723–32. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100(25):14875–80. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R. et al. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7(11):1140–6. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Chen Z. et al. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Developmental Cell. 2002;3(3):339–50. doi: 10.1016/S1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. p53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta. 2008. [DOI] [PMC free article] [PubMed]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7(12):911–24. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Loncarek J. et al. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10(3):322–8. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan TM. et al. Centrosome duplication proceeds during mimosine-induced G1 cell cycle arrest. J Cell Physiol. 2008;215(1):182–91. doi: 10.1002/jcp.21298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. et al. Abnormal centrosome amplification in the absence of p53. Science. 1996;271(5256):1744–7. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Duensing S. et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(18):10002–7. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ. 2006. [DOI] [PubMed]
- Harper JW. et al. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- Harper JW. et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6(4):387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmura K. et al. Direct evidence for the role of centrosomally localized p53 in the regulation of centrosome duplication. Oncogene. 2007;26(20):2939–44. doi: 10.1038/sj.onc.1210085. [DOI] [PubMed] [Google Scholar]
- Tarapore P. et al. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene. 2001;20(25):3173–84. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- Duensing A. et al. p21(Waf1/Cip1) Deficiency Stimulates Centriole Overduplication. Cell Cycle. 2006;5(24) doi: 10.4161/cc.5.24.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C. et al. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood. 1999;93(4):1390–8. [PubMed] [Google Scholar]
- Nishimura T. et al. Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells. 2005;10(1):75–86. doi: 10.1111/j.1365-2443.2005.00816.x. [DOI] [PubMed] [Google Scholar]
- Nakayama K. et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. Embo J. 2000;19(9):2069–81. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X. et al. ECRG2 disruption leads to centrosome amplification and spindle checkpoint defects contributing chromosome instability. J Biol Chem. 2008;283(9):5888–98. doi: 10.1074/jbc.M708145200. [DOI] [PubMed] [Google Scholar]
- Hemerly AS. et al. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323(5915):789–93. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R. et al. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130(1):105–15. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. EGFRvIII expression and PTEN loss synergistically induce chromosomal instability and glial tumors. Neuro Oncol. 2009;11(1):9–21. doi: 10.1215/15228517-2008-081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter AL. et al. The neu oncogene: an erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature. 1984;312(5994):513–6. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- Harari D, Yarden Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene. 2000;19(53):6102–14. doi: 10.1038/sj.onc.1203973. [DOI] [PubMed] [Google Scholar]
- Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61(Suppl 2):1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- Montagna C. et al. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene. 2002;21(6):890–8. doi: 10.1038/sj.onc.1205146. [DOI] [PubMed] [Google Scholar]
- Saavedra HI. et al. MAPK mediates RAS-induced chromosome instability. Journal of Biological Chemistry. 1999;274(53):38083–90. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- Saavedra HI. et al. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene. 2000;19(34):3948–54. doi: 10.1038/sj.onc.1203723. [DOI] [PubMed] [Google Scholar]
- Ussar S, Voss T. MEK1 and MEK2, different regulators of the G1/S transition. J Biol Chem. 2004;279(42):43861–9. doi: 10.1074/jbc.M406240200. [DOI] [PubMed] [Google Scholar]
- Yun C. et al. Mitotic aberration coupled with centrosome amplification is induced by hepatitis B virus X oncoprotein via the Ras-mitogen-activated protein/extracellular signal-regulated kinase-mitogen-activated protein pathway. Mol Cancer Res. 2004;2(3):159–69. [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2(2):103–12. doi: 10.1016/S1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Reddy HK. et al. Cyclin-dependent kinase 4 expression is essential for neu-induced breast tumorigenesis. Cancer Res. 2005;65(22):10174–8. doi: 10.1158/0008-5472.CAN-05-2639. [DOI] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(6841):1017–21. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Nelsen CJ. et al. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J Biol Chem. 2005;280(1):768–76. doi: 10.1074/jbc.M407105200. [DOI] [PubMed] [Google Scholar]
- Hanse EA. et al. Cdk2 plays a critical role in hepatocyte cell cycle progression and survival in the setting of cyclin D1 expression in vivo. Cell Cycle. 2009;8(17):2802–9. doi: 10.4161/cc.8.17.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. et al. Genetic and epigenetic changes in mammary epithelial cells identify a subpopulation of cells involved in early carcinogenesis. Cold Spring Harb Symp Quant Biol. 2005;70:317–27. doi: 10.1101/sqb.2005.70.051. [DOI] [PubMed] [Google Scholar]
- Holst CR. et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63(7):1596–601. [PubMed] [Google Scholar]
- McDermott KM. et al. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4(3):e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. et al. Increased gamma-tubulin expression and P16INK4A promoter methylation occur together in preinvasive lesions and carcinomas of the breast. Ann Oncol. 2009;20(3):441–8. doi: 10.1093/annonc/mdn651. [DOI] [PubMed] [Google Scholar]
- Kondo Y. et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One. 2008;3(4):e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8(5):345–57. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- Mackay A. et al. cDNA microarray analysis of genes associated with ERBB2 (HER2/neu) overexpression in human mammary luminal epithelial cells. Oncogene. 2003;22(17):2680–8. doi: 10.1038/sj.onc.1206349. [DOI] [PubMed] [Google Scholar]
- Meijer L. et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243(1-2):527–36. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M. et al. A cyclin-dependent kinase inhibitor, butyrolactone I, inhibits phosphorylation of RB protein and cell cycle progression. Oncogene. 1994;9(9):2549–57. [PubMed] [Google Scholar]
- Hanashiro K. et al. Roles of cyclins A and E in induction of centrosome amplification in p53-compromised cells. Oncogene. 2008;11;27(40):5288–302. doi: 10.1038/onc.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29(7):1760–73. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing A. et al. RNA polymerase II transcription is required for human papillomavirus type 16 E7- and hydroxyurea-induced centriole overduplication. Oncogene. 2007;26(2):215–23. doi: 10.1038/sj.onc.1209782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino F. et al. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer. 2006;5:38. doi: 10.1186/1476-4598-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini L. et al. Centrosome amplification induced by hydroxyurea leads to aneuploidy in pRB deficient human and mouse fibroblasts. Cancer Lett. 2006;238(1):153–60. doi: 10.1016/j.canlet.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Tarapore P, Okuda M, Fukasawa K. A mammalian in vitro centriole duplication system: evidence for involvement of CDK2/cyclin E and nucleophosmin/B23 in centrosome duplication. Cell Cycle. 2002;1(1):75–81. doi: 10.4161/cc.1.1.103. [DOI] [PubMed] [Google Scholar]
- Brady SN. et al. Nucleophosmin protein expression level, but not threonine 198 phosphorylation, is essential in growth and proliferation. Oncogene. 2009;28(36):3209–20. doi: 10.1038/onc.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliekelman M. et al. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69(1):45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67(21):10103–5. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28(1-2):85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B. et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes & Development. 2001;15(1):50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova OB. et al. MYC abrogates p53-mediated cell cycle arrest in N-(phosphonacetyl)-L-aspartate-treated cells, permitting CAD gene amplification. Mol Cell Biol. 1998;18(1):536–45. doi: 10.1128/mcb.18.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A. et al. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci USA. 2003;100(17):9974–9. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S. et al. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006;66(13):6598–605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- Denko NC. et al. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci USA. 1994;91(11):5124–8. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Schultz N, Helleday T. p53 protects from replication-associated DNA double-strand breaks in mammalian cells. Oncogene. 2004;23(13):2324–9. doi: 10.1038/sj.onc.1207379. [DOI] [PubMed] [Google Scholar]