Abstract
Background
Genes subject to genomic imprinting are mono-allelically expressed in a parent-of-origin dependent manner. Each imprinted locus has at least one differentially methylated region (DMR) which has allele specific DNA methylation and contributes to imprinted gene expression. Once DMRs are established, they are potentially able to withstand normal genome reprogramming events that occur during cell differentiation and germ-line DMRs are stably maintained throughout development. These DMRs, in addition to being either maternally or paternally methylated, have differences in whether methylation was acquired in the germ-line or post fertilization and are present in a variety of genomic locations with different Cytosine-phosphate guanine (CpG) densities and CTCF binding capacities. We therefore examined the stability of maintenance of DNA methylation imprints and determined the normal baseline DNA methylation levels in several adult tissues for all imprinted genes. In order to do this, we first developed and validated 50 highly specific, quantitative DNA methylation pyrosequencing assays for the known DMRs associated with human imprinted genes.
Results
Remarkable stability of the DNA methylation imprint was observed in all germ-line DMRs and paternally methylated somatic DMRs (which maintained average methylation levels of between 35% - 65% in all somatic tissues, independent of gene expression). Maternally methylated somatic DMRs were found to have more variation with tissue specific methylation patterns. Most DMRs, however, showed some intra-individual variability for DNA methylation levels in peripheral blood, suggesting that more than one DMR needs to be examined in order to get an overall impression of the epigenetic stability in a tissue. The plasticity of DNA methylation at imprinted genes was examined in a panel of normal and cancer cell lines. All cell lines showed changes in DNA methylation, especially at the paternal germ-line and the somatic DMRs.
Conclusions
Our validated pyrosequencing methylation assays can be widely used as a tool to investigate DNA methylation levels of imprinted genes in clinical samples. This first comprehensive analysis of normal methylation levels in adult somatic tissues at human imprinted regions confirm that, despite intra-individual variability and tissue specific expression, imprinted genes faithfully maintain their DNA methylation in healthy adult tissue. DNA methylation levels of a selection of imprinted genes are, therefore, a valuable indicator for epigenetic stability.
Background
DNA methylation levels at gene promoters and Cytosine-phosphate guanine (CpG) islands associated with gene regulatory regions undergo dynamic changes during differentiation and can vary between normal tissues [1]. In cancer cells epigenetic programming results in global methylation changes [2] and it is difficult to ascertain which methylation changes are abnormal without knowing what normal baseline methylation profiles are for the tissue from which the cancer originates [3]. Since aberrant DNA methylation is thought to be an early indicator of cancer, it will be useful to have a series of reporter loci to indicate the epigenetic health of a tissue sample.
Imprinted genes exhibit monoallelic parent-of-origin specific gene expression. They have roles in fetal growth and development [4] and are usually located within the genome in clusters [5] or as pairs of retrogenes [6]. At present, 64 human genes are known to be subject to genomic imprinting [7] and a further seven show some evidence of imprinted expression. Every imprinted cluster has at least one differentially methylated region (DMR), where DNA methylation is present on one parental allele. A single DMR can regulate a number of imprinted genes within a cluster and, therefore, the methylation status of one DMR can provide information about a number of genes [5]. DMRs can be sub-classified into germ-line and somatic DMRs. Germ-line DMRs are loci which exhibit differences in methylation states between the sperm and the egg. These differences are maintained post-fertilization. At somatic DMRs, DNA methylation is still parent-of-origin specific, but is acquired after fertilization. Once established, DNA methylation imprints are able to withstand genome-wide DNA methylation reprogramming events during the peri-implantation period after fertilization and also during tissue differentiation. Imprinted genes only succumb to genome-wide reprogramming in the primordial germ-line, prior to the resetting of the imprint according to the gonadal sex of the germ-line. This robust feature of maintaining DNA methylation in somatic tissue makes imprinted loci ideal indicators of the overall epigenetic health of a cell.
Many imprinted genes are themselves oncogenes or tumour suppressors [8]; their aberrant expression could drive tumourigenesis. Examples of potential oncogenic imprinted genes include paternally expressed IGF2, DLK1, PEG1/MEST, PEG3 and PEG10 which are normally expressed in early fetal kidney development and up regulated in Wilms' tumour [9,10]. Aberrant IGF2 and DLK1 expression has also been shown in adult renal cancers [11]. The down-regulation of the maternally expressed tumour suppressing non-coding H19 RNA may leads to cancer in Wilms' tumour and many adult onset cancers [12]. Additionally, the retinoblastoma gene (RB1) has recently been shown to have preferential maternal expression [13], thus adding another tumour suppressor to the repertoire of maternally expressed growth inhibiting genes. These examples illustrate that DNA methylation at imprinted regions may have functional roles in oncogenesis and could be used as a surrogate biomarker for loss of imprinting as previously proposed [14] or simply as an indicator of cancer [15].
The best characterised DMR binding protein is CTCF, an 11-zinc finger protein that binds to the H19-DMR [16]. CTCF binds the unmethylated maternal allele and mediates the insulator function that prevents the paternal IGF2 allele from accessing enhancers downstream of H19. It has been shown that mutations of CTCF binding sites within the H19-DMR lead to a gain of methylation on the maternal allele, suggesting that CTCF also protects against de novo methylation [17-19]. We have recently shown that in Beckwith-Wiedemann and Silver-Russell patients methylation changes at the H19 DMR result in concordant changes at the DMRs within the IGF2 locus [20]. These changes suggest cross talk between the DMRs in cis, which may be mediated by CTCF, and cohesin, through higher order chromatin looping at the IGF2/H19 locus [21,22].
Indications of gene imprinting networks [23] and the identification of protein factors such as ZFP57, a KRAB zinc finger protein that is important for establishing maternal imprints in the oocyte and maintaining methylation at maternal and paternal imprinted domains post-fertilization [24,25], have created the need to analyse larger numbers of imprinted genes in imprinting defects. This may provide more mechanistic clues as to the role of loss of imprinting in cancer and congenital disease than when studying single imprinted genes in isolation.
In order to understand the role that changes in methylation at imprinted genes have in pathophysiology, the normal methylation levels in a variety of tissues and the inter-individual variability needs to be known. High-throughput methylation studies currently employ several technologies but none of these are able to quantitatively identify methylation changes at imprinted genes. This is because imprinted genes often have lower methylation densities at their DMRs (especially the paternally methylated germ-line DMRs [26]) than non-imprinted genes and, therefore, may not be quantitatively detected with antibodies to methyl-CpGs. Many arrays are designed for promoter regions and the DMRs for many imprinted genes are not covered by these arrays. More importantly, however, these technologies rely on the binary detection of either 'methylated' or 'unmethylated' and allele specific methylation associated with imprinted genes means crucial information is lost in such genome wide studies. More focussed methylation studies for imprinted loci in humans are required. One recent approach utilized a microarray representing characterized murine imprinted loci to highlight the tissue specific variability of DMRs [27]. Pyrosequencing (PSQ) is preferable to other methods of methylation analysis in that it is processive and quantitative [28] and reviewed in [29]. PSQ is particularly suited for the examination of selected regions in large numbers of samples [15,30] and avoids some of the pitfalls when using methodologies not tailored to analysing specific CpGs [31].
We have, therefore, designed PSQ assays to cover all the known and potential DMRs of all human imprinted genes identified to date. The methylation levels in eight different tissue types were analysed, representing the widest spectrum of adult human tissues ever assayed for imprinted methylation. A sub-set of DMRs were also assayed in 50 different human blood samples, to establish the intra-individual variation. Our data represents the first comprehensive tissue wide comparison of differential methylation in the human.
Results
Novel methylation assays for differentially methylated regions
We designed and optimized 50 PSQ methylation assays to represent all known human imprinted loci, including imprinted clusters, paired imprinted retrogenes and orphan imprinted genes. Our assays provide a quantitative measure of imprinted methylation. Our assays covered 3-9 CpGs within regions either known to, or with the potential to, regulate 51 imprinted genes (Table 1 and Additional File 1; Table S1). We tested our 50 assays on a panel of seven adult tissues [brain, breast, colon, heart, kidney, liver and testis (containing sertoli cells which show imprinted gene expression [32] and sperm cells, which show imprint erasure)] and term placenta. For the purpose of this study, and due to the limitations involved in acquiring high quality human DNA, we have classified the brain as a single tissue, although results from analysis of the mouse brain suggests there may be region dependant parent-of-origin specific expression [33]. The exact location of all assays and the results in each tissue can be downloaded from the laboratory website [34].
Table 1.
Characteristics of each region investigated
| Assay | Chr | Location | P-O-O methylation mark | Germ-line/somatic | Average somatic methylation | Location within gene | CpG island | CTCF | CpG density |
|---|---|---|---|---|---|---|---|---|---|
| DIRAS3 (3) | 1 | 68285444 | Maternal | Somatic | 70.22 | Gene body Exonic | Yes | YV | 6.2 |
| DIRAS3 (2) | 1 | 68288969 | Maternal | Germ-line# | 50.42 | Promoter/Ex 1 | Yes | N | 5.6 |
| DIRAS3 (1) | 1 | 68290053 | Maternal | Germ-line# | 46.09 | 5' upstream (GPR) | Yes | YV | 5.4 |
| ZDBF2 | 2 | 206834066 | Paternal | Germ-line | 47.65 | Intergenic | No | Y | 1.4 |
| NAP1L5 | 4 | 89837958 | Maternal | Germ-line | 50.16 | Embeded gene promoter | No | N | 7.2 |
| ZAC | 6 | 144371274 | Maternal | Germ-line | 40.83 | Isoform promoter | Yes | N | 11.2 |
| IGF2R-2 | 6 | 160346534 | Unknown | Unknown | 81.34 | Gene body intronic | Yes | N | 6.6 |
| SLC22A1 | 6 | 160475377 | Unknown | Unknown | 82.78 | Gene body Exonic | Yes | N | 5.6 |
| SLC22A3 | 6 | 160688909 | Unknown | Unknown | 39.89 | Promoter/Ex 1 | Yes | YV | 7.6 |
| MEST (s) | 7 | 129913465 | Maternal | Somatic | 28.15 | Isoform promoter | Yes | Y | 8.2 |
| MEST | 7 | 129918562 | Maternal | Germ-line | 45.84 | Isoform promoter | Yes | Y | 7 |
| GRB10 (g) | 7 | 50817567 | Maternal | Germ-line | 45.47 | Isoform promoter | Yes | YV | 12.6 |
| GRB10 (s) | 7 | 50829108 | Maternal | Somatic | 19.19 | Isoform promoter | Yes | YV | 6 |
| PEG10 | 7 | 94123850 | Maternal | Germ-line | 55.63 | Promoter | Yes | Y | 6.4 |
| PON1 | 7 | 94791675 | Unknown | Unknown | 34.04 | Promoter/Ex 1 | Yes | N | 4.6 |
| INPP5FV2 | 10 | 121568151 | Maternal | Germ-line | 60.89 | Isoform promoter | Yes | YV | 11.2 |
| H19 DMR | 11 | 1977714 | Paternal | Germ-line | 45.23 | 5' upstream (GPR) | No | Y | 5.8 |
| IGF2 (2) | 11 | 2110885 | Paternal | Somatic | 35.41 | Gene body Exonic | Yes | N | 6.2 |
| IGF2 (0) | 11 | 2126069 | Paternal | Somatic | 54.59 | Isoform promoter | No | N | 2.2 |
| KCNQ1 | 11 | 2422247 | Unknown | Unknown | 22.27 | 5' upstream (GPR) | Yes | YV | 6.8 |
| KvDMR | 11 | 2678628 | Maternal | Germ-line | 50.68 | antisense RNA promoter | Yes | YV | 5.6 |
| KCNQ1DN | 11 | 2847182 | Unknown | Unknown | 26.22 | 5' upstream (GPR) | Yes | N | 11.4 |
| CDKN1C | 11 | 2861764 | None | None | 12.07 | Gene body Exonic | Yes | Y | 10.8 |
| OSBPL5 (3) | 11 | 3098345 | Unknown | Unknown | 82.66 | Gene body Exonic | Yes | N | 4.2 |
| RB1 | 13 | 47791152 | Maternal | Germ-line | 59.47 | Gene body intronic | Yes | N | 6.4 |
| DLK | 14 | 100262671 | Paternal | Somatic | 56.05 | Promoter/Ex 1 | Yes | N | 9.2 |
| IG-DMR | 14 | 100345582 | Paternal | Germ-line | 58.17 | Intergenic | No | N | 2.4 |
| MEG3-US | 14 | 100360453 | Paternal | Somatic | 49.55 | Intergenic | Yes | N | 6.6 |
| MEG3 | 14 | 100361829 | Paternal | Somatic | 48.99 | Promoter/Ex 1 | Yes | YV | 6.2 |
| RTL | 14 | 100419312 | None | None | 88.27 | Gene body Exonic | Yes | N | 6.4 |
| MKRN3 | 15 | 21362387 | Unknown | Unknown | 62.44 | Gene body Exonic | No | N | 4.4 |
| SNRPN | 15 | 22751911 | Maternal | Germ-line | 47.12 | Isoform promoter | Yes | N | 7.8 |
| UBE3A | 15 | 23234499 | None | None | 3.15 | Promoter/Ex 1 | Yes | Y | 9.6 |
| ATP10A | 15 | 23658720 | None | None | 13.82 | Promoter/Ex 1 | Yes | N | 5.4 |
| GABRB3 | 15 | 24425436 | Unknown | Unknown | 47.8 | Isoform promoter | Yes | YV | 7.6 |
| GABRA5 | 15 | 24663372 | Unknown | Unknown | 87.29 | Promoter/Ex 1 | Yes | YV | 13 |
| TCEB3C | 18 | 42797692 | None | None | 86.52 | Embedded gene pronoterd | Yes | YV | 7.6 |
| NLRP2 | 19 | 60186163 | Unknown | Unknown | 61.93 | Gene-body Exonic | Yes | N | 7 |
| PEG3/ZIM2 | 19 | 62043527 | Maternal | Germ-line | 49.92 | Gene body intronic | Yes | N | 9.2 |
| USP29 | 19 | 62322335 | Unknown | Unknown | 43.27 | Intergenic | Yes | N | 7.4 |
| ZIM3 | 19 | 62348129 | Unknown | Unknown | 74.21 | Promoter/Ex 1 | No | N | 1 |
| MCTS2 | 20 | 21441323 | Maternal | Germ-line | 44.72 | Embedded gene promoter | Yes | Y | 6.6 |
| NNAT | 20 | 35582455 | Maternal | Germ-line | 77.93 | Embedded gene promoter | Yes | YV | 5.2 |
| BLCAP | 20 | 35589631 | None | None | 8.37 | Promoter/Ex 1 | Yes | YV | 7.6 |
| L3MBTL | 20 | 41576732 | Maternal | Germ-line | 51.23 | Gene body Exonic (Ex1) | Yes | Y | 7.6 |
| NESP | 20 | 56848977 | Paternal | Somatic | 45.7 | Isoform promoter | Yes | Y | 9.2 |
| NESPAS | 20 | 56860398 | Maternal | Somatic | 47.53 | Isoform promoter | Yes | N | 6.4 |
| GNAS XL | 20 | 56864018 | Maternal | Germ-line | 43.98 | Isoform promoter | Yes | N | 5.6 |
| GNAS 1A | 20 | 56898576 | Maternal | Somatic | 25.1 | Isoform promoter | Yes | N | 12 |
Characteristics of each assay: Average somatic methylation is the average level for Brain, Breast, Colon, Heart, Kidney, Liver and Placenta. Genomic localization was analysed by BLASTing the amplicon against UCSC genomic sequence. The location of the amplicon in context of each gene sequence was recorded.
GPR = general promoter region; CpG Island amplicon is within an UCSC annotated CpG island; CTCF binding is determined by analysis of ENCODE tracts on UCSC, (Y = Yes); binding in a tissue specific manner (YV = Yes variable); N = not binding; CpG density was calculated by taking the mid-point of the amplicon used for each assay and analysing the sequence 250bp each side (500bp in total); density is given as No. of CpGs per 100bp; # = the DMR for DIRAS3 could be either DMR1 or DMR2.
Chr = chromosome
The specificity and linearity of the assays were measured using standard templates of known ratios of methylated:unmethylated DNA. The known amount of methylated DNA in the input sample could be compared with the methylation reported by the assay (Additional File 2; Figure S1a). A normal DMR on our standard template contained 50% methylated input DNA and gave an average PSQ read of 50.38%. We therefore defined a DMR as reporting a methylation level by PSQ within the range of 35.73%-65.03% (1.5 times the standard deviation of the mean known DMRs). The assays all report linearity with the increasing amount of methylated DNA in the standard template.
We validated the assays for sensitivity by testing the smallest amount of input DNA required to accurately report levels of 50% methylation in normal samples. As little as 1ng of bisulphite treated DNA could accurately report normal levels of methylation in 23/24 assays. At this low concentration of template, allelic drop-out was only observed in one single assay (L3MBTL-DMR). The template DNA of 2.5ng per assay was found to be the lowest amount of DNA required to confidently run a full set of 50 assays that report methylation levels. This is a significant improvement on previous PSQ methodology [28].
The intra-assay reproducibility was assessed by plotting two technical replicates of each assay against one another. The linear relationship between the technical replicates confirmed that intra-assay reproducibility was reliable (regression analysis gave an r2 = 0.96, Bland-Altman correlation showed a bias of just 1.60%; Additional File 2, Figure S1b).
Five of our assays were in regions that did not show 35%-65% methylation in any of the tissues tested. Of these UBE3A, BLCAP and CDKN1C were hypomethylated (average methylation range of 2.96% to 13.53%), while TCB3C and RTL were hypermethylated (average methylation range of 65.99% to 91.23%). These data are consistent with previous reports that have shown UBE3A, BLCAP, RTL and CDKN1C to be imprinted genes without promoter DMRs (UBE3A [35], BLCAP [36], CDKN1C [37], RTL [38]). All the other assays showed methylation levels consistent with differential methylation in at least one tissue (Additional File 1, Table S2 and Additional File 3, Figure S2). Together, these data represent a catalogue of baseline normal methylation levels in adult tissues.
As the methylation data was analysed on a single platform, we were able to examine the differences of the DMRs when sub-classified into categories such as germ-line or somatic, CpG density and genomic position.
Germ-line versus somatic DMRs
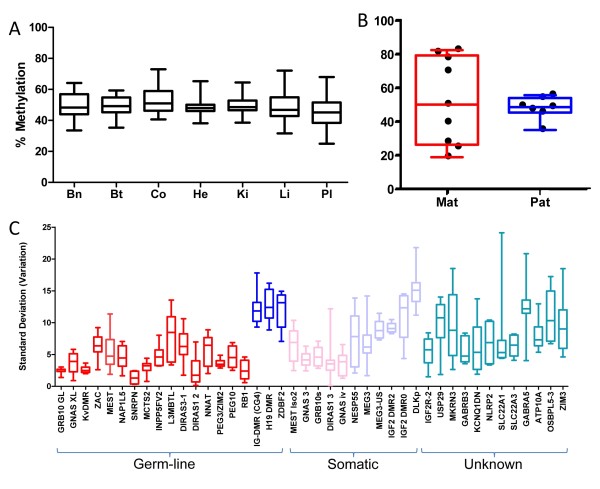
In order to assess whether germ-line DMRs are more stable than somatic DMRs and would, therefore, have uniform levels of methylation in all tissues, we compared the intra-tissue variability of methylation separately for germ-line and somatic DMRs. The average methylation levels of the 17 germ-line DMRs was 47.26% in all somatic tissues and methylation levels did not vary amongst tissues (Figure 1a). Methylation levels in testis were significantly different from the average methylation of other tissues (matched pair t-test P = 0.0009): this was expected as the sperm cells in the testis erased the maternal imprints. The germ-line DMRs were hypomethylated in the testis, with the exception of H19, IG-DMR and ZBDF2 which are known to be paternally methylated (Additional File 3, Figure S2 and Additional File 1, Table S2). DMRs that were outliers from the 35%-65% figure calculated above include NNAT and INPP5FV2. INPP5FV2 has been published as a germ-line DMR [39] but we found it to be hypermethylated in colon and liver. Although NNAT is also a published germ-line DMR [36], our data shows that in adult tissue only the brain reports methylation levels consistent with a DMR: it is hypermethylated in other tissues. NNAT has high expression levels in the brain and low levels of expression in other tissues and, therefore, this may not be a human germ-line DMR (RefExA-[40]).
Figure 1.
Average methylation levels for germ-line differentially methylated regions (DMRs; n = 17 in each tissue). Box plots show median, inter-quartile range and maximum/minimum methylation (Bn = brain; Bt = breast; Co = colon; He = heart; Ki = kidney; Li = liver; Pl = placenta). (b) Average tissue methylation levels of somatic DMRs when analysed by parent-of-origin of the methylation. Each data point represents an individual C-phosphate guanine (CpG). Box plots show median, inter-quartile range and maximum/minimum methylation. (c) Intra-CpG variability of DMR methylation. The standard deviation of all CpGs assayed was calculated for each tissue (n = 8). Box plots show median, inter-quartile range and maximum/minimum methylation. Red = maternal germ-line DMRs; pink = maternal somatic DMRs; dark blue = paternal germ-line DMRs; light blue = paternal somatic DMRs; green = parent-of-origin and/or germ-line somatic status unknown.
Analyses of somatic DMRs showed more variation in the average methylation per DMR in different tissues as expected. Many somatic DMRs are only allele specifically methylated in selected tissues - for example, GNAS 1A in brain, MEST isoform 2 in placenta [41], MKRN3 in liver and SLC22A1 in placenta - and our data confirms this. We also observed that, for the seven somatic DMRs where the parent of origin is published as paternally methylated, the average methylation levels were between 35%-65% across all tissues, independent of expression and are not tissue specifically variable. In contrast 4/5 maternally methylated somatic DMRs are tissue specific - that is, only reporting methylation levels consistent with a DMR in specific tissues (Figure 1b; Additional File 1, Table S2). However, this does not correlate with expression in adult tissues [40] and imprinted differential methylation is not a good indicator of expression levels in human adult tissue. For example, PON1 reports methylation levels consistent with a DMR in brain and kidney but is only highly expressed in the liver. The somatic DMR at the MEG promoter reports levels consistent with a DMR in all tissues but is only expressed in brain, placenta and testis. Tissue specific expression is also often isoform dependent.
As somatic DMRs are thought to be more variable and less stable than germ-line DMRs, we analysed the intra CpG variability of each DMR and compared somatic and germ-line DMRs in all tissues (Figure 1c). The major difference observed was that paternally methylated DMRs showed greater intra-CpG variability across the region assayed than maternally methylated DMRs. Any individual CpG from a maternally methylated DMR is, therefore, more indicative of the rest of the island.
Effects of CpG density and CTCF binding on variability of differential methylation
We identified which of the DMRs contained CTCF binding sites by matching their genome coordinates for enrichment of CTCF binding in a data set from human liver tissue (M Wilson, unpublished data). We then correlated methylation levels obtained with our assays in liver DNA with CTCF binding within the DMR. Overall, 11/43 DMRs had CTCF binding in liver and, of these, six were germ-line and five were somatic DMRs. There was no evidence of a difference between methylation levels reported in DMRs with CTCF binding sites compared to CTCF binding in the liver (t-test, P = 0.17; Additional File 4, Figure S3Ai).
In a similar analysis, the average methylation levels reported for all somatic tissues were correlated with published CTCF binding data in four separate cell lines (ENCODE data set [42,43]). Twelve were found to bind CTCF in some cell lines (variable CTCF) and eight bound CTCF in all cell lines. There was also no evidence of difference in methylation levels between DMRs that bound CTCF and those that did not (t-test, P = 0.99; Additional File 4, Figure S3Aii).
CTCF only binds non-methylated sequences, so we assumed that the somatic DMRs that were hypermethylated in liver (n = 10) could still have CTCF binding sites that would be occupied in other tissues but be negative in liver. One surprising result was the CTCF binding to the GABRA5 locus in liver, which was 83% methylated. This may be explained by binding of CTCF to a different part of the CpG island to that assayed or it may have been due to the CTCF binding observed being exclusively on the 17% of the GABRA5 that DNA strands within the population that are unmethylated.
Most (38/44) of the regions assayed are CpG islands as defined on the University of California, Santa Cruz (UCSC) genome browser. We also investigated the correlation between CpG density and methylation levels. CpG density was calculated by counting the number of CpG sites 250bp each side of the mid-point of the amplicon used in the assay (Additional File 1, Table S3). The density of CpGs in each amplicon was similar to the CpG density in the overall region CpG island, as annotated by the UCSC genome browser. There was no correlation between the CpG density and average somatic methylation level reported (r2 = 0.090; Additional File 4, Figure S3b)
Effect of genomic position on DMR methylation stability
DMRs can be located intergenically, at promoters or at other regions within the gene body. Additional File 4, Figure S3c, shows no difference between location of the DMR and average somatic methylation levels (ANOVA, P = 0.29). The gene body DMRs also most likely have variable methylation levels in somatic tissues. Most of the gene body DMRs were somatic DMRs.
Germ-line DMRs within promoters maintained 35%-65% methylation levels independently of gene expression (for example, KVDMR, ZAC1, GRB10). This was also true for known paternally methylated somatic DMRs. In contrast, known maternal somatic DMRs were hypo/hypermethylated in most tissues and only reported levels consistent with a DMR in one or two tissues. However, this was independent of adult expression levels [40] but may reflect expression during different developmental stages.
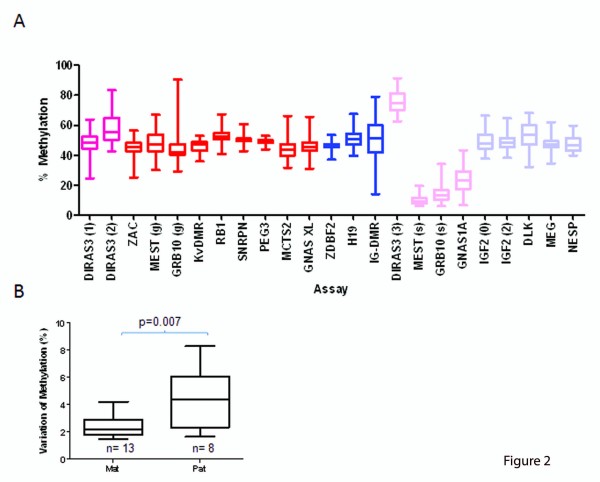
Intra-individual variation of methylation levels at imprinted DMRs
Methylation levels can vary between individuals due to a number of environmental and genetic reasons. In order to determine the extent of inter-individual variation in methylation levels at imprinted genes in a population, we analysed 23 DMRs in peripheral blood samples from 50 healthy humans. The 23 DMRs chosen represent a range of germ-line and somatic DMRs, incorporating maternally and paternally methylated DMRs. Figure 2a shows the average and range of methylation at each DMR analysed in 50 individuals. Outliers (methylation levels falling outside of the 95% and 99% CI (confidence interval; Additional File 1, Table S4) were observed for 17/23 DMRS (99% CI). However, there were no significant differences in the frequencies of either a germ-line or a somatic DMR being an outlier (P = 0.88). This is true for genes that are expressed in the blood (ZIM2/PEG3, GNAS and SNRPN) as well as those expressed at very low levels (IGF2, ZAC and MEST [40]).
Figure 2.
Analysis of 23 differentially methylated region (DMR) assays in 50 individual blood samples. (a) Average methylation levels of 50 different individuals. Red = maternal germ-line DMRs; pink = maternal somatic DMRs; dark blue = paternal germ-line DMRs; light blue = paternal somatic DMRs. Box plots show the median, inter-quartile range and maximum/minimum methylation B standard deviation of all C-phosphate guanines (CpGs) assayed for maternal and paternal DMRs. A significant difference was observed between the intra-assay variability reported (matched pair t-test, P = 0.007).
Nineteen blood samples had outliers for one or more (95% and 99% CI) DMRs. However, no individual is over-represented within the outliers or shows a consistent gain or loss in methylation across several DMRs (repeated measure ANOVA; P = 0.60).
Four of the DMRs were consistently either hypermethylated (DIRAS3 - 3) or hypomethylated (MEST (s), GRB10 (s) and GNAS1A) in all samples. These were all maternally methylated DMRs and repeated the observation seen in tissues; paternally methylated somatic DMRs are not tissue specific, whereas somatic maternally methylated DMRs can be tissue specific.
Where DMRs are sequentially present on the same chromosome, loss or gain of methylation at one DMR did not affect the other DMRs in cis (comparison of DMRs within the same cluster showed no correlation between methylation levels; max r2 = 0.086, data not shown).
Individual CpGs within the maternally methylated DMRS were tightly clustered near the mean methylation level for each individual, whereas the CpGs within the paternally methylated DMRs had a wider range of intra-assay CpG variability. This can be seen for each DMR for all 50 individuals in Additional File 5, Figure S4. This variability for maternal and paternally methylated DMRs was significantly different (Figure 2b, P = 0.007). This indicates that in maternal DMRs each CpG changes concurrent with the rest of the island whereas the individual CpGs within paternally methylated DMRs are less likely to be reflective of the rest of the region. This is similar to the trend seen for paternal DMRs which have an increased intra-DMR variability of methylation in blood.
Detection of methylation changes at Imprinted loci in cell-lines
Cell lines are known to accumulate methylation in passage and are epigenetically unstable, providing a model system with which to examine the plasticity of imprinted DMRs. We examined the methylation levels of nine germ-line DMRs (six maternal and three paternal) and six somatic DMRs (three maternal and three paternal) in a panel of normal and cancer cell lines. Additional File 6, Figure S5 shows a comparison of the methylation levels in cell lines to the baseline levels of methylation as established in blood. All cell lines, even the three cell lines reported to be from normal tissue, had changes in methylation levels. As expected, the cancer cell lines had more variable methylation, some of which could be attributable to karyotype abnormalities (Additional File 1, Table S5 [44]). In this limited sample set we observed that more change was seen in somatic DMRs than in the germ-line DMRs (Additional File 1, Table S5). When evaluated against the tissue specific data (Additional File 3, Figure S2) this was found to be independent from the tissue of origin of the cell line.
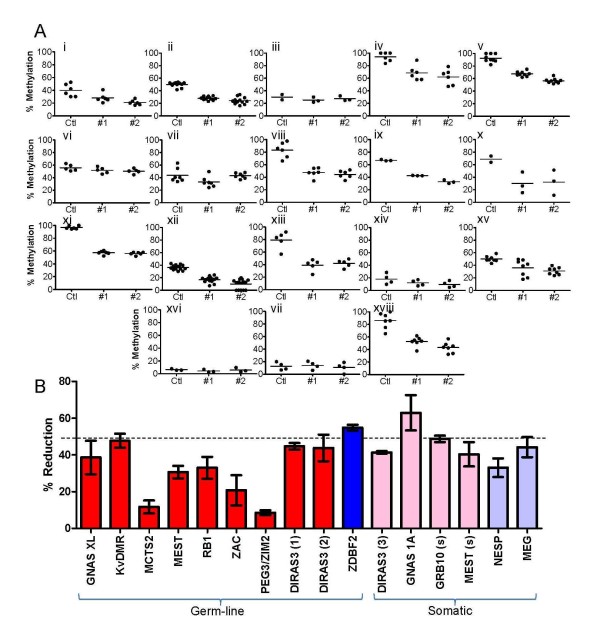
Sum159 had normal methylation at most DMRs, some assays [MEST (g), Retinoblastoma (RB1), DIRAS3 (1), GRB10 (s) and MEG3] reported hypermethylation and hypomethylation was observed at the IGF2 assays (Sum159 contains a rearrangement at the RB1 locus [44]). We treated this cell line with 5 Azacytidine (a demetylating agent) to see how the plastic the methylation is at loci reporting normal methylation and to see how reversible the hypermethylation is. A significant decrease in methylation was observed at all loci, except the IGF2 DMRs (which already were hypomethylated). Not all DMRs lost methylation at the same rate (Figure 3 and Additional File 1, Table S5). Approximately 30%-60% (average 50%) methylation was lost in 13/18 of the loci (Figure 3a). ZAC, ZIM2/PEG3 and MCTS2 showed more a modest 10% - 25% reduction in methylation.
Figure 3.
Methylation changes at differentially methylated regions (DMRs) after treatment with 5 Azacytidine. (A) Methylation levels of each loci in Sum159 (control) and after treatment with 5 Azacytidine. #1 = 1 nM treatment, #2 = 3 nM treatment. Each data point represents an individual CpG: (i) GNAS XL; (ii) KvDMR; (iii) MCTS2; (iv) MEST (g); (v) RB1; (vi) ZIM2/PEG3; (vii) ZAC; (viii) DIRAS3 (1); (ix) DIRAS3 (2); (x) ZDBF2; (xi) DIRAS3 (3); (xii) GNAS 1A; (xiii) GRB10 (s); (xiv) MEST (s); (xv) NESP55; (xvi) IGF2 (0); (xvii) IGF2 (2); (xviii) MEG3. (b) Percent reduction of methylation at each DMR analysed. Red = maternal germ-line DMRs; pink = maternal somatic DMRs; dark blue = paternal germ-line DMRs; light blue = paternal somatic DMRs. Error bars represent the range of difference between the two experiments.
Discussion
Differential DNA methylation has a particular importance in establishing and maintaining mono-allelic imprinted gene expression. Many DMRs are located within defined gene regulatory elements - such as promoters and chromatin insulators. Where the DMR is located within a regulatory element, it is assumed that parent-of-origin specific methylation regulates the binding of transcription factors in an allele specific manner (reviewed in [5]). Most imprinted genes have developmental and tissue restricted expression patterns but, remarkably, many DMRs maintain allele specific methylation in adult somatic tissues independent of absolute levels of expression. In addition, the H19 DMR can function as an ectopic somatic DMR when inserted into a non-imprinted locus. Therefore, the methylation levels and stability of the ectopic DMR is not determined by the parent-of-origin specific marks in the germ-line [45]. When studying cancer, accurate analysis of DNA methylation and the understanding of normal levels are imperative for assessing whether changes in imprinting in complex heterogeneous tissues are physiological or pathophysiological.
We have developed high-throughput, sensitive, quantitative PSQ assays for DNA methylation at all characterized human imprinted loci and have produced the first comprehensive analysis of tissue specific methylation at human imprinted loci. Validation of the PSQ assays proved that the assays were both quantitative and reproducible. The small amount of DNA required and the multiplexing of the assays by using the biotin tag [46,47] means the assays are both efficient and economic for screening large numbers of clinical samples for a variety of different imprinted loci. Recent improvements in ~PSQ technology allows up to 15 different CpG dinucleotides within 80 bp of sequence to be sequentially and accurately assayed for subtle changes in methylation. Therefore, we have meticulously placed the assays within the most suitable part of the DMR and have avoided effects that may be introduced at methylation 'shores' [48]. However, due to the variable CpG content observed within the DMRs it may have been necessary to place an assay in a region with a similar CpG density to these shores.
We analysed known and potential DMR sequences associated with imprinted loci in eight different human adult tissues. We also observed that the methylation levels of most DMRs are maintained in adult somatic tissues and confirmed that germ-line DMRs are particularly stable with no tissue specific gain or loss of methylation. Paternally methylated somatic DMRs are also stable, reporting similar levels across all tissues. Maternally methylated somatic DMRs are more plastic with tissue specific differences observed in adult tissues. This could suggest a difference in the mechanism or developmental stage during which the two types of somatic imprint are established.
We also investigated how methylation may vary within different individuals when the same tissue is studied. For this 23 different DMRs (representative of both maternally and paternally transmitted, germ-line and somatic) in 50 different individual blood samples were analysed. Human samples have varied genetic backgrounds, unlike mice that are inbred onto a limited range of genetic backgrounds. Therefore, it was not surprising that we found locus specific effects in most DMRs, where one or more individuals fell outside of the inter-quartile ranges with maximum or a minimum values bordering on the set thresholds of differential methylation. However, no single individual in our population had disruption of DNA methylation at multiple DMRs, thus excluding an environmental or genetic predisposition to imprinted methylation defects in this sample set.
We noticed that, for paternally methylated DMRs, the methylation at individual CpGs is variable within the DMR. In contrast, maternal DMRs have similar levels of methylation for all CpGs across the regions assayed. This reflects the different make-up of maternal and paternal DMRs, where maternal DMRs are CpG islands, while paternal DMRs do not usually have a CpG density that constitutes the threshold for an island [26]. Our analysis of the effect that CpG density had upon average methylation levels in a DMR showed no specific trends and suggests that, provided enough CpGs are included in an assay, the CpG density at the DMR will not confound the assay.
As our collection of DMRs had a variety of genomic features, we also examined whether normal baseline methylation levels were influenced by CTCF binding sites or genomic position. No significant differences in the average methylation across all tissues were observed between DMRs that contained ubiquitous CTCF binding sites compared to DMRs that contained tissue specific CTCF sites or no CTCF binding in normal tissue. It will be interesting to see whether CTCF affects the ability of DMRs to maintain their unmethylated state in cancer or if changes in CTCF binding lead to changes in gene expression via methylation changes.
Many cell lines have aberrant methylation and, in this study, we found it to be the case for a selection of imprinted genes in cancer cell lines. As methylation is potentially reversible, we examined how effective our assays were in determining quantitative methylation changes after treatment with the demethylating agent, 5 azacytidine. As expected, all methylated DMRs lost methylation after treatment with a demethylation agent but we found that not all loci lost methylation with equal efficiency. While it is uncertain what this means in the context of a single cell line, these results suggest that additional chromatin factors influence the interaction with the maintenance methyltransferases in a locus specific manner.
Conclusions
Our data provide the first comprehensive catalogue of methylation at imprinted human loci and the level of the variability in methylation in normal samples. In this role, the covalent bond of the methyl group to DNA is important because it is stable and, therefore, the DNA can be easily extracted and investigated in clinical samples. These assays will be valuable in future studies of imprinted regions in disease and in understanding gene regulation at imprinted regions.
Materials and methods
DNA samples
Eight different tissue DNAs (from three pooled individuals; brain, breast, colon, heart, kidney, liver, placenta or testis (containing sertoli cells and sperm)) and 50 different blood DNAs were purchased from Amsbio (Abingdon, UK).
Assay protocol
We designed the assays so they could be easily multiplexed. First, all primers were designed to have melting temperatures of 56°C and to produce a robust polymerase chain reaction (PCR product. We confirmed that the primers were not overlapping annotated single nucleotide polymorphisms (SNPs). Secondly, we opted for two rounds of PCR: the first with gene specific primers (Additional File 1, Table S1), and the second with a common biotinylated primer. The common biotinylated primer both reduced the cost of assay optimization and enabled high throughput second round amplification.
Human genomic DNA was bisulphite converted using the EZ Bisulphite conversion kit (Zymo, CA, USA) following the manufacturer's instructions and eluting in 10 ng/μL (assuming 100% conversion and recovery). DNA was then amplified in a two step amplification using the primers in Additional File 1, Table S1. PCR first round reactions used Thermostart ABGene MasterMix (Fisher, Loughborough, UK), 0.25 μM forward primer, 0.25 μM reverse primer and 10 ng of converted DNA. The assays were amplified in a standard PCR reaction with a melting temperature of 56°C for 25 cycles. This first round PCR was diluted 1:6 and used as a template for a second round PCRs using the same conditions but with 40 cycles and a common biotinylated reverse primer 5' B-CGCCAGGGTTTTCCCAGTCACGAC 3' [46,47]. 10 μL of final PCR product was bound to Streptavidin-sepharose beads (GE Healthcare, Buckinghamshire, UK) and prepared using the Pyromark vacuum tool and buffers (Qiagen, Düsseldorf, Germany) following the manufacturer's protocol. Each sample was run on the Pyromark Q96 machine (Qiagen) using the PyroMark Gold Q96 SQA Reagents (Qiagen). Pyrograms were analysed using the PyroQ CpG software. Graphs were prepared in Graphpad Prism 5.
Assay validation
In order to validate the assay hypermethylated and hypomethylated DNA (human methylated and non-methylated DNA kit - Zymo Research) was mixed in the following ratios prior to bisulphite treatment as above. DNA was mixed as follows:(1) 100% methylated DNA (M-DNA); (2) 3 M-DNA:1 unmethylated DNA (U-DNA); (3) 2:M-DNA:2:U-DNA; (4) 1M-DNA:3 U-DNA; and (5) 100% U-DNA. This was used as input as described above.
In order to validate the amount of input DNA required, 100 ng, 50 ng, 25 ng and 10 ng of template DNA was used as an input into the Zymo conversion (as above) in final concentrations of 10 ng/μL, 5 ng/μL, 2.5 ng/μL and 1 ng/μL. In order to validate the reproducibility of the assay, duplicate aliquots of 500 ng of template DNA were converted and used in the assays as described above.
Investigating methylation levels in tissue, blood and cell line samples
1 μg of DNA (tissue samples) or 400 ng (blood) was bisulphite converted using the EZ Bisuphite conversion kit (Zymo) following the manufacturer's instructions and eluting in 10 ng/mL (assuming 100% conversion and recovery). In order to assess DNA levels in different cell lines, DNA was prepared from cells using a DNeasy kit (Qiagen): 500 ng of eluted DNA was bisulphite converted and analysed as described above.
In order to investigate the effect treatment of 5-azacytidine had on the methylation of imprinted regions, Sum159 cells were plated at 40% confluence and treated with 1 nM or 3 nM of 5-azacytidine for 24 h. Cells were then washed three times with phosphate buffered saline and incubated in normal media for a further 24 h. The cells were harvested and DNA extracted with a DNeasy kit (Qiagen). DNA was bisulphite converted and assayed as described above.
Data analysis
For correlations with genomic features the following data analysis was used.
CTCF binding data
Liver specific CTCF binding data was obtained from Mike Wilson (Unpublished data, Duncan Odom Laboratory, Cambridge Research Institute, Cambridge, UK). Briefly, chromatin from prepared from human Liver was chromatin immunoprecipitated with a CTCF antibody and sequenced by Solexa sequencing. CTCF binding sites and DMRs were aligned using Galaxy [49].
Tissue wide CTCF binding data was obtained from UCSC Encode tracks for four different released cell lines (GM12878-lymphoblastoid, K562-leukaemia, NHEK-ectoderm, HUVEC-mesoderm, [42,43]).
Statistical analysis
Statistical analysis was performed in Graphpad Prism as indicated in the text. Student T-tests were used to determine whether there were statistically significant differences between: (1) Tissue specific methylation levels and the average methylation level reported by all somatic tissues; (2) for individual methylation levels in peripheral blood DNA samples and the average methylation level of all 50 blood samples; (3) for cell line methylation levels compared to average blood methylation levels; and (4 to compare methylation levels of untreated 5-azacytidine treated cell lines matched pair T tests were performed. Graphpad Prism was used for all comparison.
Abbreviations
DMR: differentially methylated region; PCR: polymerase chain reaction; PSQ: pyrosequencing; SNP: single-nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KW and AM conceived and designed the study and wrote the manuscript. KW and JH performed the experiments and analysed the data. All authors have read and approved the final manuscript.
Supplementary Material
Supplementary Tables. Table S1: Regions assayed and primer sequences for each pyrosequencing assay. (Top) Actual primers designed to bisulphate treated DNA. (Bottom) Corresponding genomic DNA sequence of primers for location within genome.
Table S2: Methylation levels of each region assayed in eight adult tissues. Shaded cells show assays that report levels consistent with a differentially methylated region (DMR). Blue/bold text indicate novel DMRs.
Table S3: Characteristics of the differentially methylated region (DMR) assays. Amplicon co-ordinates and genomic locations are shown. CTCF binding sites and C-phosphate guanine (CpG) density are also calculated.
Table S4: Twenty-three differentially methylated regions were analysed in 50 different blood samples.
Table S5: Changes of methylation in cell lines and after 5-azacytidine treatment.
Figure S2: Quality control on methylation assays. (a) Prior to bisulphite treatment, unmethylated and methylated DNA were mixed together in the ratios described. (i) ZAC differentially methylated region (DMR); (ii) GRB10 germ-line DMR; iii: GNAS germ-line DMR; (iv) MCTS2 DMR; (v) KvDMR; (vi) SNRPN DMR. (b) Reproducibility of experiments. The same DNA was independently bisulphite converted and the pyrosquencing assay run. Individual C-phosphate guanines (CpGs) for replicate 1 were plotted against replicate 2. The r2 of the correlation was 0.86 and the gradient of the trend-line 0.99. When this is plotted per DMR, r2 is 0.96 and x = 1.05. Limits of agreement calculated by a Bland-Altman correlation show a difference of 1.60 between the two replicates. This is not significant.
Figure S2: Methylation levels at eight different adult tissues for 50 regions assayed (in chromosome order). Bn = brain; Bt = breast; Co = colon; He = heart; Ki = kidney; Li = liver; Pl = placenta; Te = testis; Bl = blood. Each data point represents an individual C-phosphate guanine (CpG). Bars represent the mean methylation level.
Figure S3: Comparison of region characteristics with methylation levels. (a) Effect of CTCF binding on methylation levels. (i) Average methylation levels of differentially methylated region (DMR) assays in all tissues (CTCF binding determined from Encode Data on UCSC database). (ii) Methylation levels of DMR assays in liver. (b) Correlation between C-phosphate guanine (CpG) densities of each assay with average somatic methylation level reported. Closed circles = germ-line DMRs; open circles = somatic DMRs. (c) Effect of genomic position of DMR on methylation levels.
Figure S4: Methylation levels reported by individual C-phosphate guanines (CpGs) in 50 different individuals at 23 different differentially methylated regions (DMRs. Red = maternal germ-line DMRs; pink = maternal somatic DMRs; dark blue = paternal germ-line DMRs; light blue = paternal somatic DMRs. Each data point represents an individual CpG. Bars represent the mean methylation levels. A = DIRAS3 (1); B = DIRAS3 (2); C = DIRAS3 (3); D = ZDBF2; E = ZAC; F = MEST (g); G = MEST (s); H = GRB10 (g); I = GRB10 (s); J = H19; K = IGF2-0; L = IGF2-2; M = KvDMR; N = RB1; O = DLK; P = IG-DMR; Q = MEG; R = SNRPN; S = PEG3; T = MCTS2; U = NESP; V = GNAS XL; W = GNAS 1A.
Figure S5: Methylation levels in eight different cell lines and the average blood methylation levels. Cell lines to the left of the dashed line are normal, whereas cell lines to the right of the line are derived from cancerous samples. Each data point represents an individual C-phosphate guanine (CpG). (a) Maternal germ-line differentially methylated regions (DMRs). (b) Paternal germ-line DMRs. C = maternal somatic DMRs; D = maternal somatic DMRs.
Contributor Information
Kathryn Woodfine, Email: Kathryn.woodfine@cancer.org.uk.
Joanna E Huddleston, Email: joanna.huddleston@cancer.org.uk.
Adele Murrell, Email: amm207@cam.ac.uk.
Acknowledgements
We would like to thank: Yoko Ito for technical advice and for providing data on H19 DMR in cell line; Sarah Vowler for her advice on statistical analysis; and Claire Fielding for maintenance of the pyrosequencer. We are also grateful to Mike Wilson, Dominic Schmidt and Duncan Odom for sharing their human liver specific CTCF binding data with us. This work was supported by: a Cancer Research UK Senior Cancer Research Fellowship; an Association for International Cancer Research Project grant [06-0589]; and the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited.
References
- Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25(3):129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Warnecke PM, Bestor TH. Cytosine methylation and human cancer. Curr Opin Oncol. 2000;12(1):68–73. doi: 10.1097/00001622-200001000-00012. [DOI] [PubMed] [Google Scholar]
- Novik KL, Nimmrich I, Genc B, Maier S, Piepenbrock C, Olek A, Beck S. Epigenomics: genome-wide study of methylation phenomena. Curr Issues Mol Biol. 2002;4(4):111–128. [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19(3):281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2(11):e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneimprint. http://www.geneimprint.com
- Murrell A. Genomic imprinting and cancer: from primordial germ cells to somatic cells. ScientificWorldJournal. 2006;6:1888–1910. doi: 10.1100/tsw.2006.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel B, Metsuyanim S, Schmidt-Ott KM, Fridman E, Jacob-Hirsch J, Simon A, Pinthus J, Mor Y, Barasch J, Amariglio N. et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66(12):6040–6049. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D, Grundy R, Ferguson-Smith AC, Maher ER. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms' tumour. Br J Cancer. 2005;92(8):1574–1580. doi: 10.1038/sj.bjc.6602478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chano T, Minami K, Okabe H, Okada Y, Okamoto K. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum Mol Genet. 2006;15(6):821–830. doi: 10.1093/hmg/ddl001. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H. et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105(34):12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanber D, Berulava T, Ammerpohl O, Mitter D, Richter J, Siebert R, Horsthemke B, Lohmann D, Buiting K. The human retinoblastoma gene is imprinted. PLoS Genet. 2009;5(12):e1000790. doi: 10.1371/journal.pgen.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299(5613):1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S. et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17(17):2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003;17(5):586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303(5655):238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2003;33(1):66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- Murrell A, Ito Y, Verde G, Huddleston J, Woodfine K, Silengo MC, Spreafico F, Perotti D, De Crescenzo A, Sparago A. et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PLoS One. 2008;3(3):e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103(28):10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters J-M, Murrell A. Cohesin Is required for higher-order chromatin conformation at the imprinted IGF2-H19l locus. PLoS Genet. 2009;5(11):e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A. et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15(4):547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP. et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet. 2008;40(8):949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Origins of extreme sexual dimorphism in genomic imprinting. Cytogenet Genome Res. 2006;113(1-4):36–40. doi: 10.1159/000090813. [DOI] [PubMed] [Google Scholar]
- Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19(8):1374–1383. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejeux E, El abdalaoui H, Gut IG, Tost J. Identification and quantification of differentially methylated loci by the pyrosequencing technology. Methods Mol Biol. 2009;507:189–205. doi: 10.1007/978-1-59745-522-0_15. full_text. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134(22):3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- Murrell A, Rakyan VK, Beck S. From genome to epigenome. Hum Mol Genet. 2005;14(Spec No 1):R3–R10. doi: 10.1093/hmg/ddi110. [DOI] [PubMed] [Google Scholar]
- Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2010;6(1) doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Mann JR. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9(15):1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. pp. 682–685. [DOI] [PMC free article] [PubMed]
- Cancer Research UK. Epigenetics and imprinting. http://www.cambridgecancer.org.uk/research/loc/cambridge/ccri/murrella/?view=CRI&source=research
- Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A. 2008;146A(16):2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- Evans HK, Wylie AA, Murphy SK, Jirtle RL. The neuronatin gene resides in a 'micro-imprinted' domain on human chromosome 20q11.2. Genomics. 2001;77(1-2):99–104. doi: 10.1006/geno.2001.6612. [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA. 2006;103(17):6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Mungall AJ, Matthews L, Ryder E, Gray DJ, Pask AJ, Shaw G, Graves JA, Rogers J, Dunham I. et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008;6(6):e135. doi: 10.1371/journal.pbio.0060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JD, Underkoffler LA, Wood AJ, Collins JN, Williams PT, Golden JA, Schuster EF Jr, Loomes KM, Oakey RJ. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol. 2005;25(13):5514–5522. doi: 10.1128/MCB.25.13.5514-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- University of Tokyo Laboratory for Systems Biology and Medicine. Expression data base. http://www.lsbm.org/site_e/index.html [PubMed]
- McMinn J, Wei M, Sadovsky Y, Thaker HM, Tycko B. Imprinting of PEG1/MEST isoform 2 in human placenta. Placenta. 2006;27(2-3):119–126. doi: 10.1016/j.placenta.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM. et al. Unlocking the secrets of the genome. Nature. 2009;459(7249):927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SKY Karyotypes and FISH analysis of Epithelial Cancer Cell Lines. http://www.path.cam.ac.uk/~pawefish/
- Gebert C, Kunkel D, Grinberg A, Pfeifer K. H19 imprinting control region methylation requires an imprinted environment only in the male germ line. Mol Cell Biol. pp. 1108–1115. [DOI] [PMC free article] [PubMed]
- Guo DC, Milewicz DM. Universal primer applications for pyrosequencing. Methods Mol Biol. 2007;373:57–62. doi: 10.1385/1-59745-377-3:57. [DOI] [PubMed] [Google Scholar]
- Royo JL, Hidalgo M, Ruiz A. Pyrosequencing protocol using a universal biotinylated primer for mutation detection and SNP genotyping. Nat Protoc. 2007;2(7):1734–1739. doi: 10.1038/nprot.2007.244. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;Chapter 10 doi: 10.1002/0471250953.bi1005s19. Unit 10 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables. Table S1: Regions assayed and primer sequences for each pyrosequencing assay. (Top) Actual primers designed to bisulphate treated DNA. (Bottom) Corresponding genomic DNA sequence of primers for location within genome.
Table S2: Methylation levels of each region assayed in eight adult tissues. Shaded cells show assays that report levels consistent with a differentially methylated region (DMR). Blue/bold text indicate novel DMRs.
Table S3: Characteristics of the differentially methylated region (DMR) assays. Amplicon co-ordinates and genomic locations are shown. CTCF binding sites and C-phosphate guanine (CpG) density are also calculated.
Table S4: Twenty-three differentially methylated regions were analysed in 50 different blood samples.
Table S5: Changes of methylation in cell lines and after 5-azacytidine treatment.
Figure S2: Quality control on methylation assays. (a) Prior to bisulphite treatment, unmethylated and methylated DNA were mixed together in the ratios described. (i) ZAC differentially methylated region (DMR); (ii) GRB10 germ-line DMR; iii: GNAS germ-line DMR; (iv) MCTS2 DMR; (v) KvDMR; (vi) SNRPN DMR. (b) Reproducibility of experiments. The same DNA was independently bisulphite converted and the pyrosquencing assay run. Individual C-phosphate guanines (CpGs) for replicate 1 were plotted against replicate 2. The r2 of the correlation was 0.86 and the gradient of the trend-line 0.99. When this is plotted per DMR, r2 is 0.96 and x = 1.05. Limits of agreement calculated by a Bland-Altman correlation show a difference of 1.60 between the two replicates. This is not significant.
Figure S2: Methylation levels at eight different adult tissues for 50 regions assayed (in chromosome order). Bn = brain; Bt = breast; Co = colon; He = heart; Ki = kidney; Li = liver; Pl = placenta; Te = testis; Bl = blood. Each data point represents an individual C-phosphate guanine (CpG). Bars represent the mean methylation level.
Figure S3: Comparison of region characteristics with methylation levels. (a) Effect of CTCF binding on methylation levels. (i) Average methylation levels of differentially methylated region (DMR) assays in all tissues (CTCF binding determined from Encode Data on UCSC database). (ii) Methylation levels of DMR assays in liver. (b) Correlation between C-phosphate guanine (CpG) densities of each assay with average somatic methylation level reported. Closed circles = germ-line DMRs; open circles = somatic DMRs. (c) Effect of genomic position of DMR on methylation levels.
Figure S4: Methylation levels reported by individual C-phosphate guanines (CpGs) in 50 different individuals at 23 different differentially methylated regions (DMRs. Red = maternal germ-line DMRs; pink = maternal somatic DMRs; dark blue = paternal germ-line DMRs; light blue = paternal somatic DMRs. Each data point represents an individual CpG. Bars represent the mean methylation levels. A = DIRAS3 (1); B = DIRAS3 (2); C = DIRAS3 (3); D = ZDBF2; E = ZAC; F = MEST (g); G = MEST (s); H = GRB10 (g); I = GRB10 (s); J = H19; K = IGF2-0; L = IGF2-2; M = KvDMR; N = RB1; O = DLK; P = IG-DMR; Q = MEG; R = SNRPN; S = PEG3; T = MCTS2; U = NESP; V = GNAS XL; W = GNAS 1A.
Figure S5: Methylation levels in eight different cell lines and the average blood methylation levels. Cell lines to the left of the dashed line are normal, whereas cell lines to the right of the line are derived from cancerous samples. Each data point represents an individual C-phosphate guanine (CpG). (a) Maternal germ-line differentially methylated regions (DMRs). (b) Paternal germ-line DMRs. C = maternal somatic DMRs; D = maternal somatic DMRs.