Abstract
Natural killer (NK) cells are regulated by interactions between polymorphic killer immunoglobulin-like receptors (KIR) and human leukocyte antigens (HLA). Genotypic combinations of KIR3DS1/L1 and HLA Bw4-80I were previously shown to influence HIV-1 disease progression, however other KIR genes have not been well studied. In this study, we analyzed the influence of all activating and inhibitory KIR, in association with the known HLA inhibitory KIR ligands, on markers of disease progression in a West African population of therapy-naïve HIV-1 infected subjects. We observed a significant association between carriage of a group B KIR haplotype and lower CD4+ T cell counts, with an additional effect for KIR3DS1 within the frame of this haplotype. In contrast, we found that individuals carrying genes for the inhibitory KIR ligands HLA-Bw4 as well as HLA-C1 showed significantly higher CD4+ T cell counts. These associations were independent from the viral load and from individual HIV-1 protective HLA alleles. Our data suggest that group B KIR haplotypes and lack of specific inhibitory KIR ligand genes, genotypes considered to favor NK cell activation, are predictive of HIV-1 disease progression.
Introduction
Natural killer (NK) cells play an important role in the innate immune response against viruses and tumors, and in the regulation of the subsequent adaptive immune responses [1]. Their activity is controlled by an integration of signals from many inhibitory and activating receptors, including the killer immunoglobulin-like receptors (KIR) [2], [3]. KIR contain two or three external immunoglobulin-like domains (2D, 3D) with either long (L) or short (S) cytoplasmic tails corresponding to their function as inhibitory or activating receptors, respectively. Several inhibitory KIR have well-defined human leukocyte antigen (HLA) class I ligands. Mutually exclusive groups of HLA-C molecules with asparagine or lysine at position 80, termed C1 and C2, ligate inhibitory KIR2DL1, KIR2DL2, and KIR2DL3 [4]. A group of HLA-B molecules expressing the serologically defined Bw4 epitope recognize inhibitory KIR3DL1, with those with an isoleucine at position 80 (Bw4-80I) showing stronger inhibition than those with a threonine at this position (Bw4-80T) [5]. Both KIR and HLA loci show extreme population diversity and rapid evolution, suggesting that they are under pathogen-mediated selection and that they influence disease outcome at the individual level [6]. Indeed, several recent epidemiological studies have associated KIR/HLA compound genotypes with diseases as diverse as infection, autoimmune and inflammatory conditions, cancer, and reproductive failure [7].
HIV-1 infected patients show a large variation in disease courses [8]. A number of recent studies provide evidence that KIR and HLA loci play an important role in this. Flores-Villanueva et al. first found that HIV-1 patients with homozygous Bw4 showed delayed progression to AIDS [9]. Martin et al. confirmed this but indicated that the association was derived completely from an epistatic interaction of Bw4-80I with KIR3DS1, suggesting a model in which NK cells activated through KIR3DS1 confer protection from HIV-1 disease [10]. However, subsequent studies could not confirm the KIR3DS1/Bw4-80I association [11], [12], and no study to date has been able to prove Bw4-80I as a true ligand for KIR3DS1 [13]–[16]. The interpretation of the KIR3DS1/Bw4-80I interaction was further complicated by a recent study of the same patient cohorts showing high expression alleles of inhibitory KIR3DL1 in combination with Bw4-80I to also slow down disease progression [17]. Thus, it remains unclear how exactly KIR/HLA interactions influence HIV-1 disease outcome, and how NK cells are involved in this [18].
Few studies have analyzed KIR genes other than KIR3DS1 and KIR3DL1 in the context of HIV-1 disease. Up to 14 different functional KIR genes have been identified which, as a result of strong interlocus linkage disequilibrium, segregate in two broad haplotypes termed group A and group B [19], [20]. Consequently, association analyses with single KIR genes likely depend on the KIR haplotype in which they occur. In this study, we analyzed the influence of all activating and inhibitory KIR, KIR haplotypes, and known HLA class I inhibitory KIR ligands, on markers of disease progression in a population of West African HIV-1 infected subjects.
Materials and Methods
Study subjects
Eighty one HIV-1 infected female sex workers attending a confidential clinic in Abidjan, Côte d'Ivoire between January 1997 and May 2000 were studied cross-sectionally. A subset of 20 HIV-1 infected female sex workers enrolled for follow-up and paid between 2 and 4 visits to the clinic spanning a period of up to 18 months. All subjects were therapy-naïve at the time of enrolment and during follow-up.
Ethics Statement
The study was approved by ethical committee of the Ministry of Health, Côte d'Ivoire, the ethical committee of the Institute of Tropical Medicine, Antwerp, Belgium, and by the Institutional Review Board of the Centers for Disease Control and Prevention, Atlanta, GA. All subjects gave written informed consent prior to enrolment.
Laboratory methods
Whole blood was drawn in EDTA tubes (Becton Dickinson). Plasma was tested for HIV infection by ELISA and Western blot. CD4+ T cell counts were determined in whole blood using a FACScan flow cytometer (Becton Dickinson). HIV-1 viral load was quantified in plasma by the Amplicor HIV-1 Monitor assay, version 1.5 (Roche). Peripheral blood mononuclear cells were separated from whole blood by gradient centrifugation and stored in liquid nitrogen.
KIR and HLA class I genotyping
Genomic DNA was extracted from peripheral blood mononuclear cells using a QIAamp DNA blood mini kit (Qiagen). KIR typing was performed by PCR with sequence specific primers like previously reported [21]. KIR haplotypes were assigned by using the current working definition available at the website of the European Bioinformatics Institute (http://www.ebi.ac.uk/ipd/kir). Group B haplotypes are characterized by one or more of the following genes: KIR2DL2, KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5 and KIR3DS1. Group A haplotypes are characterized by the absence of all of these genes, and contain one or more of the following genes: KIR2DL1, KIR2DL3, KIR3DL1, and KIR2DS4. KIR genotypes consist of two KIR haplotypes. AA genotypes are identified by the absence of all group B haplotype genes. AB and BB genotypes are characterized by the presence of at least one group B haplotype gene. AB and BB genotypes cannot easily be distinguished and are collectively annotated as Bx. HLA-B and HLA-C typing was performed by PCR with sequence specific oligonucleotides (Gen-Probe) on a Luminex platform, which gives a DNA-based typing result at the level of 2 digits and permits to distinguish Bw4-80I and Bw4-80T from Bw6, and C1 from C2. High resolution typing of HLA-B*58 positive samples was performed by sequencing-based typing using the AlleleSEQR HLA-B sequencing kit (Atria Genetics). DNA sequences were detected on an automated fluorescent DNA sequencer. HLA-B alleles were assigned up to six digits with the help of ASSIGN 3.5 software.
Statistical methods
The subjects' first CD4+ T cell count and HIV-1 plasma viral load at the clinic were used in the cross-sectional analyses. The effect of single KIR and HLA genes and KIR/HLA combinations was assessed by linear regression analysis using log transformed CD4+ T cell counts and viral load levels as the dependent variables. Model selection was guided by the Akaike information criterion (AIC), which assesses the fit between the data and the model with a penalty for the number of parameters (i.e. favoring the more parsimonious model). Longitudinal changes in the CD4+ T cell count were examined by mixed-effects linear regression analysis with log transformed CD4+ T cell counts as the dependent variable. Statistical analyses were performed with R version 2.11.1 [22].
Results
Study population
HIV-1 infected female sex workers included in the study had a median age of 26 years (interquartile range (IQR), 21–32) and they reported a median duration of commercial sex work of 24 months (IQR, 12–48). At the time of enrollment, the women showed a median CD4+ T cell count of 523 cells/µl (IQR, 363–775) and a median HIV-1 plasma viral load of 4.8 log10 RNA copies/ml (IQR, 4.0–5.4). All subjects were therapy-naïve at the time of enrolment and during follow-up.
Group B KIR haplotype genes are associated with lower CD4+ T cell counts
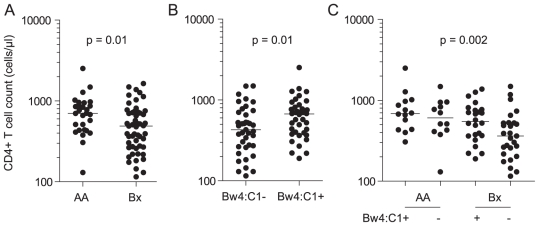
First we investigated whether the extensive variability in KIR gene content was associated with the CD4+ T cell count or plasma viral load of the HIV-1 infected subjects (Table 1a). The presence of KIR2DL2, KIR2DL5, KIR2DS3, and KIR3DS1 was associated with significantly lower CD4+ T cell counts. These genes are all specific to the group B KIR haplotype and, accordingly, subjects with a KIR Bx genotype (i.e., those carrying one or two group B haplotypes) showed a significantly lower CD4+ T cell count. No such associations were found for the viral load, although trends were seen for a number of group B KIR haplotype genes mirroring the effects on the CD4+ T cell count. To investigate whether individual group B haplotype genes showed effects on the CD4+ T cell count beyond that of the Bx genotype, we analyzed them together by multivariate linear regression (Table 2a). Only the addition of KIR3DS1 resulted in a better fit of the model relative to that with Bx alone (AIC of 156 vs. 158). These data show that HIV-1 infected subjects carrying a group B KIR haplotype have lower CD4+ T cell counts (Fig. 1A), and even more so if they also have KIR3DS1 within the frame of this haplotype.
Table 1. Univariate effects of KIR and HLA genes on the CD4+ T cell count and HIV-1 viral load level of 81 HIV-1 infected subjects.
| Frequency n (%) | CD4+ T cell count | HIV-1 viral load | |||||
| Fold difference (CI) | AIC | p | Fold difference (CI) | AIC | p | ||
| a. KIR genes and genotypes | |||||||
| 2DL1 | 77 (95) | 1.46 (0.75–2.82) | 163 | 0.260 | 4.27 (0.60–30.4) | 202 | 0.144 |
| 2DL2 | 48 (59) | 0.70 (0.53–0.93) | 158 | 0.015 | 1.36 (0.56–3.30) | 204 | 0.485 |
| 2DL3 | 63 (78) | 1.07 (0.76–1.51) | 164 | 0.702 | 1.13 (0.40–3.20) | 204 | 0.811 |
| 2DL5 | 45 (56) | 0.74 (0.56–0.98) | 160 | 0.039 | 1.99 (0.84–4.71) | 202 | 0.114 |
| 3DL1 | 80 (99) | 0.54 (0.15–1.98) | 163 | 0.345 | 10.0 (0.21–480) | 203 | 0.239 |
| 2DS1 | 9 (11) | 1.00 (0.63–1.59) | 164 | 0.990 | 0.91 (0.23–3.59) | 204 | 0.892 |
| 2DS2 | 42 (52) | 0.78 (0.58–1.03) | 161 | 0.077 | 0.98 (0.41–2.33) | 204 | 0.961 |
| 2DS3 | 25 (31) | 0.72 (0.53–0.97) | 160 | 0.031 | 1.59 (0.63–4.03) | 203 | 0.323 |
| 2DS4 | 79 (98) | 1.14 (0.45–2.87) | 164 | 0.784 | 9.75 (0.64–149) | 201 | 0.101 |
| 2DS5 | 24 (30) | 0.81 (0.59–1.10) | 162 | 0.171 | 1.64 (0.64–4.19) | 203 | 0.300 |
| 3DS1 | 5 (6.2) | 0.51 (0.29–0.92) | 159 | 0.025 | 1.96 (0.33–11.7) | 204 | 0.454 |
| AA | 28 (35) | 1.47 (1.10–1.96) | 158 | 0.010 | 0.61 (0.25–1.53) | 203 | 0.290 |
| Bx | 53 (65) | 0.68 (0.51–0.91) | 158 | 0.010 | 1.63 (0.65–4.04) | 203 | 0.290 |
| b. KIR ligand genes | |||||||
| Bw4 | 63 (78) | 1.24 (0.88–1.74) | 163 | 0.222 | 0.90 (0.32–2.53) | 204 | 0.835 |
| Bw6 | 56 (69) | 1.07 (0.78–1.46) | 164 | 0.671 | 1.01 (0.40–2.58) | 204 | 0.981 |
| Bw4-80I | 58 (72) | 1.19 (0.87–1.64) | 163 | 0.274 | 1.00 (0.38–2.60) | 204 | 1.000 |
| Bw4-80T | 7 (8.6) | 1.08 (0.65–1.81) | 164 | 0.761 | 0.69 (0.15–3.20) | 204 | 0.632 |
| C1a | 52 (71) | 1.31 (0.94–1.83) | 146 | 0.105 | 0.98 (0.35–2.76) | 188 | 0.972 |
| C2 | 66 (81) | 0.97 (0.67–1.41) | 164 | 0.880 | 0.87 (0.29–2.63) | 204 | 0.798 |
| c. HLA alleles | |||||||
| B*57 | 3 (3.7) | 1.63 (0.77–3.47) | 163 | 0.200 | 0.28 (0.03–2.73) | 203 | 0.272 |
| B*58:01 | 10 (13) | 0.99 (0.64–1.54) | 163 | 0.959 | 1.29 (0.33–5.06) | 201 | 0.711 |
Data are calculated by linear regression analysis using log transformed CD4+ T cell counts and HIV-1 viral load levels as the dependent variables. CI, 95% confidence interval; AIC, Akaike information criterion for goodness of fit; p, statistical significance.
data available for 73 of 81 subjects. p values<0.05 are in bold type.
Table 2. Multivariate effects of KIR and HLA genes on the CD4+ T cell count of 81 HIV-1 infected subjects.
| Term 1 | Term 2 | Term 3 | Model | |||||
| Fold difference (CI) | p | Fold difference (CI) | p | Fold difference (CI) | P | AIC | p | |
| a. KIR combinations | ||||||||
| Bx+2DL2 | 0.75 (0.41–1.38) | 0.358 | 0.89 (0.50–1.61) | 0.703 | 159 | 0.035 | ||
| Bx+2DL5 | 0.68 (0.41–1.12) | 0.127 | 1.00 (0.62–1.62) | 0.983 | 160 | 0.037 | ||
| Bx+2DS2 | 0.65 (0.42–1.02) | 0.061 | 1.05 (0.69–1.61) | 0.813 | 159 | 0.036 | ||
| Bx+2DS3 | 0.74 (0.53–1.04) | 0.078 | 0.83 (0.59–1.17) | 0.286 | 158 | 0.021 | ||
| Bx+2DS5 | 0.69 (0.50–0.96) | 0.030 | 0.97 (0.68–1.36) | 0.840 | 159 | 0.037 | ||
| Bx+3DS1 | 0.72 (0.54–0.96) | 0.025 | 0.58 (0.34–1.03) | 0.063 | 156 | 0.007 | ||
| b. HLA combinations a | ||||||||
| Bw4+C1 | 1.48 (1.00–2.19) | 0.049 | 1.46 (1.04–2.05) | 0.030 | 144 | 0.038 | ||
| Bw4+Bw4:C1 | 1.02 (0.66–1.56) | 0.945 | 1.46 (1.04–2.05) | 0.030 | 144 | 0.038 | ||
| C1+Bw4:C1 | 0.98 (0.64–1.52) | 0.945 | 1.48 (1.00–2.19) | 0.049 | 144 | 0.038 | ||
| Bw4:C1 | 1.47 (1.10–1.97) | 0.010 | 142 | 0.010 | ||||
| c. KIR/HLA interactions a | ||||||||
| 2DL2+C1+2DL2:C1 | 0.76 (0.43–1.36) | 0.349 | 1.35 (0.78–2.32) | 0.277 | 0.92 (0.46–1.82) | 0.797 | 145 | 0.056 |
| 2DL3+C1+2DL3:C1 | 1.12 (0.55–2.29) | 0.756 | 1.26 (0.60–2.66) | 0.535 | 1.06 (0.46–2.44) | 0.891 | 150 | 0.343 |
| 2DS2+C1+2DS2:C1 | 0.59 (0.34–1.04) | 0.071 | 1.04 (0.62–1.73) | 0.880 | 1.41 (0.73–2.74) | 0.305 | 146 | 0.076 |
| 2DS3+C1+2DS3:C1 | 0.53 (0.31–0.92) | 0.024 | 0.96 (0.62–1.50) | 0.854 | 1.76 (0.89–3.46) | 0.102 | 145 | 0.049 |
| d. KIR/HLA combinations a | ||||||||
| Bx+Bw4:C1+Bx:Bw4:C1 | 0.63 (0.41–0.98) | 0.041 | 1.29 (0.79–2.10) | 0.297 | 1.18 (0.65–2.15) | 0.585 | 140 | 0.005 |
| Bx+Bw4:C1 | 0.69 (0.51–0.93) | 0.015 | 1.44 (1.09–1.91) | 0.012 | 138 | 0.002 | ||
| Bx+3DS1+Bw4:C1 | 0.73 (0.54–0.97) | 0.033 | 0.53 (0.29–0.98) | 0.044 | 1.39 (1.05–1.84) | 0.021 | 136 | <0.001 |
Data are calculated by multivariate linear regression analysis using log transformed CD4+ T cell counts as the dependent variable. The estimated effects of the respective terms in the models are shown. Interaction terms, denoting the simultaneous occurrence of two genes, are annotated with “:”.
Data available for 73 of 81 subjects. CI, 95% confidence interval; AIC, Akaike information criterion for goodness of fit; p, statistical significance.
Figure 1. Effect of KIR and HLA genotypes on the CD4+ T cell count of HIV-1 infected subjects.
(A) Effect of AA versus Bx genotype (n = 81). (B) Combined effect of inhibitory KIR ligand genes Bw4 and C1 (n = 73). Bw4:C1 denotes the simultaneous occurrence of Bw4 and C1. (C) Combined effect of Bx and Bw4:C1 genotypes (n = 73). P values represent the statistical significance of the linear regression models using log-transformed CD4+ T cell counts as the dependent variable. Horizontal lines represent median values.
Inhibitory KIR ligand genes HLA-Bw4 and HLA-C1 are associated with higher CD4+ T cell counts
Next, we investigated whether inhibitory KIR ligand genes were associated with the CD4+ T cell count or plasma viral load of the HIV-1 infected subjects. Subjects with Bw4 or C1 showed a trend towards higher CD4+ T cell counts, with no effects on the viral load (Table 1b). Interestingly, the effects of Bw4 and C1 on the CD4+ T cell count became statistically significant when combined in a multivariate model (Table 2b). Addition of a Bw4:C1 interaction term, denoting the simultaneous occurrence of Bw4 and C1, abrogated the individual effects of Bw4 and C1, indicating that they were derived from subjects harboring both genes. On its own, the Bw4:C1 interaction term explained the variation in CD4+ T cell counts best. These data show that HIV-1 infected subjects carrying inhibitory KIR ligand genes Bw4 as well as C1 have higher CD4+ T cell counts than those lacking either Bw4 or C1 (Fig. 1B). Two member alleles of Bw4, B*57 and B*58:01, are known to display individual HIV-1 protective effects in African populations [23]. In our population, B*57 showed slightly higher CD4+ T cell counts and slightly lower viral load levels while no such effects were seen for B*58:01 (Table 1c). Adjustment for both B*57 and B*58:01 did little to abrogate the observed effects of Bw4 (1.21-fold; 95% CI, 0.85–1.74; p = 0.285) or Bw4:C1 (1.47-fold; 95% CI, 1.06–2.03; p = 0.045), suggesting that Bw4 acts at least in part independently from B*57 and B*58:01.
KIR and HLA genes have both synergistic and independent effects on the CD4+ T cell count
The protective effects of C1 and Bw4 on the CD4+ T cell counts could result from its known interactions with KIR. To test this, we investigated whether their effects could be abrogated by addition of KIR:HLA interaction terms to the multivariate linear regression models (Table 2c). Functional studies have shown that C1 binds to inhibitory KIR2DL2 and KIR2DL3, while Bw4 binds to inhibitory KIR3DL1 [4], [5], [24]. C1 and Bw4 have also been suggested to ligate activating KIR2DS2/KIR2DS3 and KIR3DS1, respectively, but this could not be confirmed to date [13]–[16], [25]. We could not detect statistical interaction of C1 with KIR2DL2 or KIR2DL3: the interaction terms did not abrogate the C1 effects. However, trends towards statistical interaction were observed for C1 with KIR2DS2 and KIR2DS3: in both cases the interaction terms showed protective effects at the cost of C1. These models indicate that the negative effects of KIR2DS2 and KIR2DS3 are dampened in the presence of C1 (KIR2DS2: 0.59-fold in the absence versus 0.83-fold in the presence of C1; KIR2DS3: 0.53-fold in the absence versus 0.93-fold in the presence of C1), and that this capacity explains most of the protective effect of C1. Because of the low numbers of subjects with KIR3DS1 or without KIR3DL1 in our study population, we were not able to calculate interactions of Bw4 with KIR3DL1 or KIR3DS1.
Next, we investigated whether the best-fitting KIR and HLA combinations were independent of each other by analyzing them together in multivariate linear regression models (Table 2d). Overall, the effects of Bx and Bw4:C1 genotypes were found to be independent, with no signs of interaction. Consequently, a better fit of the data can be expected for combinations of these genotypes. Indeed, the combination of Bx and Bw4:C1 showed a major increase in fit relative to that of Bw4:C1 alone, and even more so if KIR3DS1 was added to the model (AIC values of 142, 138 and 136, respectively). The additive effects of Bx and Bw4:C1 are shown in Fig. 1C, with HIV-1 patients possessing a KIR Bx genotype in the absence of Bw4:C1 showing the lowest CD4+ T cell counts.
Carriage of a Bx genotype in absence of Bw4:C1 is associated with faster CD4+ T cell decline
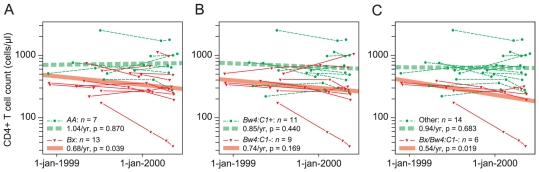
The low CD4+ T cell counts associated with a Bx genotype in absence of Bw4:C1 could result from a higher rate of CD4+ T cell decline during acute and/or chronic infection, or simply from a longer total duration of infection, a distinction that cannot be made by cross-sectional analyses. The total duration of infection is not available for this population of HIV-1 seroprevalent female sex workers, but it can be estimated by the total duration of commercial sex work which constitutes the main risk factor for HIV-1 infection in this population. No differences in duration of commercial sex work were observed between subjects with or without Bx (median values of 25 vs 22 months, p = 0.392) or Bw4:C1 (median value of 24 months for both groups, p = 0.588) and neither did adjustment for the duration of commercial sex work affect the associations between Bx or Bw4:C1 and the CD4+ T cell count (Bx: 0.68-fold; 95% CI, 0.51–0.91; p = 0.010; Bw4:C1: 1.46-fold; 95% CI, 1.08–1.97; p = 0.013). Next, we analyzed longitudinal changes in the CD4+ T cell count for a subset of 20 HIV-1 infected female sex workers with available follow up by mixed-effects linear regression analysis (Fig. 2). Subjects with a Bx genotype showed a statistically significant decline in CD4+ T cell count; this was not found for subjects with an AA genotype. Lack of Bw4:C1 did not result in a statistically significant decline in CD4+ T cell count on its own, but when combined with the presence of a Bx genotype, it provided the strongest effect. Together, these analyses suggest that the observed associations between Bx and Bw4:C1 and the CD4+ T cell count do not result from differences in the time since infection but from differences in the rate of CD4+ T cell decline during follow-up.
Figure 2. Effect of KIR and HLA genotypes on the rate of CD4+ T cell decline among HIV-1 infected subjects.
Thin lines represent individual CD4+ T cell count profiles (n = 20). Thick lines represent the fitted models calculated by mixed-effects linear regression analysis. (A) Effects of AA and Bx genotypes. (B) Effect of Bw4:C1 genotype. (C) Effect of Bx genotype in the absence of Bw4:C1. Bw4:C1 denotes the simultaneous occurrence of Bw4 and C1. The estimated fold decrease in CD4+ T cell count per year and p values are shown.
Discussion
Genotypic combinations of KIR3DS1/L1 and HLA Bw4-80I were previously shown to influence HIV-1 disease progression, however other KIR genes have not been well studied. Therefore, we analyzed all activating and inhibitory KIR, in association with the known inhibitory KIR ligands, in a population of West African HIV-1 infected subjects.
We first found that HIV-1 infected subjects with a group B KIR haplotype showed markedly lower CD4 counts and faster CD4 count decline than those without. Significant differences were observed for the individual group B haplotype genes KIR2DL2, KIR2DL5, KIR2DS3 and KIR3DS1, with KIR3DS1 showing an additional effect to that of the group B haplotype. These data confirm previous findings from Gaudieri et al., who found HIV-1 disease promoting effects for several group B haplotype genes including KIR3DS1 [11], and from Martin et al. who noted faster disease progression among carriers of KIR3DS1 in the absence of HLA Bw4-80I [10].
Next, we found that HIV-1 infected subjects carrying genes for both Bw4 and C1 showed significantly higher CD4+ T cell counts. This is in agreement with several previous reports showing associations between Bw4 or Bw4-80I and protection from HIV-1 disease progression [9], [10], [26], [27]. No previous study observed a protective effect for C1. Recent studies identified a −35C single nucleotide polymorphism near HLA-C that was associated with HIV-1 control and increased expression of HLA-C [28], [29], however this variant was not associated with either group of C1 or C2 alleles, nor could its effects be explained by one or more individual HLA-C alleles [29], [30]. Similarly, in our study, C1 did not preferentially contain HLA-C alleles in known linkage with −35C (data not shown). Remarkably, in our study, the protective effects of Bw4 and C1 appeared to be completely derived from subjects harboring both genes. This could reflect the known linkage disequilibrium between several HLA-B and HLA-C alleles and their observed joint effects on HIV-1 control [31]–[33].
Interestingly, we noted a trend towards statistical interaction of C1 with KIR2DS2 and KIR2DS3, which explained most of the C1 effect and resulted in a weakening of the negative effects of KIR2DS2 and KIR2DS3 in the presence of C1. No statistical interaction could be detected between C1 and inhibitory KIR2DL2 or KIR2DL3, despite in vitro evidence of C1 binding to KIR2DL2/3 but not KIR2DS2/3 [4], [24], [25]. It is plausible, however, that the interaction of C1 with KIR2DL2/3 appeared neutral because only 2 subjects in our study population lacked both KIR2DL2 and KIR2DL3, and thus that the presence of C1 was the only limiting factor in establishing the effect. Hence, we speculate that the weakening of the negative effects of KIR2DS2 and KIR2DS3 by C1 is mediated by its interaction with KIR2DL2/3. This conclusion concurs with previous reports showing a link between KIR2DS1 and KIR2DS2 and risk for developing psoriatic arthritis, particularly in the absence of the HLA ligands for their homologous inhibitory receptors, KIR2DL1 and KIR2DL2/3 [34], [35]. Previously, Martin et al observed a similar statistical interaction between Bw4-80I and KIR3DS1 influencing HIV-1 disease progression, however, in contrast to the tempering effects noted in the present study, the Bw4-80I/KIR3DS1 interaction term completely inversed the negative effects of KIR3DS1 now resulting in significant protection from disease progression [10]. Unfortunately, subsequent studies could not confirm this epistatic Bw4-80I/KIR3DS1 interaction [11], [12], and in the present study we did not have sufficient numbers of subjects with KIR3DS1 to replicate these calculations.
Group B KIR haplotypes are considered to be more activating than group A haplotypes due to the higher numbers of activating KIR genes that they contain [3], [6]. Indeed, subjects with a KIR AB genotype were shown to display higher NK cell responsiveness than those with an AA genotype [36]. On the other hand, HLA Bw4 and C1/2 ligands for inhibitory KIR provide the necessary inhibitory signals that set the NK cell activation threshold and guarantee self-tolerance [3], [6]. Consequently, our data suggest that KIR and HLA genotypes that favor NK cell activation, like carriage of a group B KIR haplotype and/or lack of a Bw4 or C1 inhibitory KIR ligand, play a detrimental role in the progression of HIV-1 disease. This conclusion is at variance with previous reports by Martin et al, in which a protective role for activated NK cells in HIV-1 disease progression was proposed [10], [17]. In these studies, KIR3DS1/Bw4-80I genotypes were suggested to directly activate NK cells [10], while strongly inhibitory KIR3DL1/Bw4-80I genotypes were hypothesized to display greater NK cell activation capacity due to better NK cell licensing [17]. Indeed, subsequent in vitro studies were able to show stronger NK-cell mediated virus inhibition, functional capacity, and specific NK cell subset expansion during acute HIV-1 infection for these genotypes [37]–[39]. However, other studies found that Bw4 strongly inhibited proliferation and cytotoxicity of KIR3DL1-expressing NK cells without any influence on KIR3DS1 [16], and that possession of KIR3DS1 alone was sufficient to induce strong NK cell effector functions [40]. In fact, we believe that our interpretation of a detrimental role for activating KIR/HLA genotypes makes more sense in the light of emerging data on innate immune activation during acute HIV-1 infection in establishing HIV-1 pathogenesis. For instance, acute SIV infection of non-progressing sooty mangabeys is characterized by substantially reduced levels of innate immune activation, including lower IFN-α production, IFN-regulated gene expression, and NK cell proliferation, compared to that seen in AIDS progressing rhesus macaques [41], [42]. Like rhesus macaques, HIV-1 infected humans show significant innate immune activation and expansion of NK cells during the acute phase of the infection [43], [44]. In agreement with this, a recent study found only low levels of antiviral NK cell activity together with a low frequency of KIR3DS1 in a group of rare HIV-1 controller patients [45]. Although the possible pathogenic mechanisms of carrying an activating KIR/HLA genotype remain unclear, studies have shown direct NK cell-mediated killing of CD4+ T cells that express NKp44L, a ligand for NKp44 that is induced upon exposure to a HIV-1 gp41, to correlate with HIV-1 disease progression [46], [47]. Alternatively, CD4+ and CD8+ T cells are known to express KIR and could be involved as well.
Interestingly, we found that KIR/HLA genotypes correlated best with the CD4+ T cell count and not, or at least much more weakly, with the viral load of the study subjects. This is in agreement with a previous KIR study noting better correlations with disease outcomes involving the CD4+ T cell count than the viral load [11]. In part, this could reflect the lower within-patient variation for the CD4+ T cell count, and the fact that the CD4+ T cell count shows a better clinical prognostic value than the viral load, especially during the chronic phase of the infection (the disease stage most of our study subjects are expected to be in) [48]. However, preferential correlations with the CD4+ T cell count could also reflect the specific immune responses that are involved, with activating KIR/HLA genotypes that drive cytotoxic responses being expected to directly impact on target cell numbers. In that respect, our data are in agreement with studies of SIV-infected primate species showing a strong link between rapid control of innate immune activation and lack of CD4 count depletion or disease progression, however without any influence on the often high levels of viral replication seen in these animals [41], [42]. In this model, we could hypothesize a role for viral load at the beginning of the causal pathway, with viral peptides modulating the interactions of activating and inhibitory KIR with their ligands, within the context of the inherited KIR/HLA genotype [49], [50].
Previously, we found that carriage of an AB KIR genotype and of certain inhibitory KIR combinations in the absence of their Bw4 or C1 ligands was associated with resistance to HIV-1 infection in the same population of African female sex workers [51]. These findings corroborated previous data of increased NK cell-mediated cytotoxicity in HIV-1 resistant drug users [52], and have since then been confirmed by several other studies showing phenotypic and/or genotypic activating KIR profiles in HIV-exposed seronegative populations [53]–[55]. Interestingly, the genotypes observed in our previous study are nearly the same as the ones identified here, despite differences in study design and data analysis (e.g., all AB genotypes in our previous study are actually Bx according to current guidelines, and combinations of KIR2DL2 and KIR2DL3 coincide with the presence of a AA or Bx genotype). Together, this means that in our population of African female sex workers, similar activating KIR/HLA genotypes are associated with resistance to HIV infection as well as with faster progression of the disease. This is paradoxical, yet could be logically explained by an intriguing model in which an activated NK cell profile protects the host against HIV-1 acquisition through rapid and potent elimination of HIV-1 infected target cells, but once systemically infected, promotes HIV-1 disease progression through over-activation of the innate and adaptive arms of the immune system.
The observed associations between KIR/HLA genotype and CD4+ T cell count in this study were detected by a cross-sectional analysis of therapy-naïve HIV-1 infected female sex workers. This approach is less sensitive than a longitudinal analysis of time to AIDS and it could be biased by the unknown duration of infection of the included subjects. To address this, we first used the total duration of commercial sex work, the main risk factor for HIV-1 infection in this population, as an estimate of the time since infection, and found that adjustment had no impact on the observed associations. Secondly, we calculated the longitudinal effects of the KIR/HLA genotypes on the rate of CD4+ T cell decline in a subgroup of female sex workers with available follow-up. This analysis confirmed the cross-sectional associations, however it was rather limited in number of included subjects as well as in number of follow-up samples. Thus, confirmation of our findings in larger longitudinal cohorts of untreated HIV-infected subjects remains warranted. Furthermore, future functional studies should investigate whether activating KIR profiles are expressed on NK cells, CD8+ T cells, or both, and to what extent the activating KIR/HLA genotypes proposed in this study translate into increased in vivo immune activation and enhanced in vitro target cell cytotoxicity.
In summary, we found that group B KIR haplotypes and lack of specific inhibitory KIR ligand genes, genotypes considered to favor NK cell activation, are associated with CD4+ T cell loss during HIV-1 infection. Better understanding of how genetic variation at KIR and HLA loci influences HIV-1 pathogenesis may lead to the development of immune intervention strategies aiming at controlling progression of the disease.
Acknowledgments
We thank the community of female sex workers in Abidjan for their cooperation, Odette Tossou for help with data analysis, Katrien Fransen for viral load testing, Annette Brouwer de Koning for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Belgian Fund for Scientific Research (FWO-Vlaanderen), grant number G.0660.06. Website: www.fwo.be. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 4.Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 7.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collaborative Group on AIDS Incubation and HIV Survival including the CASCADE EU Concerted Action. Time from HIV-1 seroconversion to AIDS and death before widespread use of highly-active antiretroviral therapy: a collaborative re-analysis. Lancet. 2000;355:1131–1137. [PubMed] [Google Scholar]
- 9.Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 11.Gaudieri S, DeSantis D, McKinnon E, Moore C, Nolan D, et al. Killer immunoglobulin-like receptors and HLA act both independently and synergistically to modify HIV disease progression. Genes Immun. 2005;6:683–690. doi: 10.1038/sj.gene.6364256. [DOI] [PubMed] [Google Scholar]
- 12.Barbour JD, Sriram U, Caillier SJ, Levy JA, Hecht FM, Oksenberg JR. Synergy or independence? Deciphering the interaction of HLA Class I and NK cell KIR alleles in early HIV-1 disease progression. PLoS Pathog. 2007;3:e43. doi: 10.1371/journal.ppat.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie GM, Bashirova A, Dong T, McVicar DW, Rowland-Jones SL, Carrington M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 15.O'Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 16.Morvan M, Willem C, Gagne K, Kerdudou N, David G, et al. Phenotypic and functional analyses of KIR3DL1+ and KIR3DS1+ NK cell subsets demonstrate differential regulation by Bw4 molecules and induced KIR3DS1 expression on stimulated NK cells. J Immunol. 2009;182:6727–6735. doi: 10.4049/jimmunol.0900212. [DOI] [PubMed] [Google Scholar]
- 17.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward J, Barker E. Role of natural killer cells in HIV pathogenesis. Curr HIV/AIDS Rep. 2008;5:44–50. doi: 10.1007/s11904-008-0008-2. [DOI] [PubMed] [Google Scholar]
- 19.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 20.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Hum Immunol. 2003;64:648–654. doi: 10.1016/s0198-8859(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 21.Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18:2002–2007. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; R: A language and environment for statistical computing. 2008. [Google Scholar]
- 23.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 24.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 25.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. 2010;185:4233–4237. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Vazquez A, Mina-Blanco A, Martinez-Borra J, Njobvu PD, Suarez-Alvarez B, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum Immunol. 2005;66:285–289. doi: 10.1016/j.humimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Qing M, Li T, Han Y, Qiu Z, Jiao Y. Accelerating effect of human leukocyte antigen-Bw6 homozygosity on disease progression in Chinese HIV-1-infected patients. J Acquir Immune Defic Syndr. 2006;41:137–139. doi: 10.1097/01.qai.0000195607.25262.92. [DOI] [PubMed] [Google Scholar]
- 28.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas R, Apps R, Qi Y, Gao X, Male V, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores-Villanueva PO, Hendel H, Caillat-Zucman S, Rappaport J, Burgos-Tiburcio A, et al. Associations of MHC ancestral haplotypes with resistance/susceptibility to AIDS disease development. J Immunol. 2003;170:1925–1929. doi: 10.4049/jimmunol.170.4.1925. [DOI] [PubMed] [Google Scholar]
- 32.Lazaryan A, Lobashevsky E, Mulenga J, Karita E, Allen S, et al. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J Virol. 2006;80:6056–6060. doi: 10.1128/JVI.02119-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, et al. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol. 2010;84:9879–9888. doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin MP, Nelson G, Lee JH, Pellett F, Gao X, et al. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol. 2002;169:2818–2822. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]
- 35.Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol. 2004;173:4273–4276. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- 36.Korbel DS, Norman PJ, Newman KC, Horowitz A, Gendzekhadze K, et al. Killer Ig-like receptor (KIR) genotype predicts the capacity of human KIR-positive CD56dim NK cells to respond to pathogen-associated signals. J Immunol. 2009;182:6426–6434. doi: 10.4049/jimmunol.0804224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alter G, Rihn S, Walter K, Nolting A, Martin M, et al. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol. 2009;83:6798–6805. doi: 10.1128/JVI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, et al. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol. 2010;184:2057–2064. doi: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- 40.Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, et al. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol. 2008;82:4785–4792. doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandl JN, Barry AP, Vanderford TH, Kozyr N, Chavan R, et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med. 2008;14:1077–1087. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- 42.Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alter G, Teigen N, Ahern R, Streeck H, Meier A, et al. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J Infect Dis. 2007;195:1452–1460. doi: 10.1086/513878. [DOI] [PubMed] [Google Scholar]
- 44.Li Q, Smith AJ, Schacker TW, Carlis JV, Duan L, et al. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J Immunol. 2009;183:1975–1982. doi: 10.4049/jimmunol.0803222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Connell KA, Han Y, Williams TM, Siliciano RF, Blankson JN. The Role of Natural Killer Cells in a Cohort of Elite Suppressors: Low Frequency of the Protective KIR3DS1 Allele and Limited Inhibition of HIV-1 Replication in vitro. J Virol. 2009;83:5028–5034. doi: 10.1128/JVI.02551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieillard V, Strominger JL, Debre P. NK cytotoxicity against CD4+ T cells during HIV-1 infection: a gp41 peptide induces the expression of an NKp44 ligand. Proc Natl Acad Sci U S A. 2005;102:10981–10986. doi: 10.1073/pnas.0504315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fausther-Bovendo H, Vieillard V, Sagan S, Bismuth G, Debre P. HIV gp41 engages gC1qR on CD4+ T cells to induce the expression of an NK ligand through the PIP3/H2O2 pathway. PLoS Pathog. 2010;6:e1000975. doi: 10.1371/journal.ppat.1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korenromp EL, Williams BG, Schmid GP, Dye C. Clinical prognostic value of RNA viral load and CD4 cell counts during untreated HIV-1 infection—a quantitative review. PLoS ONE. 2009;4:e5950. doi: 10.1371/journal.pone.0005950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 52.Scott-Algara D, Truong LX, Versmisse P, David A, Luong TT, et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol. 2003;171:5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 53.Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 54.Ballan WM, Vu BA, Long BR, Loo CP, Michaelsson J, et al. Natural killer cells in perinatally HIV-1-infected children exhibit less degranulation compared to HIV-1-exposed uninfected children and their expression of KIR2DL3, NKG2C, and NKp46 correlates with disease severity. J Immunol. 2007;179:3362–3370. doi: 10.4049/jimmunol.179.5.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boulet S, Sharafi S, Simic N, Bruneau J, Routy JP, et al. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS. 2008;22:595–599. doi: 10.1097/QAD.0b013e3282f56b23. [DOI] [PubMed] [Google Scholar]