Abstract
Ciliopathies represent a newly emerging group of human diseases that share a common etiology resulting from dysfunction of the cilium or centrosome. The gene encoding the retinitis pigmentosa 2 protein (RP2) is mutated in X-linked retinitis pigmentosa. RP2 localizes to the ciliary base and this requires the dual acylation of the N-terminus, but the precise mechanism by which RP2 is trafficked to the cilia is unknown. Here we have characterized an interaction between RP2 and Importin β2 (transportin-1), a member of the Importin-β family that regulates nuclear–cytoplasmic shuttling. We demonstrate that Importin β2 is necessary for localization of RP2 to the primary cilium because ablation of Importin β2 by shRNA blocks entry both of endogenous and exogenous RP2 to the cilium. Furthermore, we identify two distinct binding sites of RP2, which interact independently with Importin β2. One binding site is a nuclear localization signal (NLS)-like sequence that is located at the N-terminus of RP2 and the other is an M9-like sequence within the tubulin folding cofactor C (TBCC) domain. Mutation of the NLS-like consensus sequence did not abolish localization of RP2 to cilia, suggesting that the sequence is not essential for RP2 ciliary targeting. Interestingly, we found that several missense mutations that cause human disease fall within the M9-like sequence of RP2 and these mutations block entry of RP2 into the cilium, as well as its interaction with Importin β2. Together, this work further highlights a role of Importin β2 in regulation of the entry of RP2 and other proteins into the ciliary compartment.
Keywords: Cilia, Importin, RP2, Retinitis pigmentosa
Introduction
Cilia are highly evolutionarily conserved organelles. They can mediate many different sensory functions, which are dependent on the cell type on which they are located (Berbari et al., 2009; Silverman and Leroux, 2009). In addition, cilia function in motility at the level of both single-celled and multicellular organisms. Despite this array of functions, the basic structure of cilia appears to be highly conserved irrespective of cell type (Pedersen et al., 2008; Rosenbaum and Witman, 2002). Cilia are nucleated from a mother centriole/basal body and contain a ring of nine microtubule doublets that run along the length of the cilium, known as the ciliary axoneme. Some motile cilia also have a central microtubule pair to facilitate cilia bending. Early studies in the biflagellate Chlamydomonas reinhardtii identified two intraflagellar transport (IFT) protein complexes (Complex A and Complex B), which appear to traffic large protein aggregates or rafts from the basal body to the distal tip of the cilium and back (Rosenbaum and Witman, 2002). It has been demonstrated that these IFT proteins are highly conserved evolutionarily and many are required for cilia formation, whereas others function peripherally to direct specific cargo into the cilia.
Despite the growing understanding of IFT function and cilia assembly, there is much to learn about the regulation of ciliary trafficking. Specifically, the primary cilium is believed to be biochemically distinct from other subcellular compartments with respect to protein and lipid content. Thus the cilia are able to specifically segregate cilia proteins from non-cilia proteins (Rosenbaum and Witman, 2002). This segregation via compartmentalization provides a mechanism that allows for exquisite regulation of cilia-mediated signaling (Pazour and Bloodgood, 2008). How the cilia regulate import and export of proteins is still unclear, but it might involve an electron-dense area at the base of the cilia, which is known as the transition zone. This area might function in a similar fashion to the tight junctions of epithelia, which specifically segregate apical from basolateral plasma membrane domains by preventing the admixture by lateral diffusion of membrane proteins from each domain. The cilia transition zone might also function in a similar manner to the nuclear pore complex (NPC), a multi-protein complex that allows the regulated import and export of specific proteins between the cytosol and nucleus (Rosenbaum and Witman, 2002).
Indeed, a role for a transport mechanism to cilia and centrosomes that appears to use the nuclear transport system involving Ran and Importin proteins has recently been uncovered. One study focused on cilia and pericentrosomal trafficking by a splice variant of the Crumbs3 polarity protein (Fan et al., 2007). Another more recent work focused on Kif17, a motor that is important in IFT (Dishinger et al., 2010). These cilia-targeted proteins appear to use classical nuclear localization signals to mediate their transport. However, several other protein motifs have also been identified that facilitate trafficking of proteins to the cilia (Pazour and Bloodgood, 2008). Most notably, a VxPx motif has been identified in Polycystin-2, CNGB1b and Rhodopsin, and is necessary for localization of these proteins to cilia (Geng et al., 2006; Jenkins et al., 2006; Mazelova et al., 2009). Recent work in the retina identified that this motif might be bound by the small GTPase Arf4 (ADP-ribosylation factor 4). Furthermore, in conjunction with Rab11, FIP3 and ASAP1 (Arf-GAP with SH3 domain, ANK repeat and PH-domain-containing protein 1), Arf 4 appears to regulate the budding of cilia-destined vesicles from the trans-Golgi (Mazelova et al., 2009).
In this paper, we focus on mechanisms that mediate the trafficking of retinitis pigmentosa 2 (RP2, also known as protein XRP2) to the cilia. RP2 is an X-linked gene that, when mutated, gives rise to retinitis pigmentosa (Hardcastle et al., 1999). The RP2 gene encodes a 350 residue polypeptide that appears to be ubiquitously expressed. Previous work has demonstrated that RP2 is membrane-associated by dual acylation of the N-terminus, namely myristoylation at Gly2 and palmitoylation at Cys3 (Chapple et al., 2002). The N-terminal half of RP2 has both sequence and structural similarity to Tubulin cofactor C (TBCC), a protein that is involved in the formation of α-tubulin–β-tubulin heterodimers. Further work has shown that this domain binds and has GAP activity towards the small G-protein Arl3 (Veltel et al., 2008). Most recently RP2 has been demonstrated to traffic to the ciliary base and proposed to have a role in Golgi-to-cilia membrane delivery through its regulation of Arl3 (Evans et al., 2010). To examine the mechanism by which RP2 traffics to cilia, we performed large-scale purification of RP2 and associated proteins from renal epithelia. Using this approach, we identified Importin β2 as a binding partner of RP2 that appears to regulate RP2 trafficking into the cilia. This work adds further support to the recent findings that Importin proteins have a role in cilia trafficking.
Results
RP2 localizes to primary cilium in epithelia
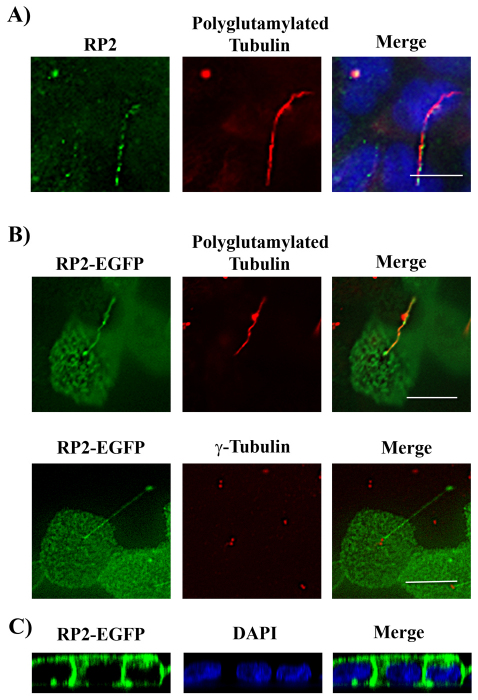
It has been demonstrated that RP2 localizes to ciliary basal bodies in several different cell types (Evans et al., 2010). Our laboratory has studied RP2 localization in MDCK cells. We consistently found both endogenous RP2 (Fig. 1A) and exogenous EGFP-tagged RP2 (Fig. 1B) localized to primary cilia in fully polarized MDCK cells. Confocal microscope images demonstrated that beside its ciliary localization, the exogenously expressed RP2–EGFP also localized to both apical and basal membranes in polarized MDCK cells (Fig. 1C).
Fig. 1.
RP2 localizes to primary cilium in epithelia. (A) Endogenous RP2 localizes to primary cilia in MDCK cells. MDCK cells were grown on Transwell filters for 7 days after confluence. Cells were then fixed and immunostained with rabbit anti-RP2 antibody (green) and mouse anti-polyglutamylated tubulin (red) and nuclei were stained with DAPI (blue). (B) RP2–EGFP localizes to primary cilia. Fully polarized MDCK cells stably expressing RP2–EGFP (green) were stained with polyglutamylated tubulin (red, top panel) or γ-tubulin (red, bottom panel). (C) RP2–EGFP localizes to both apical and basolateral membranes. Fully polarized RP2–EGFP MDCK cells were fixed and stained with DAPI to indicate nuclei. Representative z-projections are shown. Scale bars: 10 μm.
RP2 forms a complex with Importin-β
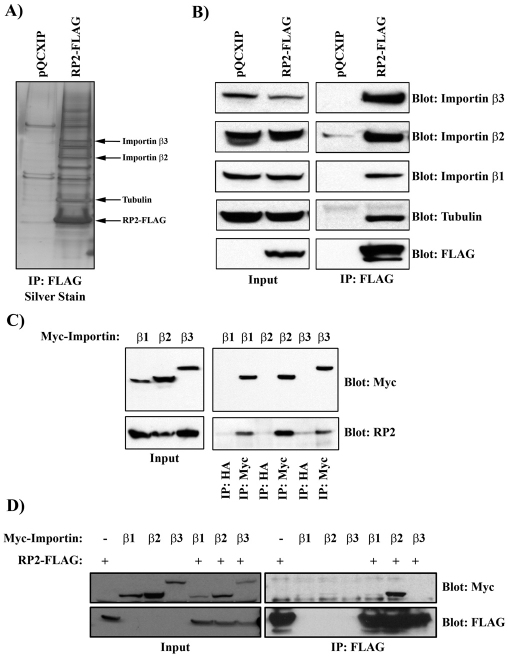
To elucidate the mechanism by which RP2 is trafficked to the cilium, a proteomic approach was adopted to identify new interacting proteins. MDCK cells stably expressing either empty vector (pQCXIP) or FLAG-tagged RP2 were generated (RP2–FLAG). Tandem mass spectrometry (MS) analysis of proteins that co-immunoprecipitated with RP2–FLAG identified several proteins including Importin β2 (Transportin-1), Importin β3 and β-tubulin (Fig. 2A and supplementary material Table S1). These interactions were confirmed by western blotting of RP2–FLAG immunoprecipitates. All three endogenous proteins, Importin β2, Importin β3 and β-tubulin, co-immunoprecipitated with RP2–FLAG (Fig. 2B). We have previously demonstrated that Importin β1 interacts with another cilia protein, Crumbs3–CLP1, so we analyzed whether Importin β1 also interacted with RP2 (Fan et al., 2007). Indeed, endogenous Importin β1 also efficiently co-immunoprecipitated with RP2–FLAG (Fig. 2B). Next, we examined whether these Importin proteins could co-immunoprecipitate endogenous RP2. MDCK cells were generated stably expressing Myc-tagged Importin β1, Importin β2 and Importin β3. Using these cells, we demonstrated that immunoprecipitation of the Importin proteins with an anti-Myc antibody, but not a control anti-HA antibody, could efficiently co-immunoprecipitate endogenous RP2 (Fig. 2C). To further test these interactions, RP2–FLAG and the Myc-tagged Importin proteins were transiently transfected into HEK293 cells and anti-FLAG immunoprecipitation performed. Interestingly, in this cell type, although none of the Importin proteins co-immunoprecipitated in the absence of RP2–FLAG, only Importin β2 co-immunoprecipitated with RP2–FLAG (Fig. 2D), suggesting that it is the major Importin β binding partner of RP2. Furthermore, GST–Importin-β2 was able to precipitate endogenous RP2 from MDCK cell lysates (see below).
Fig. 2.
RP2 forms a complex with Importin proteins. (A) Large-scale protein purification of RP2-interacting proteins. MDCK cells stably expressing either empty vector (pQCXIP) or RP2–FLAG were lysed and immunoprecipitated with immobilized FLAG antibody. Immunoprecipitates were extensively washed and separated by Bis-Tris PAGE. Resulting gels were silver stained and bands of interest excised and submitted for MS/MS analysis. (B) RP2–FLAG was immunoprecipitated from stable MDCK cells and the immunoprecipitates were subjected to Bis-Tris PAGE and blotted with the indicated antibodies. (C) MDCK cells stably expressing Myc-tagged Importin β1, Importin β2 or Importin β3 were immunoprecipitated with either anti-HA (control) or anti-Myc antibody. Immunoprecipitates were subjected to Bis-Tris PAGE and western blotted with the antibodies indicated. (D) HEK293 cells were transiently transfected with RP2–FLAG and Myc-tagged Importin β1, Importin β2 or Importin β3, respectively. Then cells were lysed and immunoprecipitated with anti-FLAG antibody and immunoprecipitates were subjected to Bis-Tris PAGE and blotted with the indicated antibodies.
Importin β2 localizes to centrosome and cilium
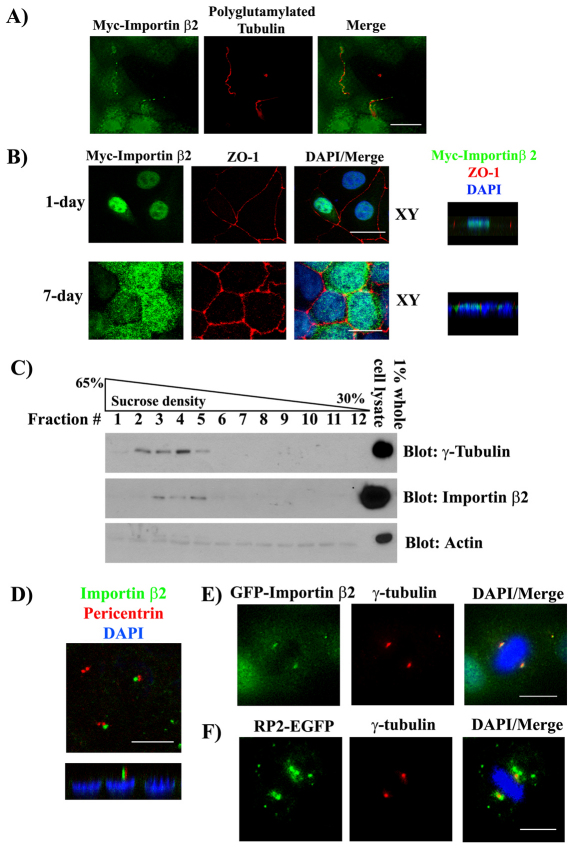
We examined the subcellular localization of Importin β2 in MDCK cells. When MDCK cells stably expressing Myc-tagged Importin β2 were grown on Transwell filters for 7 days after confluence, Myc–Importin-β2 was distributed in a punctate fashion along the length of the ciliary axoneme (Fig. 3A). This was similar to what was observed in Odora cells (Dishinger et al., 2010). Next, we examined the localization of Myc-tagged Importin β2 in subconfluent MDCK cells (1 day) and fully polarized MDCK cells (7 days). In these cells, Myc-tagged Importin β2 appeared to be almost exclusively localized to the nucleus (Fig. 3B, top) at subconfluence. However, as the cells became fully polarized, a redistribution of Myc–Importin-β2 to a subapical region was observed (Fig. 3B, bottom). It is important to note that we used 0.1% Triton X-100 to detect cilia staining of Importin β2, whereas to detect nuclear and cell body staining, we used 0.25% Triton X-100.
Fig. 3.
Importin β2 localizes to primary cilia and centrosomes in epithelia. (A) Importin β2 localizes to cilia. MDCK cells stably expressing Myc-tagged Importin β2 were grown on Transwell filters for 7 days after confluence, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 and stained with anti-Myc antibody (green) and anti-polyglutamylated tubulin antibody (red). (B) Importin β2 translocates from nuclear to subapical regions during polarization in MDCK cells. MDCK cells stably expressing Myc–Importin β2 were seeded onto Transwell filters and grown for the times indicated (1 day or 7 days). Cells were then fixed, permeabilized with 0.25% Triton X-100 and immunostained with antibodies against Myc (green) and ZO-1 (red); nuclei were stained with DAPI (blue). Images were captured by the confocal microscope to assemble X-Y and Z-stack images. (C) Importin β2 co-sediments with purified centrosomes. Centrosomes were isolated from fully polarized MDCK cells and separated on a discontinuous sucrose gradient. Gradients were fractionated and fractions subjected to Bis-Tris PAGE and western blots probed with the indicated antibodies. Lysate (1%) is shown on the right. Actin control in the bottom panel indicates centrosomal fractions have minimal contamination by the cytoskeleton. (D) Importin β2 localizes near centrosomes in confluent MDCK cells. MDCK cells were grown for 7 days after confluence on Transwell filters. Cells were then fixed with acetone and stained with antibodies against pericentrin (red) and Importin β2 (green). Nuclei were visualized with DAPI (blue). Shown are X-Y and Z projections, respectively. (E) GFP–Importin-β2 localizes to spindle poles. MDCK cells stably expressing GFP–Importin-β2 (green) were seeded at low confluence on coverslips. Cells were then fixed with acetone and centrosomes stained with anti-γ-tubulin (red); nuclei are stained with DAPI (blue). (F) RP2–EGFP localizes to a pericentrosomal region during cell division. MDCK cells stably expressing RP2–EGFP (green) were treated as in E. Scale bars: 10 μm.
We have previously shown that Importin β1 and the polarity protein Crumbs3–CLP1 interact and colocalize at centrosomes in mitotic cells and in primary cilium in fully polarized MDCK cells (Fan et al., 2007). Accordingly, we next examined whether Importin β2 is also associated with centrosomal fractions. We purified centrosomes from confluent MDCK cells and examined whether Importin β2 would co-sediment with the centrosome marker γ-tubulin using a discontinuous sucrose gradient. Indeed, in the high density γ-tubulin-positive fractions, enrichment of endogenous Importin β2 was observed (Fig. 3C). We used actin as a control to show that this high-density γ-tubulin fraction was not contaminated by cytoskeletal components. In addition, when we looked at the localization of endogenous Importin β2 in confluent MDCK cells, it appeared that a subfraction localized to centrosomes (Fig. 3D). We next examined the localization of RP2 and Importin β2 in mitotic cells. A fraction of exogenously expressed EGFP–Importin-β2 was enriched at the spindle poles of mitotic cells, as shown by colocalization with the centrosome marker γ-tubulin (Fig. 3E). In addition, EGFP-tagged RP2 was localized to a pericentrosomal region (Fig. 3F), similarly to our previously reported data for Crumbs3–CLP1 and Importin β1 (Fan et al., 2007).
RP2 interacts with the C-terminus of Importin β2
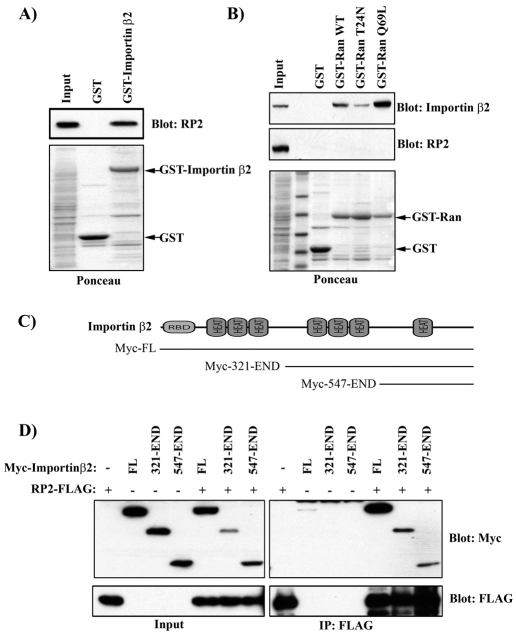
GST fused full-length Importin β2 was able to pull down endogenous RP2 (Fig. 4A). We examined which region of Importin β2 was involved in the interaction with RP2. Importin β proteins share a similar domain architecture with an N-terminal Ran-binding domain (RBD) followed by HEAT and Armadillo repeats (Fig. 4C). Because the RBD mediates binding of the GTP-bound form of the small GTPase Ran to Importin β proteins, we next tested whether constitutively active Ran (Ran Q69L) could precipitate a complex of Importin β2 and RP2. Importin β2 was efficiently precipitated by WT and Q69L Ran, but no RP2 was present in the complex (Fig. 4B). This is consistent with the ability of Ran-GTP to dissociate cargo from Importin β. To examine which region of Importin β2 is necessary for the interaction with RP2, a series of Importin β2 N-terminal truncations were generated (Fig. 4C). As previously demonstrated, full-length (FL) Myc–Importin-β2 was co-immunoprecipitated by RP2–FLAG (Fig. 4D). However, deletion of either the first 320 residues of Importin β2, encompassing the RBD and first set of HEAT repeats, or the first 546 residues did not abolish this interaction (Fig. 4D). These data suggest that the C-terminal domain of Importin β2 is sufficient to bind RP2, as has been shown with other cargoes (Stewart, 2007).
Fig. 4.
RP2 interacts with the C-terminus of Importin β2. (A) GST–Importin β2 precipitates endogenous RP2. MDCK lysates were subjected to GST pull-down with either GST alone or GST–Importin-β2 and subjected to Bis-Tris PAGE. Western blots were probed with indicated antibodies. (B) Ran does not precipitate RP2. MDCK lysates were subjected to GST pull-down with GST alone, wild-type (WT) Ran, dominant-negative (T24N) Ran or constitutively active (Q69L) Ran. Precipitates were analyzed as in A. (C) Schematic diagram showing domain organization of Importin β2 (RBD, Ran-binding domain; HEAT HEAT repeat) and N-terminal truncations. (D) The C-terminus of Importin β2 is sufficient to interact with RP2. HEK293 cells were transiently transfected with RP2–FLAG and either Myc-tagged Importin β2 full-length (FL), Importin β2 lacking the N-terminal 320 residues (321-END) or the N-terminal 546 residues (547-END). Cells were lysed and subjected to FLAG immunoprecipitation. Immunoprecipitates were subjected to Bis-Tris PAGE and blots probed with the antibodies indicated.
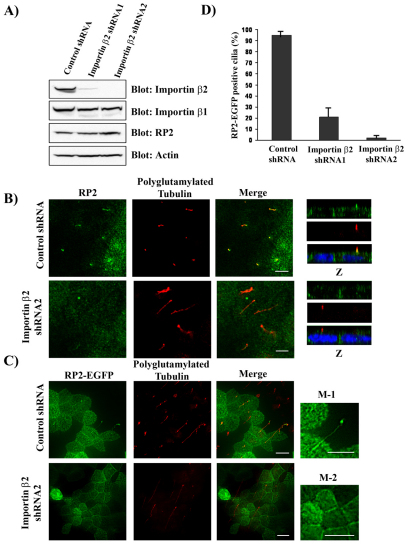
Knockdown of Importin β2 blocks RP2 ciliary targeting
Next, we wanted to test the roles of Importin β2 in targeting of RP2 to the cilium. We used two shRNA targeting sequences to knock down Importin β2 and test its role in the ciliary targeting of endogenous RP2 and RP2–EGFP in MDCK cells. Both of the shRNAs sufficiently reduced Importin β2 without affecting expression of Importin β1 and RP2 (Fig. 5A). Cells without Importin β2 grew normally, but RP2 did not travel efficiently to the cilia when Importin β2 expression levels were reduced (Fig. 5B). We next generated stably expressing RP2–EGFP cells by co-infecting RP2–EGFP into control shRNA and Importin β2 shRNA cells. After selection with G418, expression levels of Importin β2 were still suppressed, whereas expression of RP2–EGFP was similar in control and Importin β2 shRNA cells (supplementary material Fig. S1A,B). RP2–EGFP ciliary targeting was markedly reduced in cells in which Importin β2 was knocked down (Fig. 5C). This lack of cilia targeting in knockdown cells was not related to cilia length or RP2–EGFP expression (supplementary material Fig. S1C). These results are quantified in Fig. 5D, indicating significant impairment of RP2 ciliary targeting after knockdown of Importin β2.
Fig. 5.
Loss of Importin β2 inhibits RP2 ciliary trafficking. (A) Stable knockdown of endogenous Importin β2. MDCK cells were infected with retrovirus directing expression of two different canine Importin β2 shRNAs or a control shRNA (luciferase shRNA) and stable pools were selected. Lysates were subjected to Bis-Tris PAGE and blotted with the antibodies indicated. (B) Knockdown of Importin β2 significantly reduces ciliary localization of endogenous RP2. Control shRNA (top panel) and Importin β2 shRNA (bottom panel) infected MDCK cells were grown for 7 days after confluence on Transwell filters, fixed and stained with antibodies against RP2 (green) and polyglutamylated tubulin (red). (C) Ablation of Importin β2 blocks ciliary localization of exogenous RP2-EGFP. MDCK cells infected with control shRNA (upper panel) and Importin β2 shRNA-2 (bottom panel) were stably transfected with RP2–EGFP (green) and treated as in B. M-1 represents high magnification of RP2-EGFP in control shRNA cells and M-2 represents RP2–EGFP in Importin β2 shRNA cells. (D) Quantification of results from experiments depicted in C. 100 ciliated RP2–EGFP cells each from control shRNA, Importin β2 shRNA1 and shRNA2 were evaluated. Results represent the mean of three individual experiments, shown as mean ± s.d. Scale bars: 10 μm.
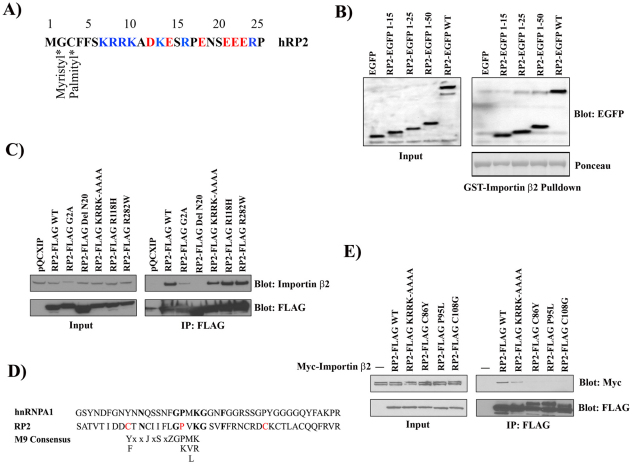
Binding sites on RP2 for Importin β2
Importin complexes regulate the trafficking of proteins between the cytosol and the nucleus. Proteins destined for the nucleus contain a nuclear localization signal (NLS) consisting of either a single cluster (monopartite) or two clusters (bipartite) of basic amino acid residues. Examination of the sequence of RP2 revealed a highly conserved basic cluster at the N-terminus resembling the consensus K-R/K-x-K/R monopartite NLS (Fig. 6A). Following this putative NLS is a string of acidic residues that have previously been shown to improve the efficacy of other NLS sequences (Fig. 6A). This type of NLS sequence can bind Importin-α proteins, but also can bind directly to Importin-β proteins (Jkel and Grlich, 1998). To test whether this sequence mediates the interaction between RP2 and Importin β2, a series of expression constructs were generated driving expression of the N-terminus of RP2 fused to EGFP. GST–Importin-β2 pull down from HEK293 lysates transfected with these constructs revealed that the first 15 residues of RP2 are sufficient to mediate the interaction with Importin β2 (Fig. 6B). In addition, the RP2–EGFP 1–15 fusion protein is efficiently trafficked to the primary cilium in MDCK cells (supplementary material Fig. S2A).
Fig. 6.
Two distinct binding sites of RP2 interact independently with Importin β2. (A) The N-terminus of RP2 contains a nuclear localization signal (NLS)-like sequence. Blue, basic residues; red, acidic residues. (B) The first 15 residues of RP2 can interact with Importin β2. HEK293 cells were transiently transfected with EGFP alone, full-length RP2–EGFP or residues 1–15, 1–25 or 1–50 of RP2 fused to EGFP. Cell lysates were subjected to precipitation with GST–Importin-β2. Precipitates were subjected to Bis-Tris PAGE and blots were probed with indicated antibodies. (C) Wild-type and mutant FLAG-tagged RP2 constructs were transfected into MDCK cells. Lysates were than immunoprecipitated with anti-FLAG antibodies and lysates (input) or FLAG immunoprecipitates were resolved on Bis-Tris gels. After transfer to nitrocellulose, the blots were probed with antibodies against Importin β2 (ImpB2) and FLAG. (D) RP2 contains an M9-like sequence. Alignment of a sequence from RP2 and the classic M9 sequence found in hnRNPA1 and the consensus M9 sequence that can mediate binding of Importin β2. The alignment and terminology is from Bogerd and colleagues (Bogerd et al., 1999) with J representing a hydrophilic amino acid and Z, a hydrophobic amino acid. (E) Point mutations of the M9 consensus (C86Y, P95L or C108G) abolish the interaction between Importin β2 and RP2. FLAG-tagged RP2 with various mutations were transfected into HEK293 cells with Myc-tagged Importin β2. After immunoprecipitation with anti-FLAG antibody, proteins were resolved on a Bis-Tris gel and transferred to nitrocellulose. Blotting was then performed with anti-FLAG or anti-Myc antibodies.
It has been previously demonstrated that targeting of RP2 to the plasma membrane requires myristoylation and palmitoylation of Gly2 and Cys3, respectively (Chapple et al., 2002). Furthermore, lipid modification is also required for targeting of RP2 to the basal body (Evans et al., 2010). Thus we examined whether these modifications are also necessary for the interaction with Importin β2. When HEK293 cells were co-transfected with Myc-tagged Importin β2 with WT, G2A point mutant (which blocks myristoylation and palmitoylation) or the C3S point mutant (which blocks only palmitoylation) and a R118H mutant (which blocks interaction with the small GTPase Arl3) (Khnel et al., 2006) of RP2–FLAG, both WT and R118H were efficiently co-immunoprecipitated with Myc–Importin-β2. However, neither the G2A nor C3S mutants co-immunoprecipitated with Importin β2 (supplementary material Fig. S2B). These results suggest that membrane association of RP2 is a prerequisite for formation of an RP2–Importin-β2 complex.
We further examined the RP2 sites required for interaction with Importin β2. MDCK cells were transfected with the following FLAG-tagged RP2 mutants: G2A, an N-terminal deletion of the first 20 amino acids, the NLS mutant that converted KRRK to AAAA, R118H and another mutation in the C-terminus, R282W (Thiselton et al., 2000). Removal of the first 20 amino acids or lipid modification (G2A) severely impaired binding to Importin β2 (Fig. 6C) as expected from the results previously obtained in HEK293 cells (supplementary material Fig. S2B).
At this point, we thought that Importin β2 would bind RP2 through the N-terminal NLS sequence. Surprisingly, we found that mutation of this polybasic NLS sequence (KRRK to AAAA) without mutating the sites of lipid modification did not abolish Importin β2 binding (Fig. 6C). This indicated that RP2 must have an alternative Importin β2 binding site. Another binding site for Importin β2 is the M9 sequence found in heteronuclear ribonucleoprotein A1 (hnRNPA1) (Pollard et al., 1996). Indeed, we found an M9-core-like sequence in RP2 (Fig. 6D), which appeared to be essential for Importin β2 binding to RP2 because point mutants of the M9 consensus residues of RP2 (C86Y, P95L or C108G) abolished the interaction with Importin β2 (Fig. 6E). Interestingly, these residues have been found to be mutated in patients with retinitis pigmentosa (Sharon et al., 2000). This suggests that the M9 sequence is part of the main Importin β2 binding site in RP2 because mutation in this site severely reduced binding and the polybasic NLS binding site was unable to compensate for this.
The interaction between Importin β2 and M9 sequence of RP2 is crucial for RP2 ciliary targeting
The results of the binding experiments correlated well when trafficking of RP2 to the cilia was studied. As shown in Fig. 7A, mutation of the polybasic NLS region did not block Importin β2 binding and did not prevent targeting to the cilia. By contrast, mutations within the M9 sequence blocked both Importin β2 binding and cilia targeting (Fig. 7B).
Fig. 7.
The interaction between Importin β2 and M9 sequence of RP2 is crucial for RP2 ciliary targeting. (A) Mutation of NLS region of RP2 does not prevent RP2 ciliary targeting. MDCK cells were transfected with RP2–EGFP (green) with the NLS motif mutated (KRRK–AAAA). Cilia are stained with anti-polyglutamylated tubulin (red). (B) Point mutations in the M9 sequence, either C89Y or P95L of RP2–EGFP blocks ciliary targeting. MDCK cells expressing EGFP–RP2 with point mutation of the M9 sequence, C89Y (top panel) or P95L (bottom panel) were grown for 7 days after confluence on Transwell filters. Cilia are stained with anti-polyglutamylated tubulin (red), whereas EGFP–RP2 is green. Scale bars: 10 μm.
Discussion
Mutation of RP2 leads to retinitis pigmentosa but the exact role of RP2 in photoreceptors is unclear (Evans et al., 2006). Recent work has demonstrated that RP2 binds to the small G protein ARL3 (Bartolini et al., 2002) and has GAP activity towards this GTPase (Veltel et al., 2008). A recent study (Evans et al., 2010) demonstrated that RP2 localizes to the basal body and is involved in Golgi to ciliary base trafficking. In addition to basal body staining, we found that RP2 localized throughout the cilia in renal epithelial cells.
In this manuscript, we identified the binding of RP2 to Importin β2 and found that this binding appears important in regulating the targeting of RP2 to cilia. Importin proteins have been classically thought to regulate nuclear traffic (for a review, see Stewart, 2007). Importin β1 uses Importin-α proteins as adaptors to bind to polybasic NLS sequences on proteins to be imported into the nucleus. Ran GTP releases the cargo as well as Importin-α proteins from Importin β1 within the nucleus (for a review, see Cook et al., 2007). Importin β2 (also known as transportin-1) does not use Importin-α proteins but rather directly interacts with cargoes, releasing them upon binding Ran GTP (Pollard et al., 1996; Bonifaci et al., 1997; Siomi et al., 1997). Importin β2 usually binds a different sequence than Importin β1, such as the M9-like sequences we describe in RP2 (Michael et al., 1995; Weighardt et al., 1995; Bogerd et al., 1999). Mutation of this M9 sequence in RP2 impaired binding of RP2 to Importin β2 and impaired RP2 trafficking to the cilia. We also identified a polybasic NLS sequence in the N-terminus of RP2. However, this polybasic sequence could not support Importin β2 binding in full-length RP2 when the M9 sequence was mutated. We did observe trafficking to the cilia of the smaller 1–15 GFP–RP2 fragment, which contains only the lipid modified sites and the polybasic NLS. The trafficking of this fragment might be different than that of the full-length RP2 because of its smaller size, which allows easier ciliary entry, or more efficient interaction of the polybasic NLS in this smaller fragment with Importin proteins.
The binding and trafficking data reported here define additional roles for Ran and its binding partners aside from nuclear trafficking. Indeed, studies have already identified additional cellular roles for Ran and its binding partners. One of the best described additional roles is the trafficking and regulation of proteins that are crucial for spindle assembly during cell division (for a review, see Ciciarello et al., 2007; Clarke and Zhang, 2008; Kalab and Heald, 2008). In this process, Importin proteins bind and inhibit factors such as HURP and TPX2, which are essential for microtubule spindle growth, and this inhibition is relieved by Ran-GTP (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Silljé et al., 2006). Although these roles have been ascribed for Importin β1 and its Importin-α binding partners, Importin β2 might also have a role in mitotic spindle assembly (Lau et al., 2009). In this report and other recent reports, we also suggest a further role for Importin proteins in centrosomal and cilia targeting (Dishinger et al., 2010; Fan et al., 2007). In the first of these studies, we demonstrated the trafficking of a Crumbs3 splice isoform to the cilia and pericentrosomal region (Fan et al., 2007). We described a role for Ran and Importin β1 in this trafficking. More recently, Verhey and co-workers have demonstrated a role for Importin β2 in the trafficking of Kif17 to the cilia through an NLS sequence in the C-terminus (Dishinger et al., 2010). These data fit well with the hypothesis that Ran and Importin proteins control cilia traffic, possibly via the previously hypothesized ciliary pore at the base of the cilia (Rosenbaum and Witman, 2002). Others have noted the similarity between IFT proteins and nuclear pore proteins (Jekely and Arendt, 2006) and the similarities of nuclear and cilia pores (Christensen et al., 2007).
Although the trafficking of RP2 to the cilia appeared to be regulated by Importin β2, we found that RP2 also bound to Importin β1 and Importin β3. Knockdown of Importin β1 affects cell division, thus it is not possible to detect its affect on cilia trafficking. Our preliminary data with knockdown of Importin β3 showed no effect on RP2 trafficking (our unpublished observations). Binding to Importin β2 was the strongest and was detected in three different assays, so this might explain why cilia defects were seen primarily with knockdown of Importin β2. We also found that the sites of myristoylation and palmitoylation on the N-terminus of RP2 are important for the binding of RP2 to Importin proteins. Recent studies from Cheetham and co-workers have also found that lipid modification is important for targeting to the cilia base (Evans et al., 2010). We have observed similar defects in cilia trafficking of RP2 mutated at the lipid modification sites in polarized epithelial cells (Hurd et al., 2010). In fact, although mutation of the NLS sequences did not affect binding of Importin β2 to RP2 we found that mutation of the myristoylation site did impair Importin β2 binding. This is somewhat consistent with our results with Crumbs3–CLP1 where we have found that the transmembrane domain is essential for binding of Importin proteins (Fan et al., 2007) (our unpublished observations). These results suggest that membrane attachment is essential for Importin binding to these targets, but it is not clear whether it is directly involved or is essential for other cofactors. In this regard, it is interesting to note that some Importin proteins have been described to be membrane associated (Hachet et al., 2004).
It is also interesting to note that although RP2 could bind Importin proteins, we could not detect RP2 in the nucleus of epithelial cells despite trying several conditions, including UV stress, previously reported to send RP2 to the nucleus in HeLa and RPE cells (Yoon et al., 2006). If we removed the lipid anchor by mutating Gly2 to alanine, RP2 moved to the nucleus instead of the cilia (Hurd et al., 2010). Whether this involves binding to other Importin proteins is unclear, because this mutation did block binding of Importin β2 to RP2. The role of lipid modification in protein trafficking to the cilia and flagella has been described (Emmer et al., 2009; Hurd and Margolis, 2009; Saric et al., 2009). Recently Pazour and colleagues have described a cilia-trafficking motif in the Fibrocystin gene (Follit et al., 2010). They identified a sequence that contains lipid modification sites, as well as a polybasic region, to be necessary for targeting a fragment of this protein to the cilia. However, in their data they did not invoke the Ran–Importin system, but rather describe interactions of this sequence with Rab8.
In summary, our data indicate an important role for the Importin proteins in cilia trafficking. Ran and Importin proteins have been found consistently in cilia and centrosomal proteomes (Andersen et al., 2003; Liu et al., 2007; Pazour et al., 2005), and combined with studies such as those presented here, leave little doubt that the Ran–Importin system is localized to the cilia and centrosome. However, further work is required to understand their complete role. As previously described, the Ran–Importin system could work similarly to its role in nuclear trafficking by releasing cargo in a Ran-dependent fashion from Importin proteins at the base of the cilia. However the Ran–Importin system might have other roles in the cilia that need to be considered. One is the finding that Importin proteins have important roles in microtubule growth and might have a role in building the cilia (Carazo-Salas et al., 2001). Our results upon knockdown of Importin β2 showed, however, that cilia formed normally. Another possible role is trafficking of proteins from the cilia to the nucleus. There is an expanding description of signaling proteins that travel from the cilia to the nucleus, and Importin proteins in the cilia might have a role in this process. Proteins travel from the cilia to the nucleus in several signaling pathways (Haycraft et al., 2005; Huangfu et al., 2003; Lal et al., 2008), suggesting that Importin proteins could also function to traffic proteins out of the cilia. Regardless, it seems likely that the role of the Ran–Importin system in cilia function is a rich area for future study.
Materials and Methods
Antibodies
Rabbit anti-RP2 antibody was generated by immunization with recombinant human RP2 (Cocalico Biologicals, Reamstown, PA). Antibody was affinity purified with immobilized recombinant human HIS-RP2. Mouse anti-FLAG, anti-acetylated tubulin, anti-α-tubulin, anti-polyglutamylated tubulin, anti-γ-tubulin and rabbit anti-Importin-β2 antibodies were purchased from Sigma. Rabbit anti-EGFP was purchased from Invitrogen. Rabbit anti-Myc was purchased from Bethyl labs and mouse anti-Importin-β1 was from Affinity Bioreagent. Rabbit anti-pericentrin was purchased from Covance. Mouse anti-Importin-β3 was purchased from Abnova. Mouse anti-Ran was purchased from BD Biosciences.
Constructs
All constructs were generated by PCR from full-length ESTs (Mouse Importin β1, β2 and β3; Open Biosystems, Huntsville, AL). RP2–EGFP and RP2–FLAG expression constructs were generated by cloning the human RP2 open reading frame into pEGFP-N1 and pQCXIP respectively. For GST fusions, open-reading frames were cloned into unique restriction sites of pGEX-4T3 (GE Healthcare). Myc-tagged Importin proteins and Importin-β2–EGFP were cloned into the multiple cloning sites of pQCXIH or pEGFP-N1, respectively (Clontech). For recombinant protein expression, ORFs of Importin and Ran were cloned into pGEX4T3 (GE Healthcare).
Cell culture
MDCKII and HEK293 cells were grown in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin sulfate and 2 mM L-glutamine. For generation of stable MDCK cell lines, HEK293T cells were transiently transfected with pGAG/POL and pVSVG plus either pQCXIH for stable protein expression or pSIREN-RetroQ for shRNA expression (Clontech). Virus-containing medium was collected, filtered though a 0.4 μm filter and mixed with an equal volume of fresh culture medium. Polybrene (Clontech) was added to 4 μg/ml and virus was added to MDCKII cells. 12 hours later, medium was replaced. 48 hours after infection, stable expressing pools were selected using medium containing 200 μg/ml Hygromycin or 5 μg/ml Puromycin, respectively (Invitrogen, Carlsbad, CA). All transient transfection experiments were performed using Fugene 6 reagent (Roche).
Immunofluorescence and confocal microscopy
We performed immunofluorescence as described previously (Fan et al., 2007). In general, cells were fixed with 4% paraformaldehyde for 15 minutes. After brief washing, 0.25% Triton X-100 in PBS was used to permeabilize the cells in most conditions. We used 0.1% Triton X-100 in PBS to permeabilize cells for ciliary staining. For γ-tubulin and pericentrin staining, we used cold acetone to fix the cells for 4 minutes. These procedures were followed by 3% goat serum in PBS blocking for 30 minutes. Primary antibodies and secondary antibodies were diluted and incubated as previously described (Fan et al., 2007). All images were obtained using either inverted fluorescence microscope (Nikon Eclipse TE2000U) or a meta laser-scanning confocal microscope (LSM 510; Carl Zeiss MicroImaging).
Protein knockdown
Canine Importin β2 sequences GCAGTGCCTTTGCTACCTTAG and GCATGTTAAGCCTTGTATAGC were selected as Importin β2 shRNA targeting sequences. These sequences were placed into the Clontech shRNA Sequence Designer Tool and the selected oligos were annealed and ligated in pSIREN-RetroQ (Clontech) to encode the shRNA. Retroviral shRNA infection was as above.
Centrosome purification
Centrosomes were isolated using a modified protocol (Zhou et al., 2006). In brief, MDCKII cells were grown for 6 days after confluence before incubation with 10 μg/ml nocodazole and 5 μg/ml cytochalasin B at 37°C for 90 minutes. Cells were washed with PBS, 8% sucrose in 0.1 × PBS, and finally 8% sucrose. Cells were lysed (1 mM HEPES, pH 7.2, 0.5% NP-40, 0.5 mM MgCl2, 0.1% 2-mercaptoethanol, and protease inhibitors) with shaking at 4°C for 10 minutes. Lysates were cleared by centrifugation at 2500 g for 10 minutes and supernatant was filtered through a 40 μm nylon mesh. HEPES (pH 7.2) was added to a final concentration of 10 mM and DNaseI added to 2 U/ml followed by incubation on ice for 30 minutes. Lysates were underlaid with 60% sucrose solution (60% wt/wt sucrose in 10 mM PIPES, pH 7.2, 0.1% Triton X-100 and 0.1% 2-mercaptoethanol) and spun at 25,000 g for 45 minutes to sediment centrosomes into the sucrose cushion. The sucrose cushion was collected and diluted with lysis buffer to a final concentration of sucrose of 30%. The centrosome preparation was then loaded onto a discontinuous sucrose gradient (0.5 ml of 70%, 0.3 ml of 50% and 0.3 ml of 40% sucrose solution) and spun at 120,000 g for 60 minutes. Fractions (0.15 ml each) were collected and diluted with 1 ml of 10 mM PIPES, pH 7.2 and sedimented by centrifugation at 16,000 g for 15 minutes. For centrosomal protein analysis, the supernatant was discarded and the pelleted protein resuspended in LDS sample buffer (Invitrogen).
Immunoprecipitation and western blotting
HEK293 and MDCK cells were rinse twice in ice-cold PBS and lysed in Triton lysis buffer [1% Triton X-100 (v/v), 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 10% (v/v) glycerol] followed by centrifugation. For anti-FLAG immunoprecipitations, FLAG M2 beads (Sigma) were added to cleared lysates and incubated overnight at 4°C. For Myc and HA immunoprecipitations, cleared lysates were incubated overnight at 4°C with 2 μg of appropriate antibody plus 20 μl Protein-A–Sepharose (Invitrogen). Immunoprecipitations were washed four times with lysis buffer and resuspended in LDS sample buffer (Invitrogen). Samples were subsequently run on 4–12% Bis-Tris Novex gels (Invitrogen) and transferred to PVDF. Membranes were blocked with 5% BSA-TBS and primary antibodies incubated for 1 hour in blocking buffer. Secondary antibodies were added in 5% non-fat milk in TBS with 0.05% Tween20 for 1 hour. For GST pull-down experiments, 4 μg glutathione Sepharose4B immobilized GST protein was incubated with cleared lysate overnight at 4°C. Beads were then washed four times with lysis buffer and resuspended in LDS sampled buffer. For MS/MS analysis FLAG immunoprecipitations were performed as described above and run on Bis-Tris gels. Gels were subsequently washed and stained with Imperial Blue colloidal Coomassie (Pierce) and positive bands excised. MS/MS analysis was performed by the Michigan Proteome Consortium.
Supplementary Material
Acknowledgments
We thank Anand Swaroop, Hemant Khanna, Kristen Verhey and Jeff Martens for support and helpful discussions. The work was supported by a grant from the PKD Foundation and by NIH DK84725. This work utilized the Microscopy Core of the Michigan Diabetes Research and Training Center funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/5/718/DC1
References
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003). Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570-574 [DOI] [PubMed] [Google Scholar]
- Bartolini F., Bhamidipati A., Thomas S., Schwahn U., Lewis S. A., Cowan N. J. (2002). Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J. Biol. Chem. 277, 14629-14634 [DOI] [PubMed] [Google Scholar]
- Berbari N., O'Connor A., Haycraft C., Yoder B. (2009). The primary cilium as a complex signaling center. Curr. Biol. 19, R526-R535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H. P., Benson R. E., Truant R., Herold A., Phingbodhipakkiya M., Cullen B. R. (1999). Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J. Biol. Chem. 274, 9771-9777 [DOI] [PubMed] [Google Scholar]
- Bonifaci N., Moroianu J., Radu A., Blobel G. (1997). Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA 94, 5055-5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Gruss O. J., Mattaj I. W., Karsenti E. (2001). Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3, 228-234 [DOI] [PubMed] [Google Scholar]
- Chapple J. P., Hardcastle A. J., Grayson C., Willison K. R., Cheetham M. E. (2002). Delineation of the plasma membrane targeting domain of the X-linked retinitis pigmentosa protein RP2. Invest. Ophthalmol. Vis. Sci. 43, 2015-2020 [PubMed] [Google Scholar]
- Christensen S. T., Pedersen L. B., Schneider L., Satir P. (2007). Sensory cilia and integration of signal transduction in human health and disease. Traffic 8, 97-109 [DOI] [PubMed] [Google Scholar]
- Ciciarello M., Mangiacasale R., Lavia P. (2007). Spatial control of mitosis by the GTPase Ran. Cell. Mol. Life Sci. 64, 1891-1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P. R., Zhang C. (2008). Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9, 464-477 [DOI] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., Conti E. (2007). Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647-671 [DOI] [PubMed] [Google Scholar]
- Dishinger J. F., Kee H. L., Jenkins P. M., Fan S., Hurd T. W., Hammond J. W., Truong Y. N., Margolis B., Martens J. R., Verhey K. J. (2010). Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat. Cell Biol. 12, 703-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer B., Souther C., Toriello K., Olson C., Epting C., Engman D. (2009). Identification of a palmitoyl acyltransferase required for protein sorting to the flagellar membrane. J. Cell Sci. 122, 867-874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. J., Hardcastle A. J., Cheetham M. E. (2006). Focus on Molecules: X linked Retinitis Pigmentosa 2 protein, RP2. Exp. Eye Res. 82, 543-544 [DOI] [PubMed] [Google Scholar]
- Evans R. J., Schwarz N., Nagel-Wolfrum K., Wolfrum U., Hardcastle A. J., Cheetham M. E. (2010). The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum. Mol. Genet. 19, 1358-1367 [DOI] [PubMed] [Google Scholar]
- Fan S., Fogg V., Wang Q., Chen X. W., Liu C. J., Margolis B. (2007). A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J. Cell Biol. 178, 387-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J., Li L., Vucica Y., Pazour G. (2010). The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J. Cell Biol. 188, 21-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., Somlo S. (2006). Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 119, 1383-1395 [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin [alpha] on TPX2 activity. Cell 104, 83-93 [DOI] [PubMed] [Google Scholar]
- Hachet V., Kcher T., Wilm M., Mattaj I. (2004). Importin alpha associates with membranes and participates in nuclear envelope assembly in vitro. EMBO J. 23, 1526-1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle A. J., Thiselton D. L., Van Maldergem L., Saha B. K., Jay M., Plant C., Taylor R., Bird A. C., Bhattacharya S. (1999). Mutations in the RP2 gene cause disease in 10% of families with familial X-linked retinitis pigmentosa assessed in this study. Am. J. Hum. Genet. 64, 1210-1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K. (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1, e53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83-87 [DOI] [PubMed] [Google Scholar]
- Hurd T., Margolis B. (2009). Cystin, cilia, and cysts: unraveling trafficking determinants. J. Am. Soc. Nephrol. 20, 2485-2486 [DOI] [PubMed] [Google Scholar]
- Hurd T., Zhou W., Jenkins P., Liu C.-J., Swaroop A., Khanna H., Martens J., Hildebrandt F., Margolis B. (2010). The retinitis pigmentosa protein RP2 interacts with polycystin 2 and regulates cilia-mediated vertebrate development. Hum. Mol. Genet. 19, 4330-4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely G., Arendt D. (2006). Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays 28, 191-198 [DOI] [PubMed] [Google Scholar]
- Jenkins P. M., Hurd T. W., Zhang L., McEwen D. P., Brown R. L., Margolis B., Verhey K. J., Martens J. R. (2006). Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr. Biol. 16, 1211-1216 [DOI] [PubMed] [Google Scholar]
- Jkel S., Grlich D. (1998). Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491-4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Heald R. (2008). The RanGTP gradient-a GPS for the mitotic spindle. J. Cell Sci. 121, 1577-1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khnel K., Veltel S., Schlichting I., Wittinghofer A. (2006). Crystal structure of the human retinitis pigmentosa 2 protein and its interaction with Arl3. Structure 14, 367-378 [DOI] [PubMed] [Google Scholar]
- Lal M., Song X., Pluznick J. L., Di Giovanni V., Merrick D. M., Rosenblum N. D., Chauvet V., Gottardi C. J., Pei Y., Caplan M. J. (2008). Polycystin-1 C-terminal tail associates with beta-catenin and inhibits canonical Wnt signaling. Hum. Mol. Genet. 17, 3105-3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. K., Delmar V. A., Chan R. C., Phung Q., Bernis C., Fichtman B., Rasala B. A., Forbes D. J. (2009). Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol. Biol. Cell 20, 4043-4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Tan G., Levenkova N., Li T., Pugh E. N., Jr., Rux J. J., Speicher D. W., Pierce E. A. (2007). The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 6, 1299-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelova J., Astuto-Gribble L., Inoue H., Tam B. M., Schonteich E., Prekeris R., Moritz O. L., Randazzo P. A., Deretic D. (2009). Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 28, 183-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W., Choi M., Dreyfuss G. (1995). A nuclear export signal in HNRNP α1- a signal-mediated, temperature-dependent nuclear-protein export pathway. Cell 83, 415-422 [DOI] [PubMed] [Google Scholar]
- Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. (2001). Importin [beta] is a mitotic target of the small GTPase ran in spindle assembly. Cell 104, 95-106 [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Bloodgood R. A. (2008). Chapter 5: targeting proteins to the ciliary membrane. In Current Topics in Developmental Biology, Vol. 85 (ed. Yoder B. K.), pp. 115-149: Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. B., Veland I. R., Schroder J. M., Christensen S. T. (2008). Assembly of primary cilia. Dev. Dyn. 237, 1993-2006 [DOI] [PubMed] [Google Scholar]
- Pollard V. W., Michael W. M., Nakielny S., Siomi M. C., Wang F., Dreyfuss G. (1996). A novel receptor-mediated nuclear protein import pathway. Cell 86, 985-994 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813-825 [DOI] [PubMed] [Google Scholar]
- Saric M., Vahrmann A., Niebur D., Kluempers V., Hehl A., Scholze H. (2009). Dual acylation accounts for the localization of {alpha}19-giardin in the ventral flagellum pair of Giardia lamblia. Eukaryot. Cell 8, 1567-1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D., Bruns G. A. P., McGee T. L., Sandberg M. A., Berson E. L., Dryja T. P. (2000). X-Linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest. Ophthalmol. Vis. Sci. 41, 2712-2721 [PubMed] [Google Scholar]
- Silljé H. H. W., Nagel S., Körner R., Nigg E. A. (2006). HURP is a ran-importin [beta]-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16, 731-742 [DOI] [PubMed] [Google Scholar]
- Silverman M., Leroux M. (2009). Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 19, 306-316 [DOI] [PubMed] [Google Scholar]
- Siomi M. C., Eder P. S., Kataoka N., Wan L., Liu Q., Dreyfuss G. (1997). Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138, 1181-1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195-208 [DOI] [PubMed] [Google Scholar]
- Thiselton D. L., Zito I., Plant C., Jay M., Hodgson S. V., Bird A. C., Bhattacharya S. S., Hardcastle A. J. (2000). Novel frameshift mutations in the RP2 gene and polymorphic variants. Hum. Mutat. 15, 580 [DOI] [PubMed] [Google Scholar]
- Veltel S., Gasper R., Eisenacher E., Wittinghofer A. (2008). The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat. Struct. Mol. Biol. 15, 373-380 [DOI] [PubMed] [Google Scholar]
- Weighardt F., Biamonti G., Riva S. (1995). Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J. Cell Sci. 108, 545-555 [DOI] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. (2001). Role of importin-beta in coupling ran to downstream targets in microtubule assembly. Science 291, 653-656 [DOI] [PubMed] [Google Scholar]
- Yoon J.-H., Qiu J., Cai S., Chen Y., Cheetham M., Shen B., Pfeifer G. (2006). The retinitis pigmentosa-mutated RP2 protein exhibits exonuclease activity and translocates to the nucleus in response to DNA damage. Exp. Cell Res. 312, 1323-1334 [DOI] [PubMed] [Google Scholar]
- Zhou C., Cunningham L., Marcus A. I., Li Y., Kahn R. A. (2006). Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell 17, 2476-2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.