Abstract
The placenta plays an important role as a regulator of fetal nutrition and growth throughout development and placental factors contribute to gestational abnormalities such as preeclampsia. This study describes the genome-wide gene expression profiles of a large (n=60) set of human placentas in order to uncover gene expression patterns associated with preeclampsia. In addition to confirming changes in expression of soluble factors associated with preeclampsia such as sFLT1 (soluble fms-like tyrosine kinase-1), sENG (soluble endoglin), and INHA (inhibin alpha), we also find changes in immune-associated signaling pathways, offering a potential upstream explanation for the shallow trophoblast invasion and inadequate uterine remodeling typically observed in pathogenesis of preeclampsia. Notably, we also find evidence of preeclampsiaassociated placental upregulation of sialic acid acetylesterase (SIAE), a gene functionally associated with autoimmune diseases.
Keywords: placenta, pre-eclampsia, preeclampsia, maternal hypertension
Introduction
Up to 8% of pregnancies are affected by hypertensive disorders [1]; most of these are due to preeclampsia, a multi-system pregnancy-associated disorder triggered by endothelial cell dysfunction and clinically characterized by the de novo onset of maternal hypertension and proteinuria [2]. About 15% of preterm births are due to preeclampsia; while early delivery can relieve maternal symptoms that can pose a risk to both mother and fetus, it also increases the risk of perinatal morbidity and mortality [3]. While infant mortality due to preeclampsia has been dramatically reduced in developed countries by clinical management, the risk for perinatal mortality is estimated to be 50% greater in pregnancies complicated by preeclampsia [4]. Preeclampsia and eclampsia are the third leading cause of maternal pregnancy-related deaths in the United States, behind embolism and hemorrhage [5]. Several risk factors have been identified including: first pregnancy (nulliparity), a family history of preeclampsia, diabetes, and multiple pregnancy [6, 7].
Pregnancy is a fascinating and unique state where a semi-allogeneic fetus survives in spite of the usual rules of immunology [8]. A number of studies have focused on the connection between pregnancy and preeclampsia as it relates to immunological function. In women without preeclampsia in their first pregnancy, changing paternity in a subsequent pregnancy resulted in a 30% increase in risk compared to those without change, supporting the idea that tolerization to paternal antigens may have an effect [9]. Immune suppression has recently been shown to improve blood pressure and endothelial cell function in a rat model of preeclampsia, supporting the idea that maternal immune activation is involved in its etiology [10]. It has been proposed on the one hand that the innate, not adaptive immune system, controls immuno-regulation during pregnancy and preeclampsia and that a special population of uterine NK cells are key players in this process [8, 11]. On the other hand, there is also evidence that regulatory T-cells are involved in mediating maternal tolerance during pregnancy [12].
Preeclampsia can arise from both feto-placental and maternal factors, as well as genetic factors [13]. The pathophysiology of preeclampsia is commonly divided into two stages: 1) abnormalities in placentation (from inadequate uterine remodeling and placental invasion, reduced or intermittent uteroplacental perfusion) and 2) the maternal syndrome associated with placental hypoxia and oxidative stress [14]. Clinical symptoms of preeclampsia generally improve and resolve after delivery suggesting the presence of a placental secreted factor that directly causes maternal endothelial dysfunction [2].
Known factors altered in preeclampsia include Activin-A and Inhibin A (INHA) [15, 16], Leptin (LEP) [17], soluble Endoglin (sENG) [18], soluble fms-like tyrosine kinase-1 (sFlt-1) [19], and placental growth factor (PGF) [20] and have been recently reviewed by Carty et al. [21]. sFLT1 and sENG are both anti-angiogenic factors that oppose that action of vascular endothelial growth factor (VEGF). Interestingly, overexpression of sFLT1 has been shown to induce preeclampsia in rats [22]. sEng is a soluble TGF-beta co-receptor, which may cooperatively act with sFlt1 to induce severe preeclampsia [18]. Both levels of sEng and sFlt1 correlate with disease severity and decline after delivery [18, 19].
While previous gene expression studies have been conducted on the preeclamptic human placenta, this is, to our knowledge, the largest of its kind. In this study, we place the detected gene expression changes in the context of an immune-mediated model of preeclamptic pathogenesis.
Materials and Methods
Study Design
23 preeclamptic, and 37 control placentas included in this study were collected during the years of 2004–2008 and hybridized in two batches to microarrays. Samples were randomized across arrays to control for array and batch variability.
Subjects
Subjects were women who received care at the Duke University Medical Center Obstetric Clinic. Enrollment began August 1, 2003. Women age 18 and older and their infants were eligible for inclusion. Women with multiple gestations, fetuses with documented congenital anomalies, fetuses with documented chromosomal abnormalities, and fetuses with prolonged premature rupture of the membranes (greater than 4 weeks) were excluded from enrollment. Inclusion was contingent upon live birth. Summary characteristics of the preeclamptic and control populations are presented in Table 1. The study was approved by the Duke University Medical Center Institutional Review Board (IRB 00016065).
Table 1.
Table of demographic information on study population.
| Control (n=37) | Pre-Eclamptic (n=23) | ||||
|---|---|---|---|---|---|
| Mean (s.d.) | Range | Mean (s.d.) | Range | p-value | |
| Fetal estimated | |||||
| Gestational Age (weeks) | 37.6 (1.93) | 29–40 | 33.6 (3.7) | 27–38 | P < .0001* |
| % Female | 43.2 | 56.5 | P < .4267 | ||
| Placental weight (g) | 491.7 (140.3) | 240–990 | 448.2 (237.8) | 130–1090 | P < .4322 |
| Birth weight (g) | 3258.4 (630.9) | 1190–4685 | 2278.4 (998.9) | 765–3860 | P < .0001* |
| Corrected percentile birth weight | 50.6 (31.0) | 4–99 | 38.4 (26.4) | 2–85 | P < .1153 |
| Maternal parity | 2.35 (1.7) | 0–7 | .42 (.69) | 0–2 | P < .0001* |
| Weight (lb) | 213.9 (51.8) | 125–358 | 209.2 (45.5) | 140–322 | P < .1474 |
| % Induced | 69.6 | 21.6 | P < 0.0002* | ||
Significant at P<0.05
Preeclampsia Clinical Inclusion Criteria
Women were diagnosed with preeclampsia if their systolic blood pressure was at least 140 mm Hg, their diastolic blood pressure was at least 90 mm Hg and they had proteinuria with an estimated 300 mg of protein or greater excreted in 24 hour measured directly or indirectly by protein creatinine ratio.
Sample collection procedures
Eligible participants were approached by the study coordinator and after giving informed consent, participants completed a questionnaire administered by the study coordinator. The questionnaire included information on demographics (height, weight, race, ethnicity), lifestyle (smoking, substance abuse), and medical and reproductive history. An approximately 2 cm3 placental sample, excluding fetal membranes, was obtained within 5 cm of the placental umbilical insertion site within an hour of delivery. Placental samples were flash frozen in liquid nitrogen and samples were homogenized under liquid nitrogen using a mortar and pestle. Maternal and infant medical records were reviewed for pertinent data including gestational age at delivery, fetal sex and weight, placental weight, ethnicity, maternal parity, and induction of labor.
RNA isolation and first-strand cDNA synthesis
Total RNA was isolated from term human placenta using the Totally RNA kit (Ambion) following the manufacturer’s instructions with slight modifications. Briefly, homogenized tissue chilled in liquid nitrogen was lysed in 10 volumes of denaturation solution (200 mg tissue, 2 ml denaturation solution). The lysed sample was pre-cleared of bulk genomic DNA and cellular debris by spinning at 4,000×g for 5 minutes. The supernatant was sequentially extracted with equal volumes phenol:chloroform, and acid phenol:chloroform (pH 4.5) using phase divider gels (Sigma), then precipitated with an equal volume of 100% isopropanol. The pellet was washed twice with 70% ethanol and resuspended in elution buffer (1 mM Tris-Cl). Total RNA was treated with Turbo DNA-free to remove traces of genomic DNA contamination (Ambion). RNA integrity was assessed using a Bioanalyzer (Agilent Biotechnologies) and only samples with a RIN > 7 were used. Total RNA was converted into 1st cDNA using the AffinityScript Multitemperature cDNA Synthesis Kit (Stratagene) according to manufacturer’s instructions.
Microarray Hybridization
Labeling and hybridization for Illumina Human6-v2 BeadArrays was performed according to manufacturer’s instructions. Briefly, the Illumina TotalPrep RNA Amplification Kit (Applied Biosystems, Foster City, CA) was used for a first and second strand reverse transcription step from 500 ng of total RNA. This was followed by an overnight in vitro transcription reaction that incorporates biotin-labeled nucleotides. 1.5 ug of biotin-labeled cRNA was mixed with an Illumina-supplied hybridization buffer containing several control oligonucleotides. Labeled cRNA was hybridized to BeadArrays at 58°C for 18 hr. Washing, blocking, and streptavadin-Cy3 staining was performed out following manufacturer’s protocol. BeadChips were scanned on the BeadArray Reader and scanned images were analyzed using BeadStudio software.
qRT-PCR Validation
Quantitative RT-PCR was performed using a multiplex Taqman-format 5’-nuclease assays (Integrated DNA Technologies) where both gene of interest and reference gene are simultaneously queried. Expression of genes of interest were interrogated using FAM6-labeled double-quenched probes. POLR2A was chosen for a reference gene on the basis of low coefficient of variation in the microarray data and recommendations from the Microarray Quality Control Consortium (MAQC) [23] and interrogated with a HEX550-labeled dual-labeled-probe (Integrated DNA Technologies). A subset of 31 patients were chosen for this confirmatory q-RT-PCR study. Thermal cycling parameters were: [95 °C, 20 s, initial denaturation], [95 °C, 15 s, denaturation; 60 °C, 20 s, annealing and extension]40 cycles. qRT-PCRs were performed using Taqman Fast Advanced Master Mix (Applied Biosystems) on a StepOnePlus Real-Time PCR instrument (Applied Biosystems). Correlation between the qRT-PCR and microarray was evaluated to quantitatively confirm the microarray data.
Microarray Statistical Analysis
Transcript data was log2 transformed, and quantile normalized [24]. Following this, transcripts which did not attain a detection p-value of <= 0.05 (compared to synthetic sequences on the array not complementary to mammalian genomes, used as negative controls) in any sample were dropped from analysis, leaving 35580 transcripts out of the original 48701. This filtered expression data was scaled to values between 0 and 10, and mean-centered around 0.
JMP Genomics 4.0 was used to perform principle variants components analysis as previously described [25]. A series of models were fitted to calculate the contribution of each of the factors to the measured transcriptional variation: classification, gender, induction of labor, their pair-wise two-way interactions, and estimated gestational age. Gene-specific analysis of variance (ANOVA) was performed using the following model using PROC MIXED in SAS (SAS Institute, Cary, NC), treating classification, gender as fixed effects:
| (model 1) |
| (model 2) |
Custom hypothesis tests were constructed to test for differential expression between pre-eclamptic and control groups, as well as male and female samples. Raw p-values were corrected for multiple comparisons via the conservative Benjamini-Hochberg FDR at α < 0.05 (for pathway analysis) and Bonferonni at α < 0.05 (heatmap visualization) methods [26] as implemented in PROC MULTTEST in SAS (SAS Institute, Cary NC).
Unsupervised hierarchical clustering of the gene expression patterns of placental differentially expressed genes between control and preeclamptic pregnancies was performed using JMP Genomics 8 (SAS Institute, Cary, NC). Differences in demographics between the control and preeclamptic groups were tested using logistic regression using JMP 8 (SAS Institute, Cary NC).
Pathways Analysis
Molecules from the data set that met the FDR<0.05, that were annotated as high-quality (“perfect” or “good”) in an updated Illumina probe set annotation [27], and were associated with a canonical pathway in Ingenuity’s Knowledge Base were considered for the analysis. Canonical pathways analysis (Ingenuity Pathways Analysis library of canonical pathways) identified the pathways that were most significant to the data set. The significance of the association between the data set and the canonical pathway was measured by using Fisher’s exact test to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
Results
Power Analysis
To determine how likely the statistical tests are to detect differential expression with varying sample sizes, we conducted a retrospective mixed model power analysis. The median effect size detected by our full experimental design (n=60) at a Bonferonni-level of significance was 0.41. At this effect size, the power to detect differential expression is 0.985, 0.318, and 0.003, for experimental designs with 60, 30, and 15 patients, respectively (See Supplementary Figure 1). Thus, the power to detect realistic changes in differential expression is dramatically decreased in the reduced-sample designs, with neither the n=30 or n=15 design meeting the conventional threshold of 80% statistical power for an effect size of 0.41, equivalent to a 1.32X fold change (See Supplementary Figure 1). Power analysis of a design analyzing a subset of births between the gestational ages of 34–37 weeks (n=23) indicates that this design is insufficiently powered to detect preeclamptic differential expression (See Supplementary Figure 1).
Demographics of Study Population
No significant differences in sex, placental weight, corrected birth weight, or maternal weight were detected between uncomplicated pregnancies and pregnancies complicated by preeclampsia. A significant effect was detected, however, for uncorrected birth weight, maternal parity, fetal estimated gestational age, and induction of labor (See Table 1.).
Variance Components Analysis and Mixed Model Selection
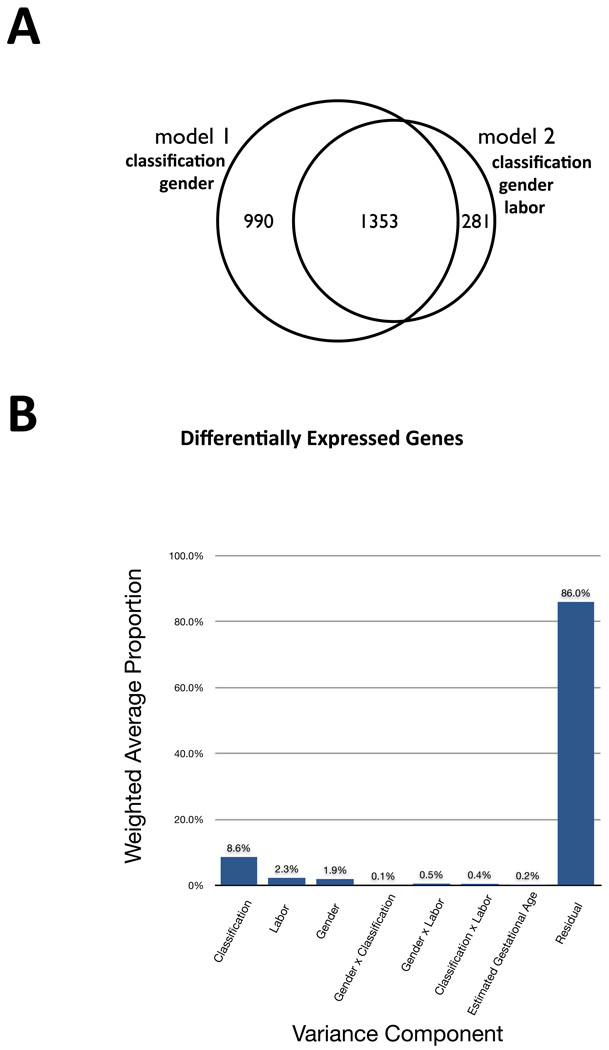
The majority of preeclamptic pregnancies are induced as part of clinical management, while the converse is true for normal pregnancies (See Table 1.) To control for this effect, we fit model 2 including gender, classification, labor, their pair-wise interactions, and a batch effect. No significant differentially expressed genes were detected for the labor effect at either a conservative Bonferonni or relaxed FDR-corrected p<0.05 significance threshold. The differentially expressed genes detected for the classification (preeclampsia or normal) effect in model 1 and model 2 were similar (See Figure 1a). As no significant effect of labor was detected, we chose to present the results of model 1 (which is higher-powered).
Figure 1. Model selection and variance component analysis.
(a) Venn diagram of overlap between placental differentially expressed genes between preeclamptic and normal pregnancies for model 1 and model 2. Model 2 controls for the effect of induction of labor. No differentially genes were detected for the effect of induction of labor. The genes detected for the classification effect (preeclampsia or normal) overlapped substantially for model 1 and model 2. 82% of the genes detected by model 2 at FDR < 0.05 are also detected by model 1. Model 1 detects a larger number of differentially expressed genes due to having a higher statistical power. The results of model 1 were used in pathway analysis. (b) Variance components analysis of the first five principle components of variation indicates that classification is the dominant contributor to transcriptional variation in this study. There are minor contributions of labor and gender, and minimal contributions of the pairwise interaction effects and estimated gestational age. This analysis suggest that most of the differentially expressed genes we detect are associated with classification and is corroborated by the results of model 2 in which we control for the effect of labor.
Additionally, to assess the independent contributions of other factors, such as estimated gestational age and induction of labor, to the observed transcriptional profiles, we conducted variance component analysis. This analysis indicates that classification (preeclampsia or normal) is the main contributing effect on expression variation, with minor contributing effects of sex and labor. Estimated gestational age has no significant contribution to the principle components of expression variation (See Figure 1b).
Quality Control: Differentially Expressed Genes Between Placentas from Male and Female Births
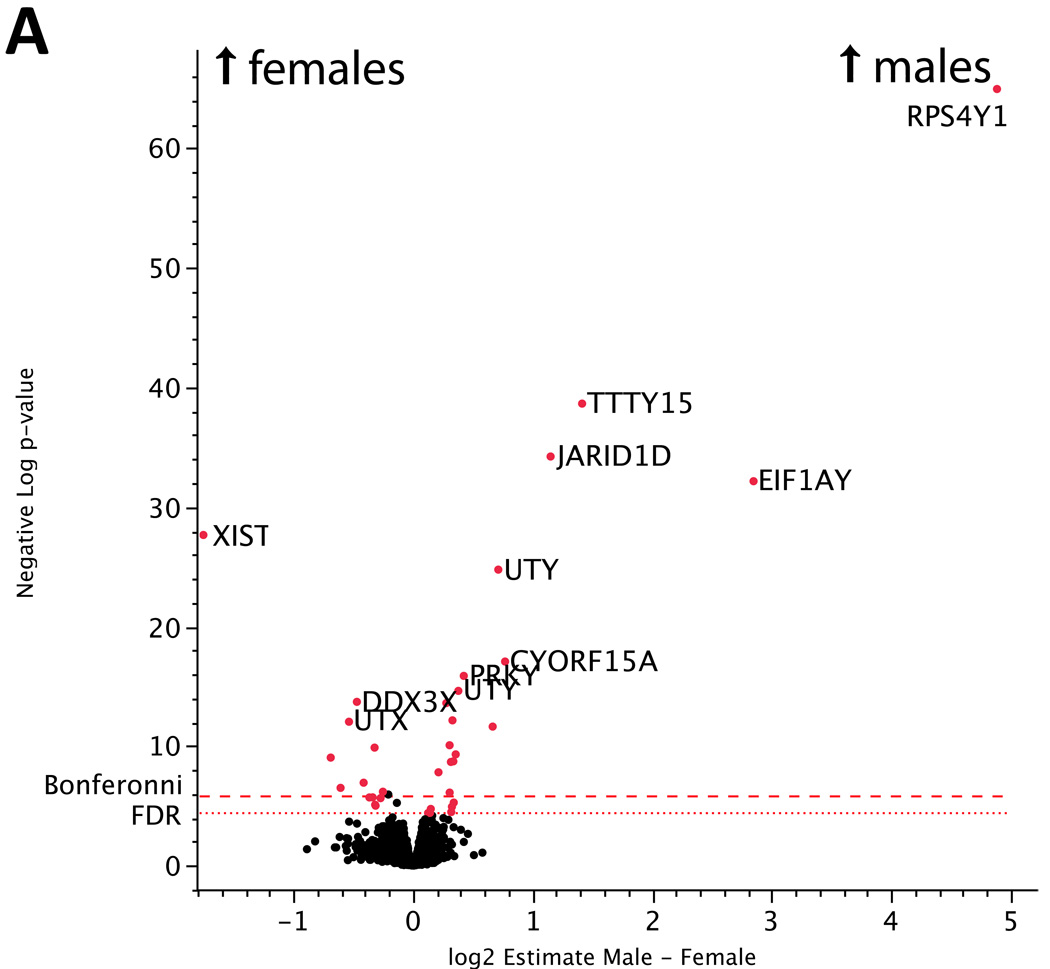
The estimate of the fixed sex effect between placentas of male and female infants from our gene-specific ANOVA found 36 genes to be differentially expressed at a conservative Bonferroni-corrected significance level of p < 1.405E-6 (See Figure 2a, Supplementary Table 1). As expected, nearly all of these differentially expressed genes (25/26, 96%) localize to the sex chromosomes, confirming the quality of the microarray data. These genes represent transcripts that are either specifically expressed from the Y chromosome or genes that escape X-inactivation, consistent with previous reports of human sex-specific expression variability [28]. The detection of these genes by using sex as a model phenotype confirms the sensitivity of our genome-wide gene expression profiling approach. The dataset is freely available at GEO repository under accession XXXX.
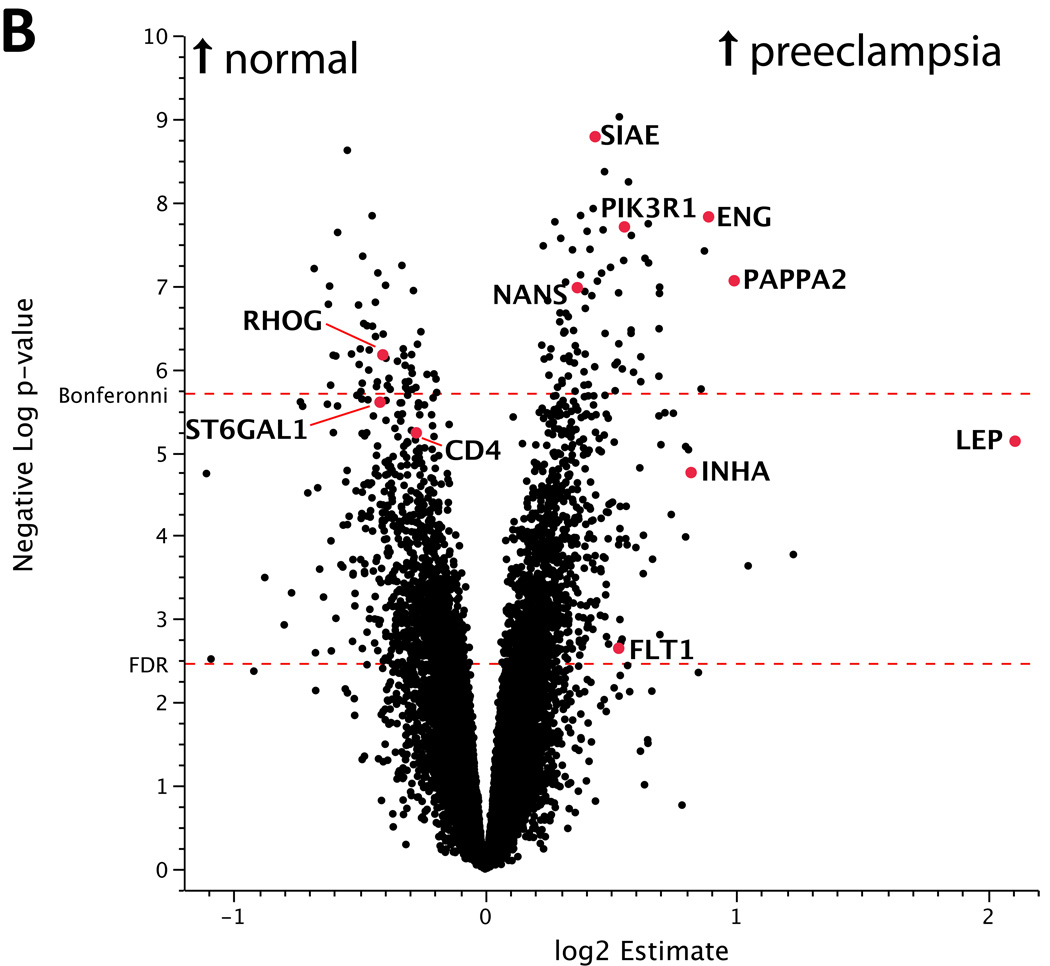
Figure 2.
(a) Volcano plot of differences in gene expression between placentas from male and female neonates. The x-axis represents the estimate of the difference in expression between males – females on a log2 scale. The y-axis represents the negative log of the uncorrected p-value on a log10 scale. Genes upregulated in placentas from male neonates are found in the upper right while those upregulated in placentas from female neonates are in the upper left. The points highlighted in red are differentially expressed genes that exceed a significance threshold of FDR<0.05 and map to the X or Y chromosome. (b) Volcano plot of expression differences between preeclamptic and control placentas. The x-axis represents the estimate of the difference in expression between preeclampsia – control on a log2 scale. The y-axis represents the negative log of the uncorrected p-value on a log10 scale. Genes upregulated in preeclamptic placentas are found in the right, while those upregulated in controls are in the left. A number of known preeclampsia associated factors such as ENG, FLT1, INHA, PAPPA were also found to be differentially expressed in our study. The differentially expressed genes at FDR<0.05 were used for pathway analysis. Only genes that were annotated as high-quality in an updated Illumina re-annotation were included in this figure and in pathway analysis.
Differentially Expressed Genes and Pathways Between Normal and Preeclamptic Placentas
We found 128 genes to be differentially expressed between normal and preeclamptic placentas at a Bonferonni-corrected significance threshold. These genes include known preeclampsia-associated genes ENG, PAPPA2 (pappalysin-2), and RDH13 (retinol dehydrogenase 13) [29]. (See Figure 2b, Supplementary Table 2.) 2,109 genes were differentially expressed between normal and preeclamptic placentas at FDR<0.05 and include INHA, LEP, and FLT1. These FDR<0.05 genes were used to search for canonical pathways enriched in differentially expressed genes. Pathways significant above FDR<0.1 are listed in Table 2.
Table 2.
Table of pathways significantly enriched for placental differentially expressed genes between control and preeclamptic pregnancies.
| Canonical Pathways | FDR (B-H p-value) |
|---|---|
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | 0.0068 |
| CXCR4 Signaling | 0.0068 |
| Leukocyte Extravasation Signaling | 0.0083 |
| Semaphorin Signaling in Neurons | 0.0219 |
| Regulation of Actin-based Motility by Rho | 0.0617 |
| N-Glycan Biosynthesis | 0.0891 |
| NRF2-mediated Oxidative Stress Response | 0.0891 |
| Germ Cell-Sertoli Cell Junction Signaling | 0.0891 |
Inhibin A is upregulated in preeclamptic placentas. Interestingly, the magnitude of upregulation is greater in females than in male preeclamptic placentas pointing to a potential link to the increased number of preeclamptic pre-term deliveries of females [30]. ENG and FLTI are also significantly upregulated and confirm observations of others [18] [31]. We also detected the significant upregulation of three factors involved with sialic acid synthesis, transfer, and modification: N-acetylneuraminic acid synthase (NANS), and sialic acid acetylesterase (SIAE). Additionally, Sialic acid binding Ig-like lectin 6 (Siglec-6), a gene identified in previous gene expression profiling work on preeclampsia [32], was also found to be differentially expressed in our study (See Table 3).
Table 3.
Table of sialic-acid associated placental differentially expressed genes between preeclampsia and normal pregnancies with FDR<0.05.
| Ranka | Illumina Probe ID |
Gene Symbol |
Gene Description | GO Molecular Function | log2 Estimate |
FDR | p-value |
|---|---|---|---|---|---|---|---|
| 4 | ILMN_1669146 | SIAE | Sialic acid acetylesterase | hydrolase activity; serine esterase activity; sialate O-acetylesterase activity | 0.44 | 1.4E-05 | 1.6E-09 |
| 44 | ILMN_1815874 | NANS | N-acetylneuraminic acid synthase | transferase activity; N-acetylneuraminate synthase activity; N-acylneuraminate-9-phosphate synthase activity; N-acylneuraminate cytidylyltransferase activity | 0.37 | 8.3E-05 | 1.1E-07 |
| 166 | ILMN_1756501 | ST6GAL1 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | beta-galactoside alpha-2,6-sialyltransferase activity | −0.42 | 5.2E-04 | 2.5E-06 |
| 1535 | ILMN_1685630 | SIGLEC6 | sialic acid binding Ig-like lectin 6 | protein binding; sugar binding | 0.21 | 2.2E-02 | 1.0E-03 |
| 1632 | ILMN_1756937 | ST8SIA4 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | sialyltransferase activity | 0.21 | 2.4E-02 | 1.2E-03 |
Genes ranked by p-value of preeclamptic differential expression from low to high.
Benjamini-Hochberg FDR
From the pathway analysis, three pathways that are dysregulated in preeclampsia are associated with the activation of an immune response: Fcγ receptor-mediated phagocytosis in macrophages and monocytes, leukocyte extravasation signaling, and CXCR4 signaling (See Supplementary Figures 2, 3).
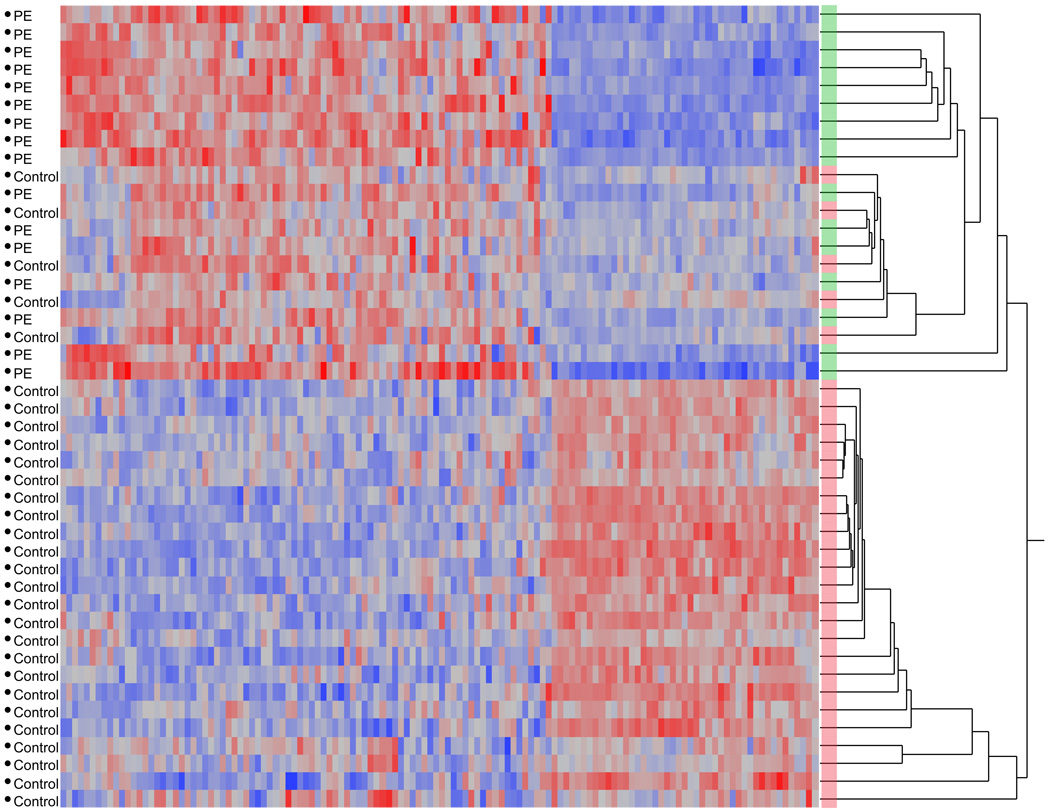
Finally, the heterogeneity in gene expression within each placental class is shown in the unsupervised two-way hierarchical-clustering heatmap in Figure 3, in which all preeclamptic samples cluster together in the first major split, as do the majority of normal samples. However, there are some normal samples that do cluster with the preeclamptic samples, indicating an interesting biological heterogeneity to the patterns of gene expression and suggesting the presence of molecular subclassifications of preeclampsia worthy of further investigation.
Figure 3. Heatmap of Bonferonni-significant differentially expressed between control and preeclamptic placentas.
While there is significant heterogeneity in human gene expression, the relatively large sample size of our microarray study allowed us to sensitively detect differential expression using a linear mixed model analysis. Reassuringly, unsupervised hierarchical clustering of the expression patterns of differentially expressed genes groups all preeclamptic and the majority of control gene expression profiles together. Some normal samples do cluster with preeclamptic samples, indicating that there may be different molecular subclassifications of preeclampsia worthy of further investigation. Preeclamptic/control status is indicated in the text to the left of the heatmap and by color (preeclampsia = green, control = red) to the right of the heatmap at each branch of the dendrogram. Patients are grouped along the Y-axis, while genes are clustered along the X-axis.
Quality Control: Taqman qPCR Validation
To validate the microarray data, we chose 6 genes exhibiting a range of differential expression between control and preeclamptic groups: SIAE, ENG, PIK3R1, RHOG, CD4, and CXCR4. ENG was selected as a positive control gene previously associated with preeclampsia [18]. Of these SIAE, ENG, PIK3R1, and CD4 were significantly correlated with the microarray data, while RHOG was not (p<0.089). There is conflicting microarray data with respect to CXCR4. There are two expressed CXCR4 probes in the array, one targeted to the 5’ UTR (ILMN_1801584) and the other targeted to the 3’ UTR (ILMN_1728927). The 5’ UTR probe reported significantly upregulated expression, while the 3’ UTR did not. Two independent qPCR assays to CXCR4 were designed and both correlated with the 3’ UTR microarray results (See Table 4) suggesting CXCR4 is not upregulated or that there are different CXCR4 isoforms, only some of which are affected. Overall, these results quantitatively confirm the gene expression patterns observed in the microarray data, with the exception of RHOG (See Table 4).
Table 4.
qPCR validation of microarray data for selected genes
| Ranka | Illumina Probe ID |
Gene | Gene Description |
GO Molecular Function | log2 Estimate |
Array p-value |
qPCR correlation p-value |
|---|---|---|---|---|---|---|---|
| 4 | ILMN_1669146 | SIAE | Sialic acid acetylesterase | hydrolase activity; serine esterase activity; sialate O-acetylesterase activity | 0.44 | 1.6E-09 | 7.1E-03* |
| 13 | ILMN_1760778 | ENG | Endoglin | protein binding | 0.89 | 1.5E-08 | 1.7E-17* |
| 16 | ILMN_1760303 | PIK3R 1 | Phosphoinositid e-3-kinase, regulatory subunit 1 (alpha) | kinase activity; protein phosphatase binding; insulin-like growth factor receptor binding; insulin binding; insulin receptor binding; phosphatidylinositol binding; ErbB-3 class receptor binding; insulin receptor substrate binding; phosphoinositide 3-kinase regulator activity | 0.55 | 2.0E-08 | 8.6E-13* |
| 91 | ILMN_1739792 | RHOG | Ras homolog gene family, member G | GTPase activity; nucleotide binding; GTP binding; protein binding | −0.41 | 6.8E-07 | 8.9E-02 |
| 236 | ILMN_1727284 | CD4 | CD4 molecule | protein homodimerization activity; protein kinase binding; transmembrane receptor activity; MHC class II protein binding; extracellular matrix structural constituent; zinc ion binding; coreceptor activity; glycoprotein binding | −0.27 | 5.9E-06 | 1.8E-03* |
| 539 | ILMN_1801584 | CXCR 4 | Chemokine (C-X-C motif) receptor 4 | C-C chemokine receptor activity; receptor activity; rhodopsin-like receptor activity; C-X-C chemokine receptor activity | −0.54 | 5.9E-05 | 1.3E-01 |
| 7138 | ILMN_1728937 | CXCR 4* | Chemokine (C-X-C motif) receptor 4 | C-C chemokine receptor activity; receptor activity; rhodopsin-like receptor activity; C-X-C chemokine receptor activity | −0.20 | 6.5E-02 | 3.2E-03* |
Genes ranked by p-value of preeclamptic differential expression from low to high.
qPCR significant Pearson correlation p<0.05.
Discussion
One of the difficulties in interpreting expression profiling studies involved preeclamptic placentas is controlling for differences in gestational age and induction of labor. In this study, while most demographic factors were not significantly different, preeclamptic and normal groups differed in estimated gestational age and induction of labor. This is unsurprising, as early induction of labor is part of the routine clinical management of preeclampsia. Nonetheless, these clinical differences can potentially confound transcriptional profiling for preeclampsia-associated genes, as the gene expression differences detected may actually be associated with gestational age or treatments used to induce labor. To determine the effects of such demographic factors we used variance component analysis and two different linear mixed models, one including labor induction and one without it. We did not detect significant differences in gene expression associated with induction of labor. This should not be interpreted as induction of labor having no effect on placental gene expression, but rather that the study was not designed to detect these differences. Our variance component analysis found that estimated gestational age has minimal independent contribution to this study’s gene expression variation. Taken together, while our results do not exclude the possibility of an effect of gestational age or labor on placental gene expression, they support that most of the differences we detect are associated with preeclampsia and not these two comfounding factors (See Supplementary Figure 1).
Additionally, since tissue sampling for the study was from placentas obtained from live births, gene expression profiles obtained potentially include both causative factors as well as downstream symptomatic responses. One important consideration is that if preeclampsia is caused by either: 1) placental factors, 2) maternal disposition and response, or 3) an interaction of the two, gene expression profiling of the preeclamptic placenta is limited by the possibility that it can arise from the combination of a normal placenta with a hypertensive maternal disposition. A comparison with gene expression profiles obtained from chorionic villi sampling earlier in gestation may help distinguish etiological factors from late acute response. In spite of these limitations, however, our data supports the use of microarray studies to understand the basis of preeclampsia.
Preeclampsia Associated Factors and Pathways
One current of model of the development of preeclampsia is that it is an exaggerated systemic maternal inflammatory response to oxidative stress from a placenta that has failed to sufficiently invade and remodel the maternal vasculature releasing placental factors into maternal circulation [14]. Through our transcriptional profiling we identified a number of transcripts encoding factors previously associated with preeclampsia such as ENG, FLT1, INHA [15, 18, 19] as well as novel factors such as SIAE.
In the analysis of the set of differentially expressed genes between placentas of uncomplicated control and pre-eclamptic pregnancies, we identified several pathways that were enriched in differentially expressed genes. Of major interest are pathways involved in an immune response: Fcy Receptor Mediated Phagocytosis in Macrophages and Monocytes, CXCR4 signaling, and leukocyte extravasation signaling (the process by which white blood cells migrate out of capillary circulation). It has been previously reported that the placenta co-opts adhesion molecules involved in the leukocyte extravasation pathway for uterine remodeling [33], and our study suggests that downregulation of these leukocyte extravasation signals are associated with the clinical manifestation of preeclampsia. The CXCR4 pathway is composed of 169 genes, of which 31 are differentially expressed at FDR < 0.05. Our qPCR data support a model where expression levels of the CXCR4 receptor itself is unchanged, but where binding of the receptor activates downstream G-protein mediated signaling including upregulation of PIK3R1. PIK3R1 is the top differentally expressed gene in each of the three immune-associated pathways.
Novel Sialic Acid Associated Factors
Recently, work from Surolia et al. [34] has described the association of functional rare genetic variation in sialic acid esterase (SIAE) with the regulation of human immune tolerance. SIAE catalyzes the removal of O-acetyl moities from the 9-OH position of sialic acids that decorate the surface of cells [35, 36]. ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (ST6GAL1) transfers sialic acid to galactose in an α2–6 linkage [37]. ST6GAL1 ablation in mice leads to a hyper-immune phenotype [38] although the mechanisms behind this is not well understood [39]. Together, ST6GAL1 and SIAE produce the natural ligands for CD22 (an inhibitor of B-cell receptor signaling and immune activation): 9-O-deacetylated, α2–6-linked sialic acid. Defects in the ST6GL1-SIAE-Siglec pathway could lead to the “unmasking” of CD22 due to the unavailability of its natural cis-ligands as proposed in a model by Pillai et al [39]. Interestingly, our microarray data indicates that SIAE is upregulated, and ST6GAL1 downregulated in the preeclamptic placenta (See Table 4.) We also corroborate the placental upregulation of SIGLEC6 in preeclampsia previously reported by others [40] (See Table 4). The functional consequence of SIAE, ST6GAL1, and SIGLEC6 transcriptional disregulation in preeclamptic placentas remains to be understood, particularly in the context of genetic variation of these transcripts.
Immune-mediated Model of Preeclamptic Pathogenesis
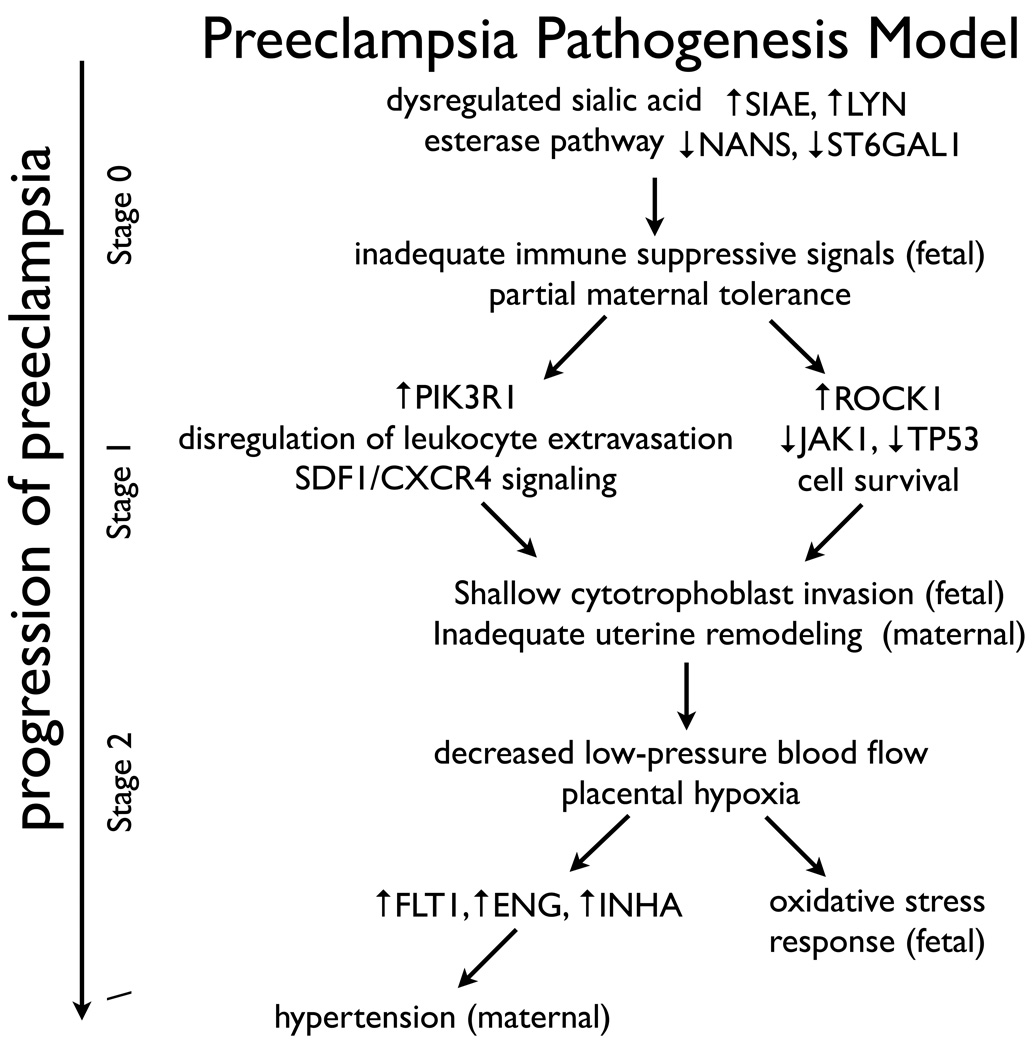
Our data is consistent with a model where trophoblast invasion is defective as a consequence of an immune-associated mechanism affecting cell migration, invasion, and survival. We propose that Sialic acid modifications play a role in either initiating or maintaining maternal immune tolerance, eventually leading to the clinical onset of preeclampsia. Failure of extravillous cytotrophoblasts to invade and remodel the maternal vasculature and replace the endothelial cells lining the spiral arteries are then predicted to be sufficient to decrease the amount of low-pressure blood flow, induce placental hypoxia and oxidative stress, and increase the expression of factors such as sFLT1, sENG, and INHA that lead to the clinical manifestation of preeclampsia in maternal hypertension and proteinuria. (See Figure 4.)
Figure 4. Preeclampsia pathogenesis model.
Our data supports a model where dysregulation of immune-associated signaling pathways, associated with atypical sialic acid modification, leads to shallow trophoblast invasion and inadequate uterine vascular remodeling. These changes are predicted to be sufficient to decrease the amount of low-pressure blood flow, induce placental hypoxia and oxidative stress, and increase the expression of factors such as sFlt1, sEng, and INHA that lead to the clinical manifestation of preeclampsia in maternal hypertension and proteinuria.
Future
This work presents the molecular characteristics of the preeclamptic placenta, by identifying a number of both known and novel factors and pathways. Molecular classification of these expression profiles might help to elucidate fetal versus maternal causes of the pregnancy-associated syndrome. While we have identified a number of genes and pathways we believe to be implicated in the etiology of preeclampsia, it is our hope that researchers in the field will use this comprehensive gene expression dataset to help guide their own efforts towards understanding the pathogenesis of this disorder. Finally, while many preeclampsia studies have focused on maternal genetic factors for association analysis, genetic analysis of the interactions between fetal and maternal genetics for disregulated genes such as SIAE could prove to be insightful.
Supplementary Material
Supplementary Figure 1. Mixed model power analysis. Full design models 1 & 2 (n=60) are sufficiently powered (>80%) to detect classification (preeclampsia – control) effect sizes > 0.4, which correspond to a 30% change in transcript levels. Reduced design models (n=30 and n=15), as well as estimated gestational age (34–37) subset models (n=23), are inadequately powered to detect these same effects.
Supplementary Figure 2. Leukocyte extravasation signaling pathway is disregulated in preeclamptic placentas. Green = upregulation in controls, Red = upregulation in preeclampsia.
Supplementary Figure 3. SDF1/CXCR4 signaling pathway is disregulated in preeclamptic placentas. Green = upregulation in controls, Red = upregulation in preeclampsia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author’s Contributions
ST performed all the experiments, and drafted the manuscript. NH and ST analyzed the data. BT, ST, and SB coordinated and collected patient samples. JP, AJ, and ST conceived of and designed the study; JP and AJ are the co-principal investigators. All authors read and approved of the final manuscript.
References
- 1.Roberts JM, Pearson G, Cutler J, Lindheimer M. Pregnancy NWGoRoHD. Summary of the NHLBI Working Group on Research on Hypertension During Pregnancy. Hypertension. 2003;41(3):437–445. doi: 10.1161/01.HYP.0000054981.03589.E9. [DOI] [PubMed] [Google Scholar]
- 2.Roberts JM, Redman CW. Pre-eclampsia: more than pregnancy-induced hypertension. Lancet. 1993;341(8858):1447–1451. doi: 10.1016/0140-6736(93)90889-o. [DOI] [PubMed] [Google Scholar]
- 3.Kanasaki K, Kalluri R. The biology of preeclampsia. Kidney International. 2009;76(8):831–837. doi: 10.1038/ki.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basso O, Rasmussen S, Weinberg C, Wilcox A, Irgens L, Skjaerven R. Trends in Fetal and Infant Survival Following Preeclampsia. JAMA: The Journal of the American Medical Association. 2006;296(11):1357. doi: 10.1001/jama.296.11.1357. [DOI] [PubMed] [Google Scholar]
- 5.MacKay AP, Berg CJ, Atrash HK. Pregnancy-related mortality from preeclampsia and eclampsia. Obstet Gynecol. 2001;97(4):533–538. doi: 10.1016/s0029-7844(00)01223-0. [DOI] [PubMed] [Google Scholar]
- 6.Odegard RA, Vatten LJ, Nilsen ST, Salvesen KA, Austgulen R. Risk factors and clinical manifestations of pre-eclampsia. BJOG. 2000;107(11):1410–1416. doi: 10.1111/j.1471-0528.2000.tb11657.x. [DOI] [PubMed] [Google Scholar]
- 7.Broughton Pipkin F. Risk factors for preeclampsia. N Engl J Med. 2001;344(12):925–926. doi: 10.1056/NEJM200103223441209. [DOI] [PubMed] [Google Scholar]
- 8.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nature Reviews Immunology. 2006;6(8):584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 9.Li DK, Wi S. Changing paternity and the risk of preeclampsia/eclampsia in the subsequent pregnancy. Am J Epidemiol. 2000;151(1):57–62. doi: 10.1093/oxfordjournals.aje.a010122. [DOI] [PubMed] [Google Scholar]
- 10.Tinsley JH, Chiasson VL, South S, Mahajan A, Mitchell BM. Immunosuppression improves blood pressure and endothelial function in a rat model of pregnancy-induced hypertension. American Journal of Hypertension. 2009;22(10):1107–1114. doi: 10.1038/ajh.2009.125. [DOI] [PubMed] [Google Scholar]
- 11.Sargent IL, Borzychowski AM, Redman CWG. NK cells and human pregnancy--an inflammatory view. Trends Immunol. 2006;27(9):399–404. doi: 10.1016/j.it.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Aluvihare VR, Kallikourdis M, Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. 2004;5(3):266–271. doi: 10.1038/ni1037. [DOI] [PubMed] [Google Scholar]
- 13.Cross JC. The genetics of pre-eclampsia: a feto-placental or maternal problem? Clin Genet. 2003;64(2):96–103. doi: 10.1034/j.1399-0004.2003.00127.x. [DOI] [PubMed] [Google Scholar]
- 14.Redman CWG, Sargent IL. Placental stress and pre-eclampsia: a revised view. Placenta. 2009;30 Suppl A:S38–S42. doi: 10.1016/j.placenta.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Muttukrishna S, Knight PG, Groome NP, Redman CW, Ledger WL. Activin A and inhibin A as possible endocrine markers for pre-eclampsia. Lancet. 1997;349(9061):1285–1288. doi: 10.1016/s0140-6736(96)09264-1. [DOI] [PubMed] [Google Scholar]
- 16.Lindheimer MD, Woodruff TK. Activin A, inhibin A, and pre-eclampsia. Lancet. 1997;349(9061):1266–1267. doi: 10.1016/S0140-6736(05)62502-0. [DOI] [PubMed] [Google Scholar]
- 17.Mise H, Sagawa N, Matsumoto T, Yura S, Nanno H, Itoh H, Mori T, Masuzaki H, Hosoda K, Ogawa Y, Nakao K. Augmented placental production of leptin in preeclampsia: possible involvement of placental hypoxia. J Clin Endocrinol Metab. 1998;83(9):3225–3229. doi: 10.1210/jcem.83.9.5117. [DOI] [PubMed] [Google Scholar]
- 18.Venkatesha S, Toporsian M, Lam C, Hanai J-i, Mammoto T, Kim YM, Bdolah Y, Lim K-H, Yuan H-T, Libermann TA, Stillman IE, Roberts D, D'amore PA, Epstein FH, Sellke FW, Romero R, Sukhatme VP, Letarte M, Karumanchi SA. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med. 2006;12(6):642–649. doi: 10.1038/nm1429. [DOI] [PubMed] [Google Scholar]
- 19.Maynard SE, Min J-Y, Merchan J, Lim K-H, Li J, Mondal S, Libermann TA, Morgan JP, Sellke FW, Stillman IE, Epstein FH, Sukhatme VP, Karumanchi SA. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111(5):649–658. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torry DS, Wang HS, Wang TH, Caudle MR, Torry RJ. Preeclampsia is associated with reduced serum levels of placenta growth factor. American journal of obstetrics and gynecology. 1998;179(6 Pt 1):1539–1544. doi: 10.1016/s0002-9378(98)70021-3. [DOI] [PubMed] [Google Scholar]
- 21.Carty DM, Delles C, Dominiczak AF. Novel biomarkers for predicting preeclampsia. Trends Cardiovasc Med. 2008;18(5):186–194. doi: 10.1016/j.tcm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, Libermann TA, Morgan JP, Sellke FW, Stillman IE, Epstein FH, Sukhatme VP, Karumanchi SA. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111(5):649–658. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L, Goodsaid FM. Evaluation of DNA microarray results with quantitative gene expression platforms. Nature Biotechnology. 2006;24(9):1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 25.Idaghdour Y, Czika W, Shianna KV, Lee SH, Visscher PM, Martin HC, Miclaus K, Jadallah SJ, Goldstein DB, Wolfinger RD, Gibson G. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2009 doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 27.Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, Darot JFJ, Ritchie ME, Lynch AG, Tavaré S. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Research. 2010;38(3):e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 29.Sitras V, Paulssen RH, Grønaas H, Leirvik J, Hanssen TA, Vårtun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30(5):424–433. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Brettell R, Yeh PS, Impey LWM. Examination of the association between male gender and preterm delivery. Eur J Obstet Gynecol Reprod Biol. 2008;141(2):123–126. doi: 10.1016/j.ejogrb.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 31.López-Novoa JM. Soluble endoglin is an accurate predictor and a pathogenic molecule in pre-eclampsia. Nephrol Dial Transplant. 2007;22(3):712–714. doi: 10.1093/ndt/gfl768. [DOI] [PubMed] [Google Scholar]
- 32.Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K, Kramer A, Rumer KK, Haimov-Kochman R, Yeh RF, Overgaard MT, Varki A, Oxvig C, Fisher SJ. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150(1):452–462. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Genbacev O, Fisher SJ. The human placenta remodels the uterus by using a combination of molecules that govern vasculogenesis or leukocyte extravasation. Ann N Y Acad Sci. 2003;995:73–83. doi: 10.1111/j.1749-6632.2003.tb03211.x. [DOI] [PubMed] [Google Scholar]
- 34.Surolia I, Pirnie SP, Chellappa V, Taylor KN, Cariappa A, Moya J, Liu H, Bell DW, Driscoll DR, Diederichs S, Haider K, Netravali I, Le S, Elia R, Dow E, Lee A, Freudenberg J, De Jager PL, Chretien Y, Varki A, Macdonald ME, Gillis T, Behrens TW, Bloch D, Collier D, Korzenik J, Podolsky DK, Hafler D, Murali M, Sands B, Stone JH, Gregersen PK, Pillai S. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature. 2010 doi: 10.1038/nature09115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takematsu H, Diaz S, Stoddart A, Zhang Y, Varki A. Lysosomal and cytosolic sialic acid 9-O-acetylesterase activities can Be encoded by one gene via differential usage of a signal peptide-encoding exon at the N terminus. J Biol Chem. 1999;274(36):25623–25631. doi: 10.1074/jbc.274.36.25623. [DOI] [PubMed] [Google Scholar]
- 36.Stoddart A, Zhang Y, Paige CJ. Molecular cloning of the cDNA encoding a murine sialic acid-specific 9-O-acetylesterase and RNA expression in cells of hematopoietic and non-hematopoietic origin. Nucleic Acids Research. 1996;24(20):4003–4008. doi: 10.1093/nar/24.20.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin LT, Marth JD, Varki A, Varki NM. Genetically altered mice with different sialyltransferase deficiencies show tissue-specific alterations in sialylation and sialic acid 9-O-acetylation. J Biol Chem. 2002;277(36):32930–32938. doi: 10.1074/jbc.M203362200. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh S, Bandulet C, Nitschke L. Regulation of B cell development and B cell signalling by CD22 and its ligands alpha2,6-linked sialic acids. Int Immunol. 2006;18(4):603–611. doi: 10.1093/intimm/dxh402. [DOI] [PubMed] [Google Scholar]
- 39.Pillai S, Cariappa A, Pirnie SP. Esterases and autoimmunity: the sialic acid acetylesterase pathway and the regulation of peripheral B cell tolerance. Trends Immunol. 2009;30(10):488–493. doi: 10.1016/j.it.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K, Kramer A, Rumer KK, Haimov-Kochman R, Yeh R-F, Overgaard MT, Varki A, Oxvig C, Fisher SJ. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150(1):452–462. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Mixed model power analysis. Full design models 1 & 2 (n=60) are sufficiently powered (>80%) to detect classification (preeclampsia – control) effect sizes > 0.4, which correspond to a 30% change in transcript levels. Reduced design models (n=30 and n=15), as well as estimated gestational age (34–37) subset models (n=23), are inadequately powered to detect these same effects.
Supplementary Figure 2. Leukocyte extravasation signaling pathway is disregulated in preeclamptic placentas. Green = upregulation in controls, Red = upregulation in preeclampsia.
Supplementary Figure 3. SDF1/CXCR4 signaling pathway is disregulated in preeclamptic placentas. Green = upregulation in controls, Red = upregulation in preeclampsia.