Abstract
Residing within the intestine is a large community of commensal organisms collectively termed the microbiota. This community generates a complex nutrient environment by breaking down indigestible food products into metabolites that are used by both the host and the microbiota. Both the invading intestinal pathogen and the microbiota compete for these metabolites, which can shape both the composition of the flora, as well as susceptibility to infection. After infection is established, pathogen mediated inflammation alters the composition of the microbiota, which further shifts the makeup of metabolites in the gastrointestinal tract. A greater understanding of the interplay between the microbiota, the metabolites they generate, and susceptibility to enteric disease will enable the discovery of novel therapies against infectious disease.
Metabolic function of the host microbiota
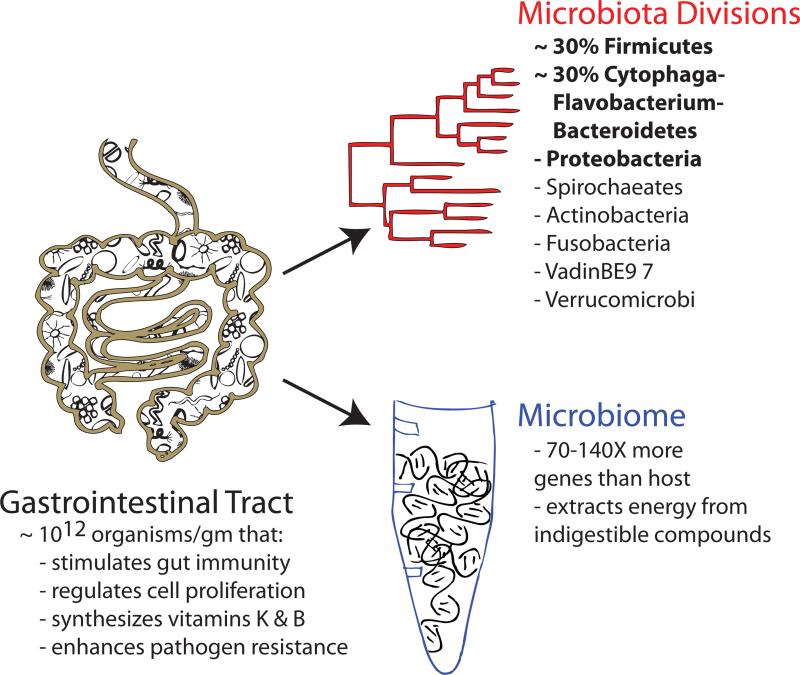
The resident microbiota of the human gastrointestinal (GI) tract is incredibly dense and diverse, containing as many as 1012 organisms per gram [1]. While all three domains of life (bacteria, archaea and eukarya) have been identified in the adult human GI tract, only 8 out of 55 known bacterial divisions have been found within this environment [2]. The composition of this enormous community provides the host with a core set of microbial genes that encode the gut microbiome [3]. It is estimated that this microbial community has 70–140 times more total genes than the human host, which encode biochemical pathways that humans have not evolved, enabling the break-down of proteins and indigestible polysaccharides into essential amino acids, vitamins, and short chain fatty acids (SCFAs) (Figure 1) [4]. Shifts in the composition of the microbiota and microbiome that correlate with obesity are associated with an increased presence of organisms and genes that ultimately increase energy extraction, storage and usage of consumed nutrients [3,5]. As the microbiota between two individuals can deviate by hundreds of species and thousands of strain differences, the corresponding metabolite environment in the GI tract for each person is unique [2,6].
Figure 1. Functions of the host microbiota.
Within the gastrointestinal (GI) tract is a community of commensal organisms, the microbiota, with an estimated density as high as 1012 organisms per gram of content [1]. Out of a total of 55 bacterial divisions identified thus far, only 8 have been identified within the human GI tract (dominant divisions are in bold) [2]. The genes encoded by this massive community are collectively termed the microbiome, which encodes an estimated 70–140 times more genes than encoded by its the human host [3,4]. Together, the organisms that reside in the GI tract and the genes they encode are necessary for the completion of essential tasks for the host, including stimulating gut immunity, regulating cell proliferation, vitamin synthesis, and mediating resistance to pathogen invasion and colonization.
Barrier function of the host microbiota
The microbiota can act as a barrier against incoming pathogens; this phenomenon has been described as colonization resistance [7]. There are several theories why the microbiota prevents pathogen colonization; some members of the microbiota can physically occupy pathogen attachment sites, some members can stimulate the mucosal immune system to alter tolerance of an invading pathogen, while others can consume nutrients the pathogen requires to proliferate.
One strategy to alter colonization resistance includes exogenously adding nutrients, also known as prebiotics, to promote the growth of individual microbiota species. An increased presence of Bifidobacteria and Lactobacilli, for example, has been shown to suppress bacterial infections caused by Salmonella enterica serovar Typhimurium (S. Typhimurium) [8,9]. However, prebiotic supplementation with nutrients thought to promote in vivo growth of Bifidobacteria and Lactobacilli did not inhibit S. Typhimurium colonization in vivo, but instead increased pathogen colonization when compared to mice fed a standard diet [10]. These studies indicate that identification of beneficial microbiota communities, and the nutrients that shape their composition, may be incomplete or not specific enough.
Host microbiota nutrient competition
Members of the GI microbiota have acquired specific mechanisms to exploit their environment and the nutrients available to them [11–13]. A recent global analysis of the microbiome encoded by 124 individuals revealed that there is a core set of genes that are likely required by any bacterium to thrive in the GI tract, including genes involved in the biodegradation of complex sugars and glycans present in the intestinal lining [14]. Evidence of the adaptability of the Bacteroides genus to host glycans was recently demonstrated when germ-free mice were co-colonized with B. thetaiotaomicron and a member of another common microbial community phyla, the Firmicutes' Eubacterium rectale. As a consequence of competition for dietary nutrients, B. thetaiotaomicron up-regulated glycoside hydrolases and signaled the host to produce mucosal glycans, presumably so that it could access a nutrient source its competitor E. rectale could not utilize [15].
Competition for nutrients is a strong force among the Bacteroides genus. During competition for dietary fructans, B. thetaiotaomicron uses a hybrid two-component signaling sensor to enable degradation and usage of fructans [16]. Additionally, genes that encode porphyranases and agarases, which enable some microbes commonly found in Japanese community members to digest seaweed, may have been transferred to the gut bacterium Bacteroides plebeius from the seaweed associated bacterium Zobellia galactanivorans, allowing B. plebeius to extract energy from otherwise indigestible food products [17]. A mutant library of B. thetaiotaomicron revealed that this microbe is highly adaptive to the microbiota composition and the availability of nutrients such as vitamin B12 [18].
Just what is the nutrient environment to which these commensal bacteria are adapting? During colonization of germ-free mice, commensal Escherichia coli were shown to utilize arginine, asparagine, aspartate, glucose/galactose, ribose, maltose, glucuronate, galacturonate and gluconate as substrates [19]. However, the nutrient environment of the GI tract is likely to be more complex with the addition of other microbiota members. For example, the levels of SCFAs found within germ-free animals are lower than in mice that had received a gut microbiota transplant from conventionally raised donors [20]. SCFA production has further been linked to the Firmicutes, as following antibiotic treatment of conventional mice, SCFAs decreased along with the diversity of the Firmicutes [21]. The production of some metabolites may even be a collaborative effort between distant community members, as observations indicate that E. rectale uses B. thetaiotaomicron produced acetate to generate the SCFA butyrate [15].
Furthermore, in soil bacterial communities, physical fungal-bacterial interactions lead to the activation of fungal secondary metabolism genes that are normally silent under laboratory conditions [22].
The composition of the host microbiota alters the outcome of enteric pathogens
The composition of the host microbiota influences the susceptibility to enteric pathogen colonization, as microbiota communities with low complexity are increasingly prone to pathogenic colonization [23]. Evidence of this phenomenon was recently demonstrated when susceptibility to S. Typhimurium was increased after administration of clinically relevant doses of antibiotics that did not change the overall bacterial load of the microbiota, but did change the ratio of Firmicutes to Bacteroidetes [24,25]. The influence of the host microbiota upon an invading pathogen is not restricted to prokaryotes, as hatching of the parasitic nematode Trichuris muris in the large intestine of mice is dependent upon physical contact of the parasitic egg with microorganisms present in the gut microbiota [26]. The composition of the host microbiota has also recently been linked to eventual pathogen clearance [27]. Why the composition of the host microbiota is critical for initial colonization and eventual clearance by these pathogens is unclear. One theory, amongst many, is that the microbiota provides metabolites that can hinder or enhance virulence of enteric pathogens.
Enteric pathogens compete for carbon within the GI tract
Primary metabolites in the GI tract are in high demand, with many of them absorbed by the host, or consumed or converted into secondary metabolites by the microbiota [28]. The composition of one primary class of nutrients, carbohydrates, is controlled by members of the host microbiota. B. thetaiotaomicron alone contains over four times the number of genes involved in acquiring and metabolizing carbohydrates than the human host, and other Bifidobacterium strains secrete polysaccharide-hydrolyzing enzymes that ferment primary fructooligosaccharides into the secondary disaccharide lactate in the GI tract [2,29].
The ability of enterohemmorrhagic E. coli (EHEC) O157:H7 to catabolize the disaccharide maltose and other secondary carbon sources helps it compete with commensal strains of E. coli for colonization of the GI tract in a streptomycin-treated mouse model of infection [30]. The ability to exploit carbon sources to enhance virulence is not limited to EHEC. A recently constructed genome scale metabolic model for S. Typhimurium and S. Enteritidis revealed that these pathogens diverge from a commensal strain of E. coli, with the majority of the compounds that the pathogenic strains preferentially utilize over the commensal strain being carbon substrates [31]. Energy generation and colonization by the food-borne bacterial pathogen Campylobacter jejuni, which resides in the GI tract of its avian reservoir, depends upon scavenging of free amino and keto acids and chemotaxis towards the carbon sources asparagine, formate, lactate and chicken mucus [32–34]. In Vibrio cholerae, passage through the intestinal tract induces genes involved in succinate, glycine, and chitin utilization that enhance the ability of the pathogen to persist within aquatic environments, an important trait that enhances transmission and propagation of this water-borne pathogen [35]. These observations suggest that multiple enteric pathogens have the ability to respond to different carbon environments, and this response is beneficial for a pathogenic lifecycle. As the carbon environment is modulated by the host microbiota, understanding how the microbiota composition controls carbon sources that pathogens exploit will almost certainly lead to unique strategies to control colonization and virulence.
The microbiota can alter virulence properties of enteric pathogens
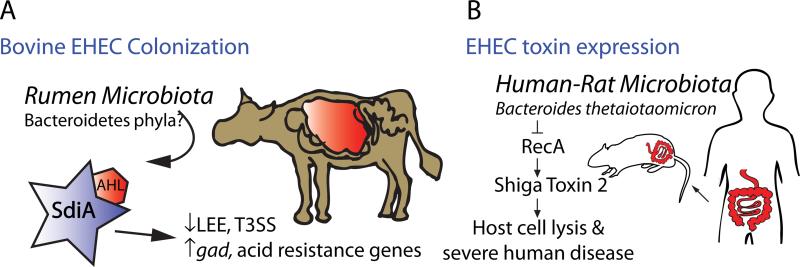
EHEC O157:H7 responds to metabolites secreted by the host microbiota to induce or repress virulence genes [36,37] (Figure 2). EHEC is transmitted to humans primarily through ingestion of foods contaminated by colonized cattle [38]. To colonize cattle, EHEC requires SdiA, a regulator that senses acyl-homoserine lactones (AHLs), which are produced by some members of the Bacteroidetes phyla [36] (Figure 2). Additionally, a virulence factor of EHEC, Shiga toxin 2 (Stx2), is produced and released into the environment by activated RecA, which induces the lytic cycle of the bacteriophage that encodes Stx2. Germ-free rats that are then colonized with human microbiota secrete molecules that both repress stx2 mRNA expression, and inhibit the RecA mediated lytic cycle independent of known quorum-sensing pathways (involving SdiA, QseA, QseC or AI-3). B. thetaiotaomicron was shown to produce this inhibitory factor, implicating a member of the human microbiota in repressing a bacterial virulence factor [37] (Figure 2). Together, these two studies demonstrate that metabolites secreted by the host microbiota may modulate EHEC colonization and virulence gene expression in two distinct hosts, its cattle reservoir, as well as its human host.
Figure 2. Chemical sensing between the microbiota and EHEC.
(A) Members of the Bacteroidetes phyla produce acyl-homoserine lactones (AHLs). These signaling molecules are prominent within the rumen of the bovine gut, but not in other areas of the GI tract. AHLs isolated from the rumen stabilize folding of SdiA, an EHEC regulator that is necessary for colonization of cattle. Specifically, the rumen AHL-SdiA complex represses transcription and protein production by the locus of enterocyte effacement (LEE), a pathogenicity island that enables EHEC to colonize and promote disease in its human host, an undesirable phenotype for commensal colonization of cattle. Conversely, the AHL-SdiA complex activated the expression of gad acid-resistance genes and promoted survival in low pH, a phenotype necessary for EHEC survival within the acidic stomachs of the cow [36]. (B) Shiga Toxin 2 (Stx2) is a major virulence factor of EHEC O157:H7, which causes protein synthesis inhibition and ultimately cell death in the human host. Prokaryotes of conventionalized rats colonized with human microbiota produced molecules which repressed RecA mediated stx2 mRNA expression and Stx2 production. Subsequent analysis revealed that these inhibitory prokaryotic molecules are produced in part by Bacteroides thetaiotaomicron, a member of the normal human intestinal microbiota [37].
Spatial nutrient differences could lead to pathogen tropism
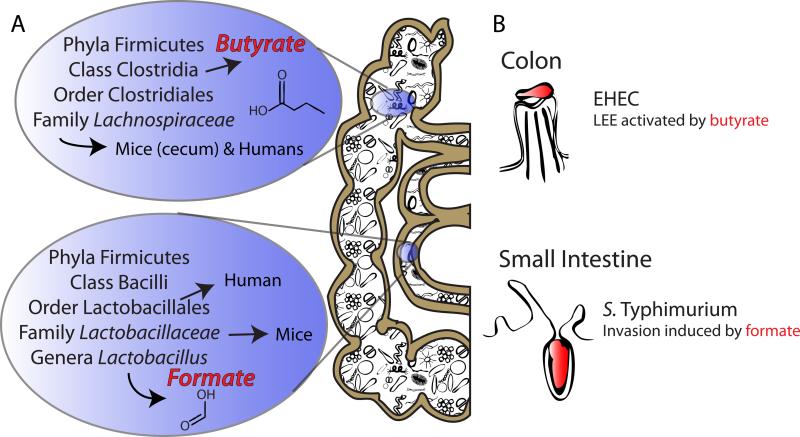
Enteric pathogens preferentially colonize different regions of the GI tract, such as S. Typhimurium in the small intestine, and EHEC in the distal ileum and colon [39]. One potential reason for tissue tropism may be because metabolites that influence pathogen replication and virulence are differentially available in these regions. SCFAs are known to have significant influence upon enteric pathogenicity, and HIV is just one pathogen of many that have recently been demonstrated to respond to SCFAs, as they cue HIV activation in the gut [40]. SCFAs are also known to influence the inflammatory host immune response [41]. The composition of SCFAs in the GI tract is modulated by the microbiota. For example, the majority of the SCFA butyrate is produced by the resident microbiota [42], with butyrate amounts varying depending on the activity and composition of the microbiota [21,43]. Additionally, the levels of individual SCFAs vary in different regions of the GI tract, with formate and acetate predominating in the small intestine, while propionate and butyrate are higher in the colon [21,44,45] (Figure 3).
Figure 3. Short-chain fatty acid (SCFA) influence upon pathogen tropism.
(A) The Firmicutes are a principal phyla in both the small intestine and the colon, with the family Lachnospiraceae dominating the colon [21,53]. The Lachnospiraceae are members of the Clostridia class, which are major producers of butyrate in the human colon [43,53]. The Lactobacillales order of the Bacillus class dominate the small intestine in humans, and upon further examination in mice, the family Lactobacillaceae within this order compose 24% of the total small intestine microbiota [21,53]. Genera belonging to this family include Lactobacillus, which heterofermentatively can produce formate as well as acetate and lactate. (B) EHEC primarily colonizes the colon of humans, where butyrate is a dominant SCFA [21,39,44,45]. In EHEC, butyrate activates the locus of enterocyte effacement (LEE) and enhances adherence of this pathogen in tissue culture [39,48]. Salmonella enterica serovar Typhimurium (S. Typhimurium) colonizes the small intestine, where formate is a dominant SCFA. The SCFA formate induces the expression of invasion genes in S. Typhimurium, while butyrate is known to repress these genes [46,47].
Interestingly, the SCFA formate acts as a diffusible signal to induce the expression of invasion genes in S. Typhimurium, while butyrate, a SCFA present at higher concentrations in lower regions of the GI tract, is known to repress invasion genes [46,47]. Conversely, exposure to butyrate enhances adherence of EHEC to Caco-2 cell monolayers during a tissue culture model of infection [48]. Furthermore, butyrate was also shown to influence activation of the locus of enterocyte effacement pathogenicity island of EHEC, which carries genes involved in the formation of attaching and effacing (A/E) lesions on intestinal epithelial cells [39,48]. In both S. Typhimurium and EHEC, the SCFA that enhances virulence, formate and butyrate, respectively, is highest in the region of the GI tract that these enteric pathogens preferentially colonize.
Pathogen mediated inflammation alters the microbiota and nutrient composition of the gut, further enhancing colonization
While the nutrient environment, which is modulated by the resident microbiota, influences initial colonization by an enteric pathogen, subsequent changes in the composition of the microbiota also lead to downstream changes in the nutrient environment and enteric colonization potential of the gut. Colonization by Citrobacter rodentium, a close relative of EPEC and EHEC, causes an inflammatory response in the GI tract, which corresponds to a major alteration in the composition of the microbiota [24]. The inflammatory response and corresponding alterations to the host microbiota have been linked to further increases in pathogen growth, as well as an increased release of glycan and amino-acid rich mucins [24,49]. Additionally, acute gut inflammation caused by S. Typhimurium infection has recently been demonstrated to generate a respiratory electron acceptor, tetrathionate, that provides a competitive growth advantage to the pathogen over the competing microbiotia [50]. The release of nutrient rich compounds, such as mucins and glycans, as well as other novel growth factors, such as tetrathionate, likely foster pathogen growth, signifying that pathogen mediated inflammation and microbiota perturbations could be a mechanism employed by the pathogen to enhance its ability to replicate in the host after an initial infection has already been established.
Conclusions
One underexploited opportunity to prevent enteric infections is to target the mechanisms pathogenic bacteria undertake to respond to the unique nutritional environment found within the GI tract [51]. Because the rate of passage through the GI tract is rapid, the ability to respond and compete for nutrients is likely to be one of the most important factors controlling the success or failure of an invading pathogen [52]. As this nutrient environment is shared between pathogens and the host microbiota, novel avenues to control infection before and after the onset of disease can be discovered by carefully studying the mechanisms enteric pathogens and members of the host microbiota utilize to generate, compete, and exploit the nutrients within the GI tract.
Abbreviations used
- (GI)
Gastrointestinal
- (SCFAs)
short chain fatty acids
- (S. Typhimurium)
Salmonella enterica serovar Typhimurium
- (EHEC)
enterohemmorrhagic E. coli
- (AHLs)
acyl-homoserine lactones
- (Stx2)
Shiga toxin 2
- (A/E)
attaching and effacing
- (LEE)
locus of enterocyte effacement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- **3.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors characterized the microbiota and microbiomes of lean and obese twin pairs. They demonstrated that a core microbiome of shared genes exists between all individuals they characterized, and that between the obese and lean pairs, shifts in the composition of the microbiota correlated with corresponding shifts in microbiome genes that are involved in energy uptake.
- 4.Possemiers S, Grootaert C, Vermeiren J, Gross G, Marzorati M, Verstraete W, Van de Wiele T. The intestinal environment in health and disease - recent insights on the potential of intestinal bacteria to influence human health. Curr Pharm Des. 2009;15:2051–2065. doi: 10.2174/138161209788489159. [DOI] [PubMed] [Google Scholar]
- **5.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate by 454 pyrosequencing of 3 normal weight, 3 morbidly obese, and 3 post-gastric-bypass surgery individuals that some members of the microbiota are more abundant in the obese. Specifically, they saw increased numbers of H(2)-producing Prevotellaceae and H(2)-utilizing methanogenic Archaea in obese individuals, and they hypothesized that interpecies H(2)-transfer between these populations may be an important mechanism to increase energy uptake in the obese.
- *6.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS ONE. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper showed that changes in the microbiota seen in Crohns disease patients correlated with changes in their metabolite profiles.
- 7.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiology Reviews. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Silva AM, Barbosa FHF, Duarte R, Vieira LQ, Arantes RME, Nicoli JR. Effect of Bifidobacterium longum ingestion on experimental salmonellosis in mice. J Appl Microbiol. 2004;97:29–37. doi: 10.1111/j.1365-2672.2004.02265.x. [DOI] [PubMed] [Google Scholar]
- 9.Truusalu K, Mikelsaar R-H, Naaber P, Karki T, Kullisaar T, Zilmer M, Mikelsaar M. Eradication of Salmonella Typhimurium infection in a murine model of typhoid fever with the combination of probiotic Lactobacillus fermentum ME-3 and ofloxacin. BMC Microbiology. 2008;8:132. doi: 10.1186/1471-2180-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Petersen A, Heegaard PMH, Pedersen AL, Andersen JB, Sørensen RB, Frøkiaer H, Lahtinen SJ, Ouwehand AC, Poulsen M, Licht TR. Some putative prebiotics increase the severity of Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 2009;9:245. doi: 10.1186/1471-2180-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Administration of prebiotics to prevent S. Typhimurium SL1344 infection in BALB/c mice did not decrease colonization, and in some examples, resulted in significantly increased colonization by the pathogen.
- *11.Tagkopoulos I, Liu Y, Tavazoie S. Anticipatory Behavior Within Microbial Genetic Networks. Science. 2008 doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides support to the hypothesis that Escherichia coli can anticipate temperature and oxygen environments and ready itself for exploitation of these environments by altering gene transcription in advance.
- *12.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]; The paper suggests that the nutrients and stresses encountered by E. coli and Saccharomyces cerevisiae early in colonization may prepare the microorganism for exploitation and/or survival to these same nutrients and stresses later in the gastrointestinal tract.
- *13.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that activation of central metabolic pathways can quickly change in response to environmental changes by reversible lysine acetylation of metabolic enzymes.
- *14.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using Illumina-based metagenomic sequencing from fecal samples of 124 individuals, the gene set of the gut microbiota was cataloged, revealing a core set of microbial genes and corresponding functions.
- **15.Mahowald M, Rey F, Seedorf H, Turnbaugh P, Fulton R, Wollam A, Shah N, Wang C, Magrini V, Wilson R, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors determine that during colonization of germ-free mice, Bacteroides thetaiotaomicron adapts to Eubacterium rectale by upregulating polysaccharide utilization loci and also signals the host to produce mucosal glycans that E. rectale cannot utilize. In turn, E. rectale adapts to B. thetaiotaomicron by decreasing production of glyan-degrading enzymes and increasing amino acid and sugar transporters, and also produces butyrate from B. thetaiotaomicron produced acetate.
- **16.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; The ability to use fructans by the Bacteroidetes phyla confers a competitive advantage in vivo, with the gene content of a fructan utilization locus differing between different members of this dominant intestinal microbiota group.
- **17.Hehemann J-H, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]; In this unique study, the authors demonstrate that genes that encode porphyranases produced by a marine Bacteroidetes member, agarases, and associated proteins have been transferred to gastrointestinal tract Bacteroidetes microbiota members in groups of Japanese populations, but not North American populations. This study therefore suggests that consumption of non-sterile seaweed, with its associated marine bacteria, is a mechanism of gene transport in Japanese populations and this transfer may increase the ability of those populations to extract energy from dietary seaweed polysaccharides.
- **18.Goodman AL, Mcnulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors used massively parallel sequencing to monitor the abundance of transposon mutants of Bacteroides thetaiotaomicron during colonization of germ-free and conventional mice. They revealed the adaptability of B. thetaiotaomicron to the composition of the microbiota, as well as its ability to compete for nutrients such as vitamin B(12).
- **19.Alpert C, Scheel J, Engst W, Loh G, Blaut M. Adaptation of protein expression by Escherichia coli in the gastrointestinal tract of gnotobiotic mice. Environ Microbiol. 2009;11:751–761. doi: 10.1111/j.1462-2920.2008.01798.x. [DOI] [PubMed] [Google Scholar]; During colonization of formerly germ-free mice, commensal E. coli expresses proteins that likely enable the bacterium to adapt to the nutritional environment of the host intestine. Some of these expressed proteins are involved in utilization of arginine, asparagine, aspartate, glucose/galactose, ribose, maltose, glucuronate, galacturonate and gluconate.
- **20.Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that short-chain fatty acids in the gastrointestinal tract are influenced by the presence of a host microbiota, and in turn, the nutrients throughout the host are altered by this gut community.
- **21.Garner C, Antonopoulos D, Wagner B, Duhamel G, Keresztes I, Ross D, Young V, Altier C. Perturbation of the small intestinal microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect Immun. 2009 doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that short-chain fatty acids have a different composition in different locations of the gastrointestinal tract, and furthermore, they are altered by antibiotic treatment and ensuing microbiota perturbation of the host.
- *22.Schroeckh V, Scherlach K, Shelest E, SchmidtHeck W, Schuemann J, Martin K, Hertweck C, Brakhage A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cocultivation of the fungus Aspergillus nidulans with soil-dwelling actinomycetes revealed that fungal-bacterial interaction leads to activation of fungal secondary metabolism genes.
- **23.Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, Mccoy KD, von Mering C, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper suggests that a microbiota with less diversity increases host susceptibility to S. enterica colonization, but in a host that contains a more diverse microbiota, the presence of similar phylotypes increases the likelyhood of colonization by members of the same phyla.
- **24.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]; The microbiota community of the colon is dramatically altered after host-mediated inflammation is triggered from bacterial infection or DSS treatment, with post-inflammation composition shifts containing an increased proportion of anaerobic bacteria, particularly of the Enterobacteriaceae family.
- **25.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008 doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Administration of clinically relevant doses of antibiotics to mice shifted the composition of the host microbiota and increased susceptibility to S. Typhimurium intestinal colonization.
- *26.Hayes KS, Bancroft AJ, Goldrick M, Portsmouth C, Roberts IS, Grencis RK. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science. 2010;328:1391–1394. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]; Physical interaction between the host microbiota and eggs of the nematode parasite Trichuris muris influences parasitic hatching.
- **27.Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, Van Maele L, Sirard J-C, Mueller A, Heikenwalder M, et al. The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Salmonella Diarrhea. PLoS Pathogens. 2010;6 doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]; By monitoring for S. Typhimurium clearance in streptomycin-treated mice, the authors observed that abolishing the secretory antibody response did not affect the kinetics of pathogen clearance from the gut lumen. Instead, mice containing a low complexity of gut flora were only able to clear S. Typhimurium from the gut lumen after transferring a normal complex microbiota to the mice, suggesting that the microbiota plays a critical role in eventual pathogen clearance from the host.
- 28.Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 29.Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol. 2005;71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30.Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, Struve C, Krogfelt KA, Cohen PS, Conway T. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun. 2008;76:2531–2540. doi: 10.1128/IAI.00096-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; The ability to catabolize the disaccharide maltose provides a competitive advantage to EHEC and K-12 strains of E. coli during colonization of the gastrointestinal tract of mice treated with streptomycin. This paper suggests that sugars used by E. coli are present in limiting quantities in the GI tract, and furthermore implies that the ability to adapt and utilize these carbon sources is critical for successful colonization.
- *31.Abuoun M, Suthers P, Jones G, Carter B, Saunders M, Maranas C, Woodward M, Anjun M. Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J Biol Chem. 2009 doi: 10.1074/jbc.M109.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]; A genome-scale pathogenic Salmonella metabolic model was constructed and compared with a metabolic model of a commensal strain of Escherichia coli, and used to predict differences in nutrient utilization. These in silico growth predictions were then compared with phenotyping microarray data, confirming that nutrient utilization differences exist between pathogens and commensal organisms.
- 32.Velayudhan J, Jones MA, Barrow PA, Kelly DJ. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infection and Immunity. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrixson DR, Dirita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Molecular Microbiology. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- *34.Vegge CS, Brøndsted L, Li Y-P, Bang DD, Ingmer H. Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl Environ Microbiol. 2009;75:5308–5314. doi: 10.1128/AEM.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; In Campylobacter jejuni, energy taxis is a primary force in environmental navigation, underlying the importance of the carbon environment of the host in chicken colonization and virulence.
- 35.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host & Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Hughes DT, Terekhova DA, Liou L, Hovde CJ, Sahl JW, Patankar AV, Gonzalez JE, Edrington TS, Rasko DA, Sperandio V. Chemical sensing in mammalian host-bacterial commensal associations. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.1002551107. [DOI] [PMC free article] [PubMed] [Google Scholar]; EHEC acyl-homoserine lactone detection through SidA may help the pathogen recognize commensal bacteria, such as the Bacteroidetes phyla. Furthermore, SdiA is necessary for EHEC cattle colonization, a reservoir for this deadly human disease, and SdiA-AHL chemical signaling may aid in the pathogens adaptation to the commensal environment of the gastrointestinal tract.
- **37.de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77:783–790. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Production of Shiga Toxin, a toxin which EHEC releases in the human gut, is repressed by small molecules that are secreted by Bacteriodes thetaiotaomicron, a predominant member of the human microbiota.
- 38.Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci USA. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- **40.Kantor B, Ma H, Webster-Cyriaque J, Monahan P, Kafri T. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0905859106. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper raises the possibility that gut microbiota communities that are rich in short-chain fatty acid production increase gene expression and replication of nonintegrating HIV-1.
- **41.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]; Short-chain fatty acids, produced by the microbiota, influence the inflammatory state of the gut by binding the G-protein-coupled receptor 43. Furthermore, mice deficient in GPR43 are unable to resolve inflammation, similar to what is observed in germ-free mice that are deficient in production of short-chain fatty acids.
- 42.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 43.Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 44.Argenzio RA, Southworth M. Sites of organic acid production and absorption in gastrointestinal tract of the pig. Am J Physiol. 1975;228:454–460. doi: 10.1152/ajplegacy.1975.228.2.454. [DOI] [PubMed] [Google Scholar]
- 45.Laerke HN, Jensen BB, Højsgaard S. In vitro fermentation pattern of D-tagatose is affected by adaptation of the microbiota from the gastrointestinal tract of pigs. J Nutr. 2000;130:1772–1779. doi: 10.1093/jn/130.7.1772. [DOI] [PubMed] [Google Scholar]
- **46.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. Butyrate specifically down-regulates salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol. 2006;72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; In vitro, butyrate supplementation to growth medium down-regulates the expression of pathogenicity island 1 genes of S. Enteritidis and S. Typhimurium.
- **47.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. Formate acts as a diffusible signal to induce Salmonella invasion. Journal of Bacteriology. 2008;190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Intestinal epithelial invasion of S. Typhimurium mediated by Salmonella pathogenicity island 1 is inhibited by deleting acetate kinase and phosphotransacetylase genes. These mutants secreted less formate, and when exogenous formate was supplemented, invasion was restored, suggestive that formate found in the distal ileum of the host can trigger invasion.
- **48.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology (Reading, Engl) 2009;155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]; The short-chain fatty acid butyrate, at low concentrations, enhances promoter activity of a positive regulator of the locus of enterocyte effacement through a leucine-responsive regulatory protein. The authors suggest that upon entering the distal ileum, EHEC responds to increased levels of butyrate by increasing virulence expression and this enables efficient colonization of their target niche.
- 49.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- **50.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]; During the conversion of microbiota generated toxic hydrogen sulphide gas by mammalian cells, thiosulphates are oxidized to tetrathionate by reactive oxygen species that are generated during S. Typhimurium invasion. S. Typhimurium can metabolize the resulting tetrathionate, promoting its colonization of the host.
- 51.Brown S, Palmer K, Whiteley M. Revisiting the host as a growth medium. Nat Rev Microbiol. 2008 doi: 10.1038/nrmicro1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:686–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]