Abstract
It is estimated that more than 90% of human genes express multiple mRNA transcripts due to alternative splicing. Consequently, the proteins produced by different splice variants will likely have different functions and expression levels. Several genes with splice variants are known in bone, with functions that affect osteoblast function and bone formation. The primary goal of this study was to evaluate the extent of alternative splicing in a bone subjected to mechanical loading and subsequent bone formation. We used the rat forelimb loading model, in which the right forelimb was loaded axially for 3 minutes, while the left forearm served as a non-loaded control. Animals were subjected to loading sessions every day, with 24 hours between sessions. Ulnae were sampled at 11 time points, from 4 hours to 32 days after beginning loading. RNA was isolated and mRNA abundance was measured at each time point using Affymetrix exon arrays (GeneChip® Rat Exon 1.0 ST Arrays). An ANOVA model was used to identify potential alternatively spliced genes across the time course, and five alternatively spliced genes were validated with qPCR: Akap12, Fn1, Pcolce, Sfrp4, and Tpm1. The number of alternatively spliced genes varied with time, ranging from a low of 68 at 12h to a high of 992 at 16d. We identified genes across the time course that encoded proteins with known functions in bone formation, including collagens, matrix proteins, and components of the Wnt/β-catenin and TGF-β signaling pathways. We also identified alternatively spliced genes encoding cytokines, ion channels, muscle-related genes, and solute carriers that do not have a known function in bone formation and represent potentially novel findings. In addition, a functional characterization was performed to categorize the global functions of the alternatively spliced genes in our data set. In conclusion, mechanical loading induces alternative splicing in bone, which may play an important role in the response of bone to mechanical loading.
Keywords: Alternative splicing, bone formation, exon arrays, mechanical loading
1. Introduction
It is estimated that more than 90% of human genes express multiple mRNA transcripts due to alternative splicing [1-3], and the number of mRNA transcripts comprising the human transcriptome is estimated to exceed 10 million [4]. As splice variants contain a different combination of exons, the translated proteins produced by these transcripts may have differing characteristics, such as start and stop codons, which differ from the primary isoform. The diversity of the proteome is due largely to alternative splicing of pre-mRNA transcripts, and the proteins produced by different splice variants will likely vary in expression level and function.
Mechanical loading is a potent anabolic stimulus that substantially strengthens bones [5, 6], and we believe that mechanical loading can be used as a paradigm for bone anabolism. This approach has the advantage of producing a local response, so the contralateral limb can be used as a control to compare with loading effects on the loaded limb. The time course of bone formation after initiating mechanical loading is well characterized. New osteoblasts appear on the bone surface 24 to 48 hours after initiating mechanical loading [7] and bone formation is observed within 96 hours of loading [8]. Bone formation increases between 5 and 12 days after starting loading [8], but after 6 weeks of loading bone formation returns to baseline levels [9]. These data indicate that applied mechanical loading to bone results in osteoblast recruitment followed by matrix production, which lasts for around five weeks before declining to baseline levels [10]. While mechanical loading is known to affect signaling pathways and gene expression [11, 12], the extent of alternative splicing in the response of bone following loading has not yet been studied.
Several genes with splice variants are known in bone, with functions that affect osteoblast function and bone formation, including Fosb, peroxisome proliferator-activated receptor gamma (Pparg), vascular endothelial growth factor (Vegf), and runt related transcription factor 2 (Runx2). A truncated isoform of Fosb, ΔFosb, is expressed in osteoblasts and when overexpressed in transgenic mice, induces increased bone formation throughout the skeleton that leads to osteosclerosis [13]. ΔFosb is also important in mechanical loading-induced signaling pathways in bone cells [14]. Δ2ΔFosb is a second truncated isoform of Fosb, which enhances signaling through the bone morphogenetic protein (BMP) signaling pathway and also induces increased bone formation throughout the skeleton [15].
Osteoblasts and adipocytes are both derived from mesenchymal stem cells, and the primary isoform of PPARγ favors adipocyte proliferation [16] and inhibits osteoblast differentiation [17, 18]. PPARγ splice variants seem to be important in regulation of signal transduction in osteoblasts [19]. In particular, PPARγ-1 expression is correlated with extracellular matrix synthesis and mineralization [19].
Mechanical loading is known to increase expression of VEGF in osteoblasts [20]. Five VEGF splice variants exist, and in loaded osteoblasts, soluble VEGF isoforms were expressed in response to low frequency stretch while matrix-bound VEGF isoforms were expressed in response to high frequency stretch [21]. In a rat fracture healing model, two VEGF splice variants (VEGF120 and VEGF164) were detected in the fracture callus [22].
RUNX2 is a transcription factor important in osteoblast differentiation and bone formation, is expressed in mature osteoblasts, and is essential for normal bone formation [23, 24]. RUNX2 regulates expression of many bone related genes, including osteocalcin [23], type I collagen [25], and osteopontin [26]. There are three RUNX2 splice variants, and although there are some inconsistencies in the literature, all three isoforms are present in adult mouse bone and likely contribute to osteoblast differentiation [23, 27]. In addition, the RUNX2 splice variants are able to up- or downregulate target genes by expressing different isoforms [24], and the RUNX2 isoform Pebp2αA was shown to activate osteocalcin across species [28].
In addition, splice variants of Sp7 transcription factor (Sp7), periostin (Postn), collagen type I alpha 1 (Col1a1), collagen type XI alpha 2 (Col11a2), and fibroblast growth factor receptor 2 (Fgfr2) are important in bone. The Sp7 gene, also called osterix, is expressed in osteoblasts, encodes a transcription factor associated with osteoblast differentiation and bone formation [29], and has three splice variants [30]. Periostin is expressed in osteoblasts as well, plays an important role in bone formation, and has multiple isoforms [31]. Collagens are critical to the extracellular matrix, and both a major (Col1a1) and minor (Col11a2) collagen are alternatively spliced. Splice variants of Col1a1 are associated with osteogenesis imperfecta [32], while a splice variant of type XI collagen, pro-α2(XI), may play a role in matrix formation in bone [33]. Finally, a splice variant of Fgfr2, Fgfr2IIIc, is a positive regulator of bone formation [34].
The primary goals of this study were to evaluate the extent of alternative splicing in a bone subjected to mechanical loading and subsequent bone formation, validate potential important splice variants with qPCR, and identify potential biological functions of splice variants. We evaluated loading-induced alternative splicing over a time course of 4 hours to 32 days, and focused on time points of active bone formation: 4d, 6d, 8d, and 12d. We identified many alternatively spliced genes with known functions in bone formation, and we also identified alternatively spliced genes that do not have a known function in bone formation and represent potentially novel findings.
2. Materials and Methods
2.1 Animals
Adult female Lewis rats were purchased from Charles River Laboratories, Inc. The animals were fed standard rat chow and water ad libitum, and acclimated until 20 weeks of age (average weight of 209.1 g ± 12.5 g). Animals were divided into eleven groups: 4 hours (4h, n=9), 12 hours (12h, n=10), 1 day (1d, n=9), 2 days (2d, n=10), 4 days (4d, n=10), 6 days (6d, n=10), 8 days (8d, n=8), 12 days (12d, n=7), 16 days (16d, n=9), 24 days (24d, n=11), and 32 days (32d, n=12). All procedures were performed in accordance with the Institutional Animal Care and Use Committee guidelines of Indiana University.
2.2 Mechanical Loading
A standard model for bone loading was employed in which the right forelimb is loaded axially for three minutes per day, while the left forearm served as a non-loaded, contralateral control [5, 35, 36]. Prior to loading, animals were anesthetized with 3.0% isoflurane administered at a flow rate of 1.5 L/min. Compressive load was applied as an oscillating Haversine waveform for 360 cycles at a frequency of 2 Hz using a Bose ElectroForce 3200 Series electromechanical actuator (EnduraTEC). The peak load achieved during loading was 13 N, which has previously been shown to be anabolic [35]. Rats were subjected to loading sessions every day, with 24 hours between sessions. The study groups listed above are referenced to the number of days (or hours) after the first bout of bone loading was applied. For example, animals in the 4h, 12h, and 1d groups were loaded once and sacrificed 4 hours, 12 hours, or 24 hours later, respectively. Animals in the 2d group were loaded on two consecutive days and were sacrificed 24 hours after the last loading session. Animals in all other groups were loaded for the indicated number of days and sacrificed 24 hours after the last loading session. At the appropriate time point, animals were anesthetized with isoflurane and euthanized by cervical dislocation.
2.3 RNA Isolation
The shafts of the right and left ulnae were dissected, freed of all soft tissue, and snap frozen in liquid nitrogen. The ulnae were stored at −80 °C until RNA isolation. RNA was extracted using Trizol (Invitrogen) and RNeasy Mini Kits (Qiagen, Inc.). Frozen ulnae were placed into a mortar containing liquid nitrogen and crushed with a pestle. The crushed bones were immediately transferred into a 2 ml tube containing 1 ml of Trizol, homogenized for 20 seconds using a Tissue Tearor homogenizer (Cole-Parmer), incubated on the bench top for 30 minutes, and centrifuged. The supernatant was removed and used to isolate RNA. RNA was isolated using RNeasy Mini Kits according to the manufacturer's instructions, and RNA was treated with a DNA-free kit (Ambion) to remove any residual DNA. RNA quality and quantity were determined using a spectrophotometer (NanoDrop).
2.4 Exon Arrays and Analysis
Quality control for RNA included a spectrum from A220 to A350 run on a NanoDrop spectrophotometer, a native 1% agarose gel stained with ethidium bromide to detect contaminating DNA, and electrophoresis using an Agilent Bioanalyzer to examine RNA. The quality of RNA samples was assessed using 260:280 ratio, and a high quality sample was defined as having a minimum ratio of 2.00. Only four RNA samples had a 260:280 ratio of slightly less than 2.00, and these samples were chosen to optimize the quantity of total RNA as well as the quality. The range of 260:280 ratios of all samples used was 1.96 – 2.31.
Affymetrix GeneChip® Rat Exon 1.0 ST Arrays were used to analyze loading induced alternative splicing in bone. These particular exon arrays had over 4 million individual features and over 1 million probe sets, which represent the transcribed rat genome. The Rat Exon 1.0 ST exon arrays use approximately four probe sets to represent a single exon, and approximately 40 probe sets to represent a gene. The advantages of using exon arrays compared to traditional 3′ in vitro transcription (IVT) microarrays include the use of more probes to represent a single gene, the uniform distribution of probes across the gene instead of concentrating probes at the 3′ end, and study of alternative splice variants. Therefore, exon arrays estimate gene expression more accurately than traditional 3′ IVT microarrays, as probes from all exons in a single gene are summarized into one value that represents the expression level of the gene. As exon arrays have the capability to detect expression of each exon in a gene, exon arrays can be used to study alternative splicing events such as intron retention, exon skipping, alternative promoter usage, alternative poly-adenylation, and alternative 5′ and 3′ splicing sites.
Five matched ulna RNA samples (from the control and loaded ulnae from the same animal) from each time group were used for exon array analysis. One exception to this was the 12d group, where only four matched samples were used. The highest quality matched samples, in terms of 260:280 ratio, with the largest quantity of total RNA were chosen for exon arrays. A total of 108 exon arrays were used, as RNA from the control and loaded ulnae from 54 individual animals were analyzed on separate arrays.
The exon array hybridizations were carried out using the facilities of the Center for Medical Genomics (CMG) at Indiana University School of Medicine. Samples were labeled and hybridized using the Affymetrix WT protocol (GeneChip® Whole Transcript (WT) Sense Target Labeling Assay Manual Version 4, Affymetrix). All processing was done in balanced batches. Labeling was done in batches of 22 samples which included the RNA from both ulnae for one animal from each time group. The exon arrays were scanned using the GeneChip® Scanner 3000 using Affymetrix GeneChip® Operating System (GCOS). Data were exported for analysis in the Partek® Discovery Suite™ (Partek Inc.) [37].
Raw exon array data were obtained from the CMG and imported into Partek for further analysis. Core probe sets included RefSeq transcripts and full length mRNAs, and were chosen for analysis because they were annotated with the most confidence. Probe sets were normalized using the following settings recommended by Partek: robust multi-array average (RMA) background correction, quantile normalization, log probes using base 2, and median polish probe set summarization [38-40]. After normalization and summarization, the probe sets represented over 8,000 genes. Raw data and analyzed data were MIAME compliant [41], were deposited in NCBI's Gene Expression Omnibus database [42], and are accessible through GEO Series accession number GSE22286 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22286).
2.5 Identification of Alternatively Spliced Genes
Core probe sets from each time point were analyzed with an alternative splice ANOVA model to identify alternatively spliced genes. The main variables in the ANOVA model were loading condition (loaded or control) and animal, and the alternative splice factor was loading condition (loaded or control). The output is an alternative splicing score, which is the minimum p-value from a z-test comparing expression of all probe sets within a gene [37]. A low p-value suggests that expression of at least one probe set differs from the rest, and thus the gene has a high probability of being alternatively spliced [37]. An alternatively spliced gene was defined as having a p-value < 0.01. Probe sets with a signal value less than 3 were not reliable and were excluded from analysis.
2.6 Functional Characterization
Groups of genes were then defined based on gene and associated protein function, using Ingenuity Pathways Analysis (IPA, Ingenuity® Systems, www.ingenuity.com). IPA uses information about gene relationships from the literature to characterize gene sets, create gene networks, and identify important signaling pathways and functions in gene expression data. The IPA algorithm is based on the assumptions that highly interconnected genes represent important biological relationships and have similar functions. IPA was used to identify the top biological functions for genes that were alternatively spliced during the period of active bone formation: 4d, 6d, 8d, and 12d. The output of IPA is a p-value, and a low p-value suggests that association of a group of genes and a given biological function is not due to random chance. A p-value < 0.05 was considered significant, and the top 10 biological functions for alternatively spliced genes at 4d, 6d, 8d, and 12d are reported. A limitation of IPA is that a bone specific filter is not available in the analysis, so results cannot be limited to bone and therefore may not always be relevant.
2.7 qPCR
Expression data from exon arrays were validated with quantitative real time PCR (qPCR). Two regions from each gene were amplified with custom primers and probes. One region corresponded to an Affymetrix probe set within an exon that was not differentially expressed with loading or was differentially expressed less than the rest of the exons (i.e. internal control exon). A second region corresponded to an Affymetrix probe set within an exon that was up- or downregulated with loading (i.e. changed exon). Expression of internal control and changed exons within fibronectin 1 (Fn1), a kinase anchor protein 12 (Akap12), secreted frizzled-related protein 4 (Sfrp4), procollagen C-endopeptidase enhancer (Pcolce), and tropomyosin 1 (Tpm1) genes were assessed with PrimeTime qPCR Assays (Integrated DNA Technologies, Inc.). Forward primer, reverse primer, and probe sequences are listed in Table 1.
Table 1.
Primer and probe sequences used in qPCR experiments to validate alternatively spliced genes identified on exon arrays.
| Internal Control Exons | ||
|---|---|---|
| Gene | Primer and probe sequences (5′ → 3′) | |
|
Akap12 – Exon 1 Probe set ID: 6396395 |
Forward Primer | AGGCAGTTCCACCGAGCA |
| Reverse Primer | AGCTGCTCCCGAGGCTTCA | |
| Probe | AGCTGGTGCTCAGTGGCCAT | |
|
Fn1 – Exon 6 Probe set ID: 5726812 |
Forward Primer | CAATGATCAGGACACCAGGAC |
| Reverse Primer | CACTCTGTAGAACATGTCGCT | |
| Probe | CACACTTCCACTCCCCTCTGCC | |
|
Pcolce – Exon 4 Probe set ID: 5889513 |
Forward Primer | CAGGTGCGAGGCGG |
| Reverse Primer | CAGGCCGCCCATTCCAC | |
| Probe | AAATGGGTCATGGACCCGCG | |
|
Sfrp4 – Exon 7 Probe set ID: 6105477 |
Forward Primer | GATGTCCATCAGGTGGC |
| Reverse Primer | CTACTGGGCAGCTTCTG | |
| Probe | AGTATGCACTGCTGTGGCCACAA | |
|
Tpm1 – Exon 8 Probe set ID: 6485754 |
Forward Primer | CCAAGTTCGACAGCTGG |
| Reverse Primer | CTTATCCTCTGCAGCCATT | |
| Probe | AGTTAAGAATAATGGATCAGACCTTGAAAGC | |
| Changed Exons | ||
| Gene | Primer and probe sequences (5′ → 3′) | |
|
Akap12 – Exon 4 Probe set ID: 6427730 |
Forward Primer | TGGACAGCGAGAGTCAGAAGATGT |
| Reverse Primer | CCACATTGCTTTCTGAAGCAGGGA | |
| Probe | TTGAAGAAATGGCGGCCAACTCCACA | |
|
Fn1 – Exon 25 Probe set ID: 5687085 |
Forward Primer | CCCGCTAAACTCTTCCACC |
| Reverse Primer | TGATGTCATAGTCAATGCCGG | |
| Probe | TCCCTTCTCCTGCCGCAACTAC | |
|
Pcolce – Exon 6 Probe set ID: 5908862 |
Forward Primer | TTCGGGAAGTTTGATGTGGAGCCT |
| Reverse Primer | TGAGTCGTCACTCACAGCTCCATT | |
| Probe | CTGCCGATATGACTCTGTCAGTGTGTTC | |
|
Sfrp4 – Exon 5 Probe set ID: 6324245 |
Forward Primer | GATGATGCTTCTTGAAAATTGTT |
| Reverse Primer | TGTGGACCTTCTGCTTAG | |
| Probe | AGTTGAGAAATGGAGAGATCAACTAAGC | |
|
Tpm1 – Exon 5 Probe set ID: 6032871 |
Forward Primer | CTGACGTAGCATCTCTGAAC |
| Reverse Primer | TCTCATCTGCAGCCTTCT | |
| Probe | ATCCAGCTGGTTGAGGAGGAGTTGGAT | |
Three matched RNA samples from loaded and control ulnae from the 4d or 6d time groups were used for qPCR. Bone formation was ongoing at these time points, and exon expression patterns were consistent over multiple time points including 4d and 6d. RNA was reverse transcribed using the SuperScript III kit with oligo(dT) primers (Invitrogen). cDNA was diluted to a concentration of 2.5 ng/μl and used in qPCR reactions. The reactions were performed on an ABI 7900HT Fast Real-Time PCR System. Fold change was calculated by normalizing load:control ratio of the changed exon to load:control ratio of the internal control exon to verify the exon array data. The average fold change and standard error are reported from qPCR and exon arrays. Next, we determined if the degree of up- or downregulation of gene expression was consistent between exon arrays and qPCR. A paired t-test was used to compare load:control ratio for the changed exon and internal control exon for qPCR and exon arrays (*p<0.05, +p<0.10).
3. Results
3.1 Identification of Alternatively Spliced Genes
An alternative splice ANOVA model was used to identify alternatively spliced genes. The number of genes that were alternatively spliced in response to mechanical loading varied over time (Figure 1), ranging from a low of 68 in the 12h group to a high of 992 in the 16d group.
Figure 1.
The number of genes that were alternatively spliced in response to mechanical loading varied over time, ranging from a low of 68 at 12h to a high of 992 at 16d (n=5 matched samples at each time point except 12d, where 4 matched samples were used).
Table 2 contains a selected list of alternatively spliced genes identified during periods of active bone formation, between and including the 4d and 12d time groups. Many genes relevant to bone formation had significant alternative splicing p-values across time, including genes in the following groups: apoptosis, calcium, cell cycle, cytokine, cytoskeleton, growth factor, ion channel, matrix, muscle, neurotransmitter, solute carrier, transforming growth factor β (TGFβ) signaling, and Wnt/β-catenin signaling. The two largest groups of alternatively spliced genes were associated with extracellular matrix and solute carriers. The group of matrix genes was comprised of 24 members and included genes encoding proteoglycans (Acan and Fn1), collagens (Col1a2, Col2a1, Col3a1, Col4a4, Col5a1, Col11a2, Col16a1, and Col23a1), proteins related to collagen synthesis (Lepre1, Pcolce, and P4ha1), an osteoblast marker (Bglap), matrix-related proteins (Ibsp, Lox, and Sparc), and MMPs (Mmp7, Mmp10, and Mmp16). The group of solute carrier genes had 24 members as well, and examples of the types of molecules transported include amino acids, glucose, and various ions. The next largest groups of alternatively spliced genes encoded ion channels (n=16), calcium-related genes (n=11), cytokines (n=9), muscle-related genes (n=8), and Wnt/β-catenin signaling pathway genes (n=6).
Table 2.
Selected genes with significant alternative splicing p-values (p<0.01) across time, particularly during periods of active bone formation (4d, 6d, 8d, and 12d).
| Gene Group | Gene Name |
|---|---|
| Apoptosis | |
| Bik | BCL2-interacting killer |
| Dap | death-associated protein |
| Dapk2 | death-associated kinase 2 |
| Calcium | |
| Cadps | Ca++-dependent secretion activator |
| Calb2 | calbindin 2 |
| Calm1 | calmodulin 1 |
| Camk1g | calcium/calmodulin-dependent protein kinase IG |
| Camk2n1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| Capn3 | calpain 3 |
| Capn6 | calpain 6 |
| Casr | calcium-sensing receptor |
| S100a4 | S100 calcium-binding protein A4 |
| S100a8 | S100 calcium binding protein A8 |
| Stc2 | stanniocalcin 2 |
| Cell Cycle | |
| Cdc91l1 | CDC91 cell division cycle 91-like 1 |
| Cdc25b | cell division cycle 25 homolog B |
| Cdk5r1 | cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| Cdkn1b | cyclin-dependent kinase inhibitor 1B |
| Cytokine | |
| Il1b | interleukin 1 beta |
| Il1r2 | interleukin 1 receptor, type II |
| Il6 | interleukin 6 |
| Il6ra | interleukin 6 receptor, alpha |
| Il6st | interleukin 6 signal transducer |
| Il11 | interleukin 11 |
| Il17re | interleukin 17 receptor E |
| Il24 | interleukin 24 |
| Stat2 | signal transducer and activator of transcription 2 |
| Cytoskeleton | |
| Dnai1 | dynein, axonemal, intermediate chain 1 |
| Sdc3 | syndecan 3 |
| Vim | vimentin |
| Growth Factor | |
| Fgf23 | fibroblast growth factor 23 |
| Pdgfrb | platelet derived growth factor receptor, beta polypeptide |
| Pgf | placental growth factor |
| Ion Channel | |
| Cacna1a | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| Cacna1h | calcium channel, voltage-dependent, T type, alpha 1H subunit |
| Cacnb2 | calcium channel, voltage-dependent, beta 2 subunit |
| Cacnb3 | calcium channel, voltage-dependent, beta 3 subunit |
| Hcn4 | hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| Kcnb1 | potassium voltage gated channel, Shab-related subfamily, member 1 |
| Kcnc3 | potassium voltage gated channel, Shaw-related subfamily, member 3 |
| Kcnk2 | potassium channel, subfamily K, member 2 |
| Kcnma1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| Kcns2 | potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| Nalcn | sodium leak channel, non-selective |
| Scn1a | sodium channel, voltage-gated, type I, alpha |
| Scn1b | sodium channel, voltage-gated, type I, beta |
| Scn4a | sodium channel, voltage-gated, type 4, alpha subunit |
| Scn4b | sodium channel, type IV, beta |
| Scnn1b | sodium channel, nonvoltage-gated 1 beta |
| Matrix | |
| Acan | aggrecan |
| Bglap | bone gamma-carboxyglutamate (gla) protein |
| Cd44 | Cd44 molecule |
| Col1a2 | collagen, type I, alpha 2 |
| Col2a1 | collagen, type II, alpha 1 |
| Col3a1 | collagen, type III, alpha 1 |
| Col4a4 | procollagen, type IV, alpha 4 |
| Col5a1 | collagen, type V, alpha 1 |
| Col11a2 | collagen, type XI, alpha 2 |
| Col16a1 | collagen, type XVI, alpha 1 |
| Col23a1 | collagen, type XXIII, alpha 1 |
| Fn1 | fibronectin 1 |
| Ibsp | integrin binding sialoprotein |
| Lepre1 | leucine proline-enriched proteoglycan (leprecan) 1 |
| Lox | lysyl oxidase |
| Mmp7 | matrix metallopeptidase 7 |
| Mmp10 | matrix metallopeptidase 10 |
| Mmp16 | matrix metallopeptidase 16 |
| P4ha1 | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I |
| Pcolce | procollagen C-endopeptidase enhancer |
| Serpinb6b | serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| Serpinf1 | serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| Sparc | secreted protein, acidic, cysteine-rich (osteonectin) |
| Muscle | |
| Actn3 | actinin alpha 3 |
| Mybph | myosin binding protein H |
| Myf6 | myogenic factor 6 |
| Myh3 | myosin, heavy chain 3, skeletal muscle, embryonic |
| Myh6 | myosin, heavy chain 6, cardiac muscle, alpha |
| Myo1b | myosin Ib |
| Tpm1 | tropomyosin 1, alpha |
| Tpm2 | tropomyosin 2 |
| Neurotransmitter | |
| Chrna1 | cholinergic receptor, nicotinic, alpha 1 |
| Chrng | cholinergic receptor, nicotinic, gamma |
| Htr1a | 5-hydroxytryptamine (serotonin) receptor 1A |
| Solute Carrier | |
| Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 |
| Slc2a2 | solute carrier family 2 (facilitated glucose transporter), member 2 |
| Slc2a13 | solute carrier family 2 (facilitated glucose transporter), member 13 |
| Slc3a2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| Slc6a2 | solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| Slc6a15 | solute carrier family 6 (neutral amino acid transporter), member 15 |
| Slc7a3 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| Slc8a3 | solute carrier family 8 (sodium/calcium exchanger), member 3 |
| Slc9a2 | solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| Slc13a4 | solute carrier family 13 (sodium/sulfate symporters), member 4 |
| Slc13a5 | solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| Slc14a2 | solute carrier family 14 (urea transporter), member 2 |
| Slc17a5 | solute carrier family 17 (anion/sugar transporter), member 5 |
| Slc18a2 | solute carrier family 18 (vesicular monoamine), member 2 |
| Slc19a1 | solute carrier family 19 (folate transporter), member 1 |
| Slc22a3 | solute carrier family 22 (extraneuronal monoamine transporter), member 3 |
| Slc23a1 | solute carrier family 23 (nucleobase transporters), member 1 |
| Slc25a27 | solute carrier family 25, member 27 |
| Slc26a2 | solute carrier family 26 (sulfate transporter), member 2 |
| Slc34a2 | solute carrier family 34 (sodium phosphate), member 2 |
| Slc36a1 | solute carrier family 36 (proton/amino acid symporter), member 1 |
| Slc36a2 | solute carrier family 36 (proton/amino acid symporter), member 2 |
| Slc38a4 | solute carrier family 38, member 4 |
| Slc39a3 | solute carrier family 39 (zinc transporter), member 3 |
| TGF-β Signaling | |
| Bmp6 | bone morphogenetic protein 6 |
| Bmp15 | bone morphogenetic protein 15 |
| Chrdl1 | chordin-like 1 |
| Smad3 | SMAD family member 3 |
| Wnt/β-catenin Signaling | |
| Cdh2 | cadherin 2 |
| Cpz | carboxypeptidase Z |
| Sfrp4 | secreted frizzled-related protein 4 |
| Sost | sclerosteosis |
| Wnt4 | wingless-type MMTV integration site family, member 4 |
| Wnt7b | wingless-type MMTV integration site family, member 7B |
| Other | |
| Akap12 | A kinase (PRKA) anchor protein 12 |
| Ccl5 | chemokine (C-C motif) ligand 5 |
| Creb3l1 | cAMP responsive element binding protein 3-like 1 |
| Hif1a | hypoxia-inducible factor 1, alpha subunit |
| Lepr | leptin receptor |
| Ptgds | prostaglandin D2 synthase (brain) |
| Ptgs2 | prostaglandin-endoperoxide synthase 2 |
3.2 qPCR Validation of Alternative Splicing
Five genes that were alternatively spliced in response to loading on the exon arrays were validated using qPCR. These genes were chosen for validation because the exon expression patterns showed evidence of alternative splicing, there were known multiple gene isoforms in rat and/or human genomes, the alternative splicing p-value was less than 0.001, they exhibited consistent exon expression patterns across the time course, and they were biologically relevant. While these criteria limited the gene list substantially, imposing these restrictions improved the likelihood of validating alternatively spliced genes with qPCR [43].
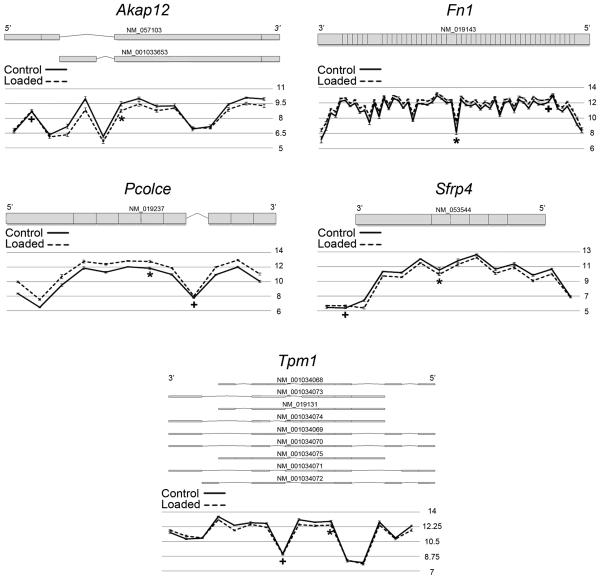
Figure 2 shows the exon expression patterns observed for Akap12, Fn1, Pcolce, Sfrp4, and Tpm1 at the 6d time point. The exon expression patterns of these genes at 6d were similar at all other time points. At the top of each image is the exon structure and mRNA transcript accession number for the known isoforms of Akap12, Fn1, Pcolce, Sfrp4, and Tpm1 in the rat genome. The bottom of each image shows a graph of the mean probe set log2 intensity level and standard error (y-axis) for each probe set in control (solid black line) and loaded (dashed black line) samples. For each gene, the difference in probe set intensity levels between control and loaded samples differed across the gene, which indicates that these genes are alternatively spliced. For example, the two probe sets on the 5′ end of Akap12 do not show different intensity levels between loaded and control conditions, but the probe sets in the middle of the gene show decreased intensity with loading. In addition, very consistent exon expression patterns were observed in all genes we studied, across the time course and in loaded and control samples.
Figure 2.
Akap12, Fn1, Pcolce, Sfrp4, and Tpm1 were alternatively spliced in response to mechanical loading. Top: the exon structure and mRNA transcript accession number for the known isoforms of Akap12, Fn1, Pcolce, Sfrp4, and Tpm1 in the rat genome. Bottom: Graph of mean probe set log2 intensity level and standard error (y-axis) across the gene in control (black solid line) and loaded (black dashed line) 6d samples (n=5 matched samples). The difference in probe set intensity levels between control and loaded samples differs across the gene, which indicates that these genes are alternatively spliced. Two probe sets corresponding to two different exons were validated with qPCR for each gene. The probe set in the internal control exon did not change with loading or changed less than the rest of the exons (+), while the probe set in the changed exon was upregulated (Fn1 and Pcolce) or downregulated (Akap12, Sfrp4, and Tpm1) with loading (*).
Figure 3 shows qPCR validation of alternatively spliced genes from exon arrays. The fold change of changed exon to internal control exon was consistent between qPCR (black bars) and arrays (white bars) for all five genes, achieving an alternative splicing validation rate of 5/5. Next, we determined if the degree of up- or downregulation of changed exon expression was consistent between exon arrays and qPCR. The changed exons in Fn1 and Pcolce were upregulated with mechanical loading compared to the internal control exons, while the changed exon in Tpm1 was downregulated. The changed exon in Sfrp4 was downregulated with qPCR, and showed slight, but not significant, downregulation with loading on exon arrays compared to the internal control exon. Exon array data showed that the changed exon in Akap12 was downregulated compared to the internal control exon, while qPCR data showed slight, but not significant, downregulation. Our validation rate for detecting consistent up- and downregulation on the exon arrays and qPCR was 3/5.
Figure 3.
qPCR validation of alternatively spliced genes from exon arrays. Portions of two Affymetrix probe sets corresponding to an internal control exon and a changed exon within the Akap12, Fn1, Pcolce, Sfrp4, and Tpm1 genes were amplified with qPCR and used to validate exon array findings at 4d or 6d (n=3 matched samples). Fold change was calculated by normalizing load:control ratio of the changed exon to load:control ratio of the internal control exon. The average fold change and standard error are reported for qPCR and exon arrays. Fold change was consistent between qPCR (black bars) and arrays (white bars) for all five genes. A paired t-test was used to compare load:control ratio of the changed exon and internal control exon for qPCR and exon array data (*p<0.05, +p<0.10).
3.3 Functional Characterization
To associate the alternatively spliced genes with underlying biological processes, we performed a functional characterization using IPA. The top 10 biological functions associated with alternatively spliced genes at 4d, 6d, 8d, and 12d, the period of active bone formation, are reported in Table 3. Of note is that three of the biological functions were common to all time points: cellular assembly and organization, cellular function and maintenance, and skeletal and muscular disorders. Three additional biological functions were common in 3 of the 4 time points: connective tissue disorders, genetic disorder, and inflammatory disease.
Table 3.
Top biological functions of alternatively spliced genes during periods of active bone formation (4d, 6d, 8d, and 12d; p<0.05).
| Biological Function | 4d | 6d | 8d | 12d |
|---|---|---|---|---|
| Cancer | X | |||
| Carbohydrate metabolism | X | |||
| Cell-to-cell signaling and interaction | X | X | ||
| Cellular assembly and organization | X | X | X | X |
| Cellular function and maintenance | X | X | X | X |
| Cellular movement | X | |||
| Connective tissue disorders | X | X | X | |
| Dermatological diseases and conditions | X | X | ||
| Developmental disorder | X | X | ||
| Endocrine system disorders | X | |||
| Genetic disorder | X | X | X | |
| Inflammatory disease | X | X | X | |
| Metabolic disease | X | |||
| Molecular transport | X | |||
| Nervous system development and function | X | |||
| Neurological disease | X | |||
| Organ morphology | X | |||
| Reproductive system development and function | X | |||
| Skeletal and muscular disorders | X | X | X | X |
| Skeletal and muscular system development and function | X | X | ||
| Tumor morphology | X |
4. Discussion
Exon arrays are very powerful tools that allow identification of alternatively spliced genes across the entire genome, and it is important to validate exon array data. We chose to validate exon array data with qPCR, as it has been used to validate exon array results with success rates between 26% and 86% [1, 44, 45]. As the reported success rates are rather variable, we included several criteria to select alternatively spliced genes to validate, in an effort to achieve a high validation success rate. The genes chosen for qPCR validation had exon expression patterns that showed evidence of alternative splicing, had known multiple gene isoforms in rat and/or human genomes, had an alternative splicing p-value less than 0.001, exhibited consistent exon expression patterns across the time course, and were biologically relevant in terms of bone formation and mechanical loading. While these criteria limited the candidate gene list substantially, imposing these restrictions improved the likelihood of validating alternatively spliced genes with qPCR [43]. We normalized the load:control ratio of the changed exon to the load:control ratio of the internal control exon to validate alternative splicing, and achieved a validation rate of 5/5. The validation rate for detecting up- and downregulation on the exon arrays and qPCR was 3/5. However, in the two cases (Akap12 and Sfrp4) where the exon array and qPCR data were not in statistically significant agreement, the expression trend for each of these genes was similar on exon arrays and qPCR.
We validated five biologically relevant genes: Akap12, Fn1, Pcolce, Sfrp4, and Tpm1. It was important to validate genes that were both up- and downregulated with loading, to ensure we could detect up- and downregulation accurately with both exon arrays and qPCR. At the whole gene level, Fn1 and Pcolce were upregulated, while Akap12, Sfrp4, and Tpm1 were downregulated.
AKAP12 is a kinase involved in protein kinase A (PKA) signaling [46], protein kinase C (PKC) signaling [46], and signal transduction [47]. Both the PKA and PKC pathways play regulatory roles in mechanotransduction [48-51]. Akap12 has long and short isoforms in both rat and human genomes, but the roles of Akap12 splice variants in bone are not known.
FN1 is a non-collagenous protein in the extracellular matrix and is important in cell adhesion, migration, growth, and differentiation [52, 53], and it has been suggested that alternative splicing of Fn1 may affect matrix remodeling [54]. Fn1 has many isoforms in human and rat genomes, and Fn1 isoforms have been studied in several rat cells and tissues including chondrosarcoma cells [55], hepatic stellate cells [56], myocardium [57], and fetal lung [58]. However, fibronectin splice variants have not been studied in rat bone.
PCOLCE is a glycoprotein that is involved with type I procollagen processing, and drives enzymatic cleavage of the amino- and carboxyl-terminal propeptides [59]. Type I collagen is a major bone matrix gene [60-62], and its expression correlates with bone formation and osteoblast recruitment in response to mechanical loading in bone [7-9]. Pcolce has multiple isoforms in both human and rat genomes, but the roles of Pcolce splice variants in bone are not known.
SFRP4 is a Wnt antagonist that binds to Wnt and suppresses both canonical and non-canonical Wnt signaling [63], and is expressed at the mRNA level in osteocytes and osteoblasts [64]. Others have shown that Sfrp4 has multiple splice variants that are expressed in a variety of tissues [65], but this study did not evaluate Sfrp4 splice variant expression in bone. We detected a long and short isoform of Sfrp4 in loaded rat bone, corresponding to the rat Sfrp4 isoforms in the UCSC Genome Browser database. The short isoform is known, but the long isoform is predicted and has not yet been validated. In our study, we were able to detect an exon in both the short and long isoform of Sfrp4.
TPM1 is an actin binding protein that is involved with muscle contraction in muscle cells and in the cytoskeleton in non-muscle cells [45, 66]. Tpm1 has multiple isoforms in both rat and human genomes. Tpm1 splice variants are specifically expressed in normal human bladder, colon, and prostate as well as in human bladder and prostate cancers [45]. While muscle-related genes seem to be highly regulated in bone cells [67], the role of Tpm1 splice variants in bone is not known.
The number of alternatively spliced genes correlated with the number of differentially expressed genes across time, and there was a spike in both the number of alternatively spliced genes and differentially expressed genes at 16d [68]. We postulate that the spike in alternatively and differentially expressed genes at 16 days corresponds to a fundamental change at the cellular and tissue levels, as the system becomes less responsive to loading over time and shifts from bone forming to baseline bone maintenance. Bone formation increases between 5 and 12 days after starting loading [8] and returns to baseline levels after 6 weeks [9]. Therefore, bone cells become desensitized to loading sometime between 12 days and 6 weeks, and we think that this transition period may occur at or near the 16d time point and contribute to the observed peak in the number of alternatively spliced and differentially expressed genes. In addition, a tissue-level change occurs. The magnitude of load applied to rat ulnae remained constant over the time course, and new bone was deposited in response to loading. Therefore, an overall tissue-level decrease in strain occurred over time, and we think this change in mechanical behavior may have affected alternative splicing as well. It is also possible that recruitment of other cell types, or other unknown factors, affected alternative splicing patterns. These possible explanations require further investigation.
The specific biological mechanisms for causing splicing pattern changes are not known. However, mechanotransduction pathways may be important in this process, as many mechanotransduction pathways are affected by loading. For example, the Wnt/β-catenin pathway is known to play an integral role in mechanotransduction and enhances the sensitivity of osteoblasts and osteocytes to loading [69]. We speculate that mechanotransduction pathways may facilitate changes in the expression and/or functions of RNA binding proteins and/or transcription factors over time, which would affect splicing patterns and gene expression, respectively.
Functional characterization showed that several biological functions were common across time, suggesting that alternative splicing may regulate groups of genes with common functions that affect bone formation. The biological functions “cellular assembly and organization”, “cellular function and maintenance”, and “skeletal and muscular disorders” were common to 4d, 6d, 8d, and 12d, while “connective tissue disorders”, “genetic disorder”, and “inflammatory disease” were common to three of the four time points. However, the relationship of some biological functions with bone formation is not clear. A limitation of Ingenuity is that there is not a bone specific filter in the analysis, so results cannot be limited to bone and may not always be relevant.
In summary, we have identified several genes that appear to be alternatively spliced in response to loading across the rat genome, and focused on time points when active bone formation was occurring. We identified many alternatively spliced genes with known functions in bone formation, including genes encoding collagens, matrix proteins, and members of the Wnt/β-catenin signaling pathway. Some of the genes we identified are known to have multiple isoforms in rat and/or human genomes, but others are thought to have only one isoform and our findings suggest that these genes may have novel splice variants. In addition, we performed a functional characterization to categorize the global functions of alternatively spliced genes during the period of active bone formation. We conclude that mechanical loading induces alternative splicing in bone, which may play an important role in the response of bone to mechanical loading.
Research Highlights.
Mechanical loading induces alternative splicing in bone
Known and potentially novel splice variants were identified
Several biological functions of alternatively spliced genes were common across time
Alternative splicing may play an important role in the response of bone to loading
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Arthritis, Musculoskeletal and Skin Diseases through grant AR46530. Exon array hybridization was carried out in the Center for Medical Genomics (CMG), which is supported in part by the Indiana Genomics Initiative at Indiana University (INGEN®, which is supported in part by the Lilly Endowment, Inc.). The authors thank the director of the CMG, Dr. Howard Edenberg, and also Dr. Jeanette McClintick for performing the hybridizations and help with analysis.
Funding sources: National Institute of Arthritis, Musculoskeletal and Skin Diseases grant AR46530.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest page
The authors declare no conflicts of interest.
References
- 1.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative premRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Affymetrix GeneChip Exon Array Design (Affymetrix Technical Note) Affymetrix White Paper. 2005 [Google Scholar]
- 3.Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–86. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P. Constructing the landscape of the mammalian transcriptome. J Exp Biol. 2007;210:1497–506. doi: 10.1242/jeb.000406. [DOI] [PubMed] [Google Scholar]
- 5.Robling AG, Hinant FM, Burr DB, Turner CH. Improved bone structure and strength after long-term mechanical loading is greatest if loading is separated into short bouts. J Bone Miner Res. 2002;17:1545–54. doi: 10.1359/jbmr.2002.17.8.1545. [DOI] [PubMed] [Google Scholar]
- 6.Warden SJ, Hurst JA, Sanders MS, Turner CH, Burr DB, Li J. Bone adaptation to a mechanical loading program significantly increases skeletal fatigue resistance. J Bone Miner Res. 2005;20:809–16. doi: 10.1359/JBMR.041222. [DOI] [PubMed] [Google Scholar]
- 7.Turner CH, Owan I, Alvey T, Hulman J, Hock JM. Recruitment and proliferative responses of osteoblasts after mechanical loading in vivo determined using sustained-release bromodeoxyuridine. Bone. 1998;22:463–9. doi: 10.1016/s8756-3282(98)00041-6. [DOI] [PubMed] [Google Scholar]
- 8.Forwood MR, Owan I, Takano Y, Turner CH. Increased bone formation in rat tibiae after a single short period of dynamic loading in vivo. Am J Physiol. 1996;270:E419–23. doi: 10.1152/ajpendo.1996.270.3.E419. [DOI] [PubMed] [Google Scholar]
- 9.Schriefer JL, Warden SJ, Saxon LK, Robling AG, Turner CH. Cellular accommodation and the response of bone to mechanical loading. J Biomech. 2005;38:1838–45. doi: 10.1016/j.jbiomech.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Turner CH, Pavalko FM. Mechanotransduction and functional response of the skeleton to physical stress: the mechanisms and mechanics of bone adaptation. J Orthop Sci. 1998;3:346–55. doi: 10.1007/s007760050064. [DOI] [PubMed] [Google Scholar]
- 11.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–77. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 12.Liedert A, Kaspar D, Blakytny R, Claes L, Ignatius A. Signal transduction pathways involved in mechanotransduction in bone cells. Biochem Biophys Res Commun. 2006;349:1–5. doi: 10.1016/j.bbrc.2006.07.214. [DOI] [PubMed] [Google Scholar]
- 13.Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med. 2000;6:985–90. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- 14.Inoue D, Kido S, Matsumoto T. Transcriptional induction of FosB/DeltaFosB gene by mechanical stress in osteoblasts. J Biol Chem. 2004;279:49795–803. doi: 10.1074/jbc.M404096200. [DOI] [PubMed] [Google Scholar]
- 15.Sabatakos G, Rowe GC, Kveiborg M, Wu M, Neff L, Chiusaroli R, Philbrick WM, Baron R. Doubly truncated FosB isoform (Delta2DeltaFosB) induces osteosclerosis in transgenic mice and modulates expression and phosphorylation of Smads in osteoblasts independent of intrinsic AP-1 activity. J Bone Miner Res. 2008;23:584–95. doi: 10.1359/JBMR.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heikkinen S, Auwerx J, Argmann CA. PPARgamma in human and mouse physiology. Biochim Biophys Acta. 2007;1771:999–1013. doi: 10.1016/j.bbalip.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecka-Czernik B, Gubrij I, Moerman EJ, Kajkenova O, Lipschitz DA, Manolagas SC, Jilka RL. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J Cell Biochem. 1999;74:357–71. [PubMed] [Google Scholar]
- 18.Rosen CJ, Ackert-Bicknell C, Rodriguez JP, Pino AM. Marrow fat and the bone microenvironment: developmental, functional, and pathological implications. Crit Rev Eukaryot Gene Expr. 2009;19:109–24. doi: 10.1615/critreveukargeneexpr.v19.i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruedigam C, Koedam M, Chiba H, Eijken M, van Leeuwen JP. Evidence for multiple peroxisome proliferator-activated receptor gamma transcripts in bone: fine-tuning by hormonal regulation and mRNA stability. FEBS Lett. 2008;582:1618–24. doi: 10.1016/j.febslet.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Wan C, Deng L, Liu X, Cao X, Gilbert SR, Bouxsein ML, Faugere MC, Guldberg RE, Gerstenfeld LC, Haase VH, Johnson RS, Schipani E, Clemens TL. The hypoxiainducible factor alpha pathway couples angiogenesis to osteogenesis during skeletal development. J Clin Invest. 2007;117:1616–26. doi: 10.1172/JCI31581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faure C, Linossier MT, Malaval L, Lafage-Proust MH, Peyroche S, Vico L, Guignandon A. Mechanical signals modulated vascular endothelial growth factor-A (VEGF-A) alternative splicing in osteoblastic cells through actin polymerisation. Bone. 2008;42:1092–101. doi: 10.1016/j.bone.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Pufe T, Wildemann B, Petersen W, Mentlein R, Raschke M, Schmidmaier G. Quantitative measurement of the splice variants 120 and 164 of the angiogenic peptide vascular endothelial growth factor in the time flow of fracture healing: a study in the rat. Cell Tissue Res. 2002;309:387–92. doi: 10.1007/s00441-002-0605-0. [DOI] [PubMed] [Google Scholar]
- 23.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 24.Makita N, Suzuki M, Asami S, Takahata R, Kohzaki D, Kobayashi S, Hakamazuka T, Hozumi N. Two of four alternatively spliced isoforms of RUNX2 control osteocalcin gene expression in human osteoblast cells. Gene. 2008;413:8–17. doi: 10.1016/j.gene.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 25.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–92. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inman CK, Shore P. The osteoblast transcription factor Runx2 is expressed in mammary epithelial cells and mediates osteopontin expression. J Biol Chem. 2003;278:48684–9. doi: 10.1074/jbc.M308001200. [DOI] [PubMed] [Google Scholar]
- 27.Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, Yamaguchi A, Komori T, Nakatsuka M. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem. 1999;274:6972–8. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 28.Pinto JP, Conceicao NM, Viegas CS, Leite RB, Hurst LD, Kelsh RN, Cancela ML. Identification of a new pebp2alphaA2 isoform from zebrafish runx2 capable of inducing osteocalcin gene expression in vitro. J Bone Miner Res. 2005;20:1440–53. doi: 10.1359/JBMR.050318. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 30.Milona MA, Gough JE, Edgar AJ. Expression of alternatively spliced isoforms of human Sp7 in osteoblast-like cells. BMC Genomics. 2003;4:43. doi: 10.1186/1471-2164-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF, Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–49. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Forlino A, Marini JC. Alternative splicing in COL1A1 mRNA leads to a partial null allele and two In-frame forms with structural defects in non-lethal osteogenesis imperfecta. J Biol Chem. 1996;271:28617–23. doi: 10.1074/jbc.271.45.28617. [DOI] [PubMed] [Google Scholar]
- 33.Urabe K, Jingushi S, Ikenoue T, Okazaki K, Sakai H, Li C, Iwamoto Y. Immature osteoblastic cells express the pro-alpha2(XI) collagen gene during bone formation in vitro and in vivo. J Orthop Res. 2001;19:1013–20. doi: 10.1016/S0736-0266(01)00043-2. [DOI] [PubMed] [Google Scholar]
- 34.Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129:3783–93. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh YF, Robling AG, Ambrosius WT, Burr DB, Turner CH. Mechanical loading of diaphyseal bone in vivo: the strain threshold for an osteogenic response varies with location. J Bone Miner Res. 2001;16:2291–7. doi: 10.1359/jbmr.2001.16.12.2291. [DOI] [PubMed] [Google Scholar]
- 36.Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam MI, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE, Turner CH. Mechanical stimulation of bone in vivo reduced osteocyte expression of sost/sclerostin. J Biol Chem. 2008;283:5866–75. doi: 10.1074/jbc.M705092200. [DOI] [PubMed] [Google Scholar]
- 37.Partek Partek Discovery Suite. Paretk Documentation. 1993-2009 [Google Scholar]
- 38.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 39.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 41.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 42.Turner CH, Akhter MP, Raab DM, Kimmel DB, Recker RR. A noninvasive, in vivo model for studying strain adaptive bone modeling. Bone. 1991;12:73–9. doi: 10.1016/8756-3282(91)90003-2. [DOI] [PubMed] [Google Scholar]
- 43.Affymetrix Identifying and Validating Alternative Splicing Events (Affymetrix Technical Note) 2006 [Google Scholar]
- 44.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorsen K, Sorensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein AM, Kruhoffer M, Laurberg S, Borre M, Wang K, Brunak S, Krainer AR, Torring N, Dyrskjot L, Andersen CL, Orntoft TF. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics. 2008;7:1214–24. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Nauert JB, Klauck TM, Langeberg LK, Scott JD. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 47.Choi MC, Jong HS, Kim TY, Song SH, Lee DS, Lee JW, Kim NK, Bang YJ. AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene. 2004;23:7095–103. doi: 10.1038/sj.onc.1207932. [DOI] [PubMed] [Google Scholar]
- 48.Li CF, Hughes-Fulford M. Fibroblast growth factor-2 is an immediate-early gene induced by mechanical stress in osteogenic cells. J Bone Miner Res. 2006;21:946–55. doi: 10.1359/jbmr.060309. [DOI] [PubMed] [Google Scholar]
- 49.Ogata T. Fluid flow induces enhancement of the Egr-1 mRNA level in osteoblast-like cells: involvement of tyrosine kinase and serum. J Cell Physiol. 1997;170:27–34. doi: 10.1002/(SICI)1097-4652(199701)170:1<27::AID-JCP4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 50.Jones DB, Nolte H, Scholubbers JG, Turner E, Veltel D. Biochemical signal transduction of mechanical strain in osteoblast-like cells. Biomaterials. 1991;12:101–10. doi: 10.1016/0142-9612(91)90186-e. [DOI] [PubMed] [Google Scholar]
- 51.Reich KM, Frangos JA. Protein kinase C mediates flow-induced prostaglandin E2 production in osteoblasts. Calcif Tissue Int. 1993;52:62–6. doi: 10.1007/BF00675628. [DOI] [PubMed] [Google Scholar]
- 52.Huang CH, Chen MH, Young TH, Jeng JH, Chen YJ. Interactive effects of mechanical stretching and extracellular matrix proteins on initiating osteogenic differentiation of human mesenchymal stem cells. J Cell Biochem. 2009;108:1263–73. doi: 10.1002/jcb.22356. [DOI] [PubMed] [Google Scholar]
- 53.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 54.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, Davies C, Williams A, Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bentolila V, Boyce TM, Fyhrie DP, Drumb R, Skerry TM, Schaffler MB. Intracortical remodeling in adult rat long bones after fatigue loading. Bone. 1998;23:275–81. doi: 10.1016/s8756-3282(98)00104-5. [DOI] [PubMed] [Google Scholar]
- 56.Pedersen EA, Akhter MP, Cullen DM, Kimmel DB, Recker RR. Bone response to in vivo mechanical loading in C3H/HeJ mice. Calcif Tissue Int. 1999;65:41–6. doi: 10.1007/s002239900655. [DOI] [PubMed] [Google Scholar]
- 57.Akhter MP, Cullen DM, Pedersen EA, Kimmel DB, Recker RR. Bone response to in vivo mechanical loading in two breeds of mice. Calcif Tissue Int. 1998;63:442–9. doi: 10.1007/s002239900554. [DOI] [PubMed] [Google Scholar]
- 58.Kodama Y, Umemura Y, Nagasawa S, Beamer WG, Donahue LR, Rosen CR, Baylink DJ, Farley JR. Exercise and mechanical loading increase periosteal bone formation and whole bone strength in C57BL/6J mice but not in C3H/Hej mice. Calcif Tissue Int. 2000;66:298–306. doi: 10.1007/s002230010060. [DOI] [PubMed] [Google Scholar]
- 59.Takahara K, Osborne L, Elliott RW, Tsui LC, Scherer SW, Greenspan DS. Fine mapping of the human and mouse genes for the type I procollagen COOH-terminal proteinase enhancer protein. Genomics. 1996;31:253–6. doi: 10.1006/geno.1996.0043. [DOI] [PubMed] [Google Scholar]
- 60.Lean JM, Jagger CJ, Chambers TJ, Chow JW. Increased insulin-like growth factor I mRNA expression in rat osteocytes in response to mechanical stimulation. Am J Physiol. 1995;268:E318–27. doi: 10.1152/ajpendo.1995.268.2.E318. [DOI] [PubMed] [Google Scholar]
- 61.Miles RR, Turner CH, Santerre R, Tu Y, McClelland P, Argot J, DeHoff BS, Mundy CW, Rosteck PR, Jr., Bidwell J, Sluka JP, Hock J, Onyia JE. Analysis of differential gene expression in rat tibia after an osteogenic stimulus in vivo: mechanical loading regulates osteopontin and myeloperoxidase. J Cell Biochem. 1998;68:355–65. doi: 10.1002/(sici)1097-4644(19980301)68:3<355::aid-jcb6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 62.Rath B, Nam J, Knobloch TJ, Lannutti JJ, Agarwal S. Compressive forces induce osteogenic gene expression in calvarial osteoblasts. J Biomech. 2008;41:1095–103. doi: 10.1016/j.jbiomech.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 64.Santos A, Bakker AD, Zandieh-Doulabi B, Semeins CM, Klein-Nulend J. Pulsating fluid flow modulates gene expression of proteins involved in Wnt signaling pathways in osteocytes. J Orthop Res. 2009;27:1280–7. doi: 10.1002/jor.20888. [DOI] [PubMed] [Google Scholar]
- 65.Yam JW, Chan KW, Ngan ES, Hsiao WL. Genomic structure, alternative splicing and tissue expression of rFrp/sFRP-4, the rat frizzled related protein gene. Gene. 2005;357:55–62. doi: 10.1016/j.gene.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 66.Varga AE, Stourman NV, Zheng Q, Safina AF, Quan L, Li X, Sossey-Alaoui K, Bakin AV. Silencing of the Tropomyosin-1 gene by DNA methylation alters tumor suppressor function of TGF-beta. Oncogene. 2005;24:5043–52. doi: 10.1038/sj.onc.1208688. [DOI] [PubMed] [Google Scholar]
- 67.Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, Kuo L, Shin DG, Rowe DW, Harris SE, Kalajzic I. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45:682–92. doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mantila Roosa SM, Liu Y, Turner CH. Gene expression patterns in bone following mechanical loading. J Bone Miner Res. 2010;23:23. doi: 10.1002/jbmr.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson JA, Chatterjee-Kishore M, Yaworsky PJ, Cullen DM, Zhao W, Li C, Kharode Y, Sauter L, Babij P, Brown EL, Hill AA, Akhter MP, Johnson ML, Recker RR, Komm BS, Bex FJ. Wnt/beta-catenin signaling is a normal physiological response to mechanical loading in bone. J Biol Chem. 2006;281:31720–8. doi: 10.1074/jbc.M602308200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.