Abstract
We examined single nucleotide polymorphisms (SNP) in the APOBEC3 locus on chromosome 22, paired to population sequences of pro-viral HIV-1 vif of peripheral blood mononuclear cells (PBMC), from 96 recently HIV-1 infected treatment naïve adults. We found evidence for the existence of an APOBEC3H linkage disequilibrium (LD) block associated with variation in GA->AA, or APOBEC3F signature, sequence changes in pro-viral HIV-1 vif sequence (top significant 10 SNPs with a top-significant p=4.8×10−3). We identified a common 5 position risk haplotype distal to APOBEC3H (A3Hrh). These markers were in high LD (D′ = 1; r2=0.98) to a previously described A3H ‘RED’ haplotype containing a variant (E121) with enhanced susceptibility to HIV-1 Vif (Zhen et al 2009 [1]). This association is confirmed by a haplotype analysis: Homozygote carriers of the A3Hrh had lower GA->AA (A3F) sequence editing on pro-viral HIV-1 vif sequence (p = 0.01), and lower HIV-1 RNA levels over time during early, untreated HIV-1 infection, (p = 0.015 mixed effects model). This effect may be due to enhanced susceptibility of A3H forms to HIV-1 Vif mediated viral suppression of sequence editing activity, slowing viral diversification and escape from immune responses.
Keywords: APOBEC3, HIV-1, vif, innate immunity, haplotype
1. Introduction
The APOBEC3 locus on chromosome 22 consists of a family of genes coding for cytidine deaminases that cause cytidine (C) to uridine (U) changes in the DNA minus strand sequence at preferred dinucleotide motifs. For example, APOBEC3G (A3G) acts primarily at ‘CC’ in the minus strand DNA (‘GG’ sense strand dinucleotide motif). This induces a change in the first position G, yielding an ‘AG’ in the resulting genomic sense strand DNA sequence [2–4]. By comparison, the APOBEC3F (A3F) member of the APOBEC3 family, recognizes the ‘GA’ plus strand dinucleotide motif and yields an ‘AA’ sense strand sequence.
The HIV-1 Vif protein binds and traffics A3G and A3F for proteolytic degradation, suppressing APOBEC3 editing activity on the HIV-1 genome. Absent Vif, A3G and A3F act to restrict viral replication by targeting the nascent HIV-1 minus strand DNA [5, 6] during pre-integration steps, inducing extensive hypermutation at preferred proviral DNA dinucleotide sites. The A3G and A3F have been heavily studied for association with HIV-1 infection [4, 7], although evidence exists that other members of the A3 family may edit and restrict HIV-1 [8, 9]. Variation in other A3 gene family members may also influence HIV-1 sequence diversification and clinical outcomes such as APOBEC3H [8, 10, 11], which may have an APOBEC3F (‘GA’) like dinucleotide target preference.
We surveyed genetic variation in the human APOBEC3 cassette (A3A, A3B, A3C, A3D(DE), A3F, A3G, A3H) in a cohort of treatment naïve recently HIV-1 infected adults, paired to measures of APOBEC3 sequence editing in the DNA sequence of pro-viral HIV-1 vif. We assayed pro-viral HIV-1 vif DNA in order to survey the reservoir of archived HIV-1 genomes, including those that may be replication incompetent, or replication impaired due to extensive sequence editing by A3G or A3F, or other A3 members [12–14]. We sought to determine if variation in A3H associated with HIV-1 sequence editing, and clinical markers independent of A3G and A3F associations.
2. Materials and Methods
2.1 Study Population
We selected 96 patients for study from the OPTIONS cohort of adults in early HIV-1 infection, which has been described elsewhere [15]. These patients were chosen based on having at least 1 aliquot of viably preserved, frozen peripheral blood mononuclear cells (PBMCs) from a time-point prior to anti-retroviral treatment from which we extracted host DNA. The OPTIONS study was approved by the University of California San Francisco Institutional Review Board, and all patients gave written, informed consent to participate in this study.
2.2 APOBEC3 Single Nucleotide Polymorphism Panel
We assayed a set of APOBEC3 loci single nucleotide polymorphisms (SNP) reported in the HapMap (http://hapmap.ncbi.nlm.nih.gov/) [16] found across the entire APOBEC3 region including APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3DE (A3DE), APOBEC3F (A3F), APOBEC3G (A3G) and APOBEC3H (A3H). We assayed all coding and non-coding SNPs within a 10KB window up and downstream of the first (A3A) and last member (A3H) of the APOBEC3 locus on chromosome 22. SNPs were genotyped using the GoldenGate SNP array assay system (Illumina, San Diego, CA, USA, Methods Supplement 1). The candidate SNPs selected for assaying variation in the APOBEC3 loci were identified via the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/) based on the HapMap 128 build [16]. We refer to SNPs by their ‘rs’ value in this report [17].
2.2 Viral Sequencing
Details on HIV-1 proviral vif DNA amplification, sequencing and the identification and counting of A3F or A3G signature sequence changes are described in the Methods Supplement.
2.3 Statistical Analysis
2.3.1 Viral sequence statistical analysis
For each potential A3G or A3F dinucleotide motif target, the number of G->A or G->G/A mixture sequence changes were identified, based on A3G and A3F dinucleotide targets mapped within the HXB2 vif reference sequence (http://www.hiv.lanl.gov/content/sequence/HIV/). The A3G or A3F dinucleotide target changes per individual were divided by the number of potential A3G and A3F targets within HXB2 to derive a proportion, which was the principal outcome measure in this study. (Methods Supplement 1 for more details).
2.3.2 Host sequence statistical analysis
We employed a minor allele frequency threshold of 2 % (values below excluded). We adopted a conservative HWE threshold of alpha = 0.05, and further excluded SNPs where greater than 5 % of the population had failed assay values. We tested the association between SNPs and the A3G and A3F signature fractions variables, using an ANOVA test as implemented in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) [18]. Similar results were obtained when implemented in R (http://cran.r-project.org/). Linkage disequilibrium analysis and graphical displays were achieved using D′ and r2 metrics and GOLD color scheme in Haploview [19]. Haplotype estimation frequencies were achieved using Estihaplo [20] and Fastphase [21]. All other statistical analyses are performed using Stata v10 (College Station, Texas, USA) and R. To relate APOBEC3 positions or haplotypes to HIV-1 clinical markers we employed longitudinal mixed effects modeling, with random effects specified for the individual and time, is SAS System Version 9.1 for Windows XP.
3. RESULTS
3.1 Patient Population Characteristics
The cohort was a median 34.6 years old (Interquartile Range (IQR) 30.6–40.8), 95 % Male, with a median HIV-1 RNA of 4.67 log10 HIV-1 RNA (copies/mL) (IQR 3.7–5.1), and a median CD4+ T cell count of 560 (IQR 472, 705). The subset of patients studied here did not differ from the larger OPTIONS cohort [22, 23].
3.2 HIV-1 Vif Sequence Editing Phenotype Estimation
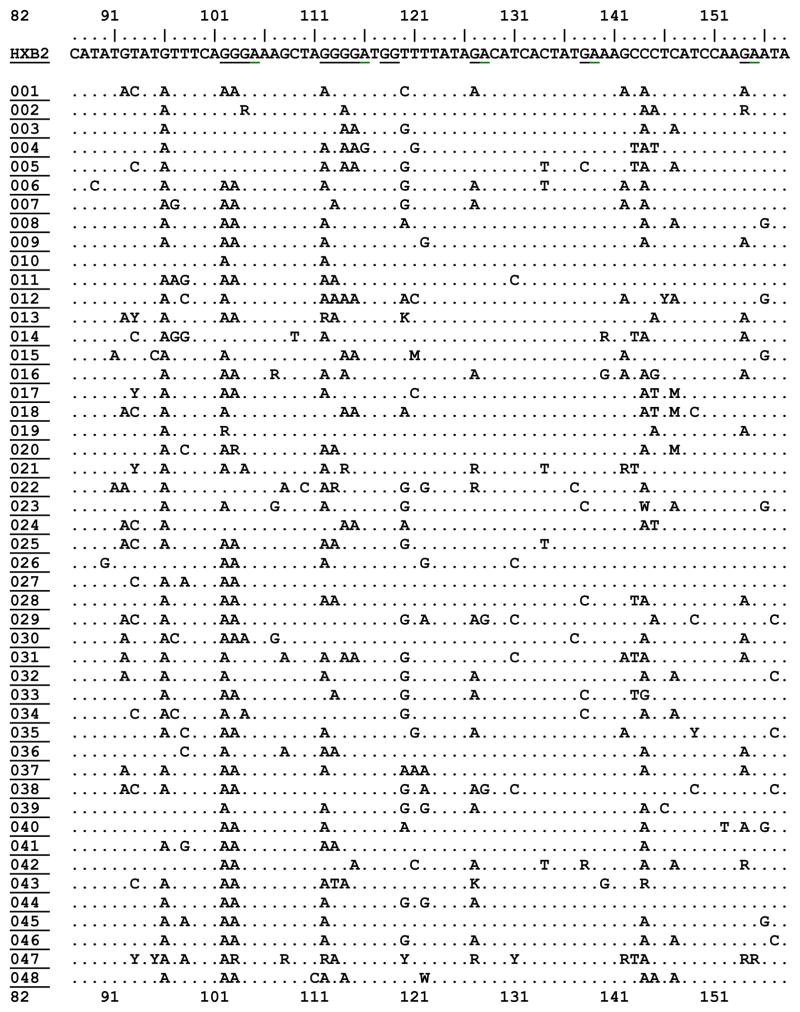
We recovered complete proviral vif DNA sequence from 94 patients. The fraction of A3F candidate motifs (‘GA’) in HXB2 that appeared as an ‘AA’ or ‘RA’ was a median 0.06 with an interquartile range of 0.04–0.08. The fraction of A3G candidate motifs (‘GG’) in HXB2 that appeared as ‘AG’ or ‘RG’, in patient specimens was a median of 0.095 with an interquartile range of 0.07, 0.12. Both the AA/GA (A3F signature) and GA/GG (A3G signature) ratios were normally distributed (Shapiro-Wilk W test for normal data p=0.5 and p=0.2 respectively). Table 1 displays a representative portion of the HIV-1 vif DNA alignment for all study participants.
Table 1. Characteristics of HIV-1 Vif DNA Sequences (82–158) from Study Subjects in Recent HIV-1 Infection.
Displaying nucleotide positions 82–158 of HIV-1 VIF DNA sequence for HXB2 viral reference strain, plus all 94 subjects for whom sequence was available for this study. Changes in sequence are shown relative to HXB2 reference strain on the top row. APOBEC3G (GG) and APOBEC3F/H (GA) dinucleotide motif positions are shown underlined in the HXB2 strain. Prepared in BioEdit. Full vif sequence for all participants will be deposited in GenBank and other open access databases.

|
3.3 APOBEC3 Genetics Genotyping Quality Control
In APOBEC3, 167 SNP markers were genotyped in 94 samples. Two individuals were removed due to high genotyping failure rate (>19 % of SNPs failed), with an average overall sample call rate of 93.7%. Thirteen SNPS were excluded due to high failure rate (9 missing in all subjects and 4 that failed in 26 % of subjects). Seventy-seven SNPs with minor allele frequencies below 2 % were removed from the analysis. Nine SNPs with a p-value below 0.05 were excluded by the exact Hardy-Weinberg Equilibrium test. After these quality control steps, 68 SNPs were analyzed, with a successful genotyping rate in the 92 remaining individuals of 99.6%. Lists of all SNPs considered in the association study analysis are in Supplemental Table 1 and 2.
3.4 APOBEC3 Loci Associations with proviral vif DNA Editing Patterns
We found a series of APOBEC3 SNPs that were each significantly associated with mutated fractions for the GA->AA (A3F) signature mutated fraction but not for GG->AG (A3G) signature mutated fraction. Table 2 displays p-values from the test of association of top-ranked SNPs with A3F- and A3G-associated signature mutation patterns. Ten out of 13 significant SNPs (p<0.05) were found within a 25kb span at the telomeric end of the APOBEC3 cluster (rs139339; rs139336; rs139323; rs139317; rs139316; rs139302 (top significant SNP p=4.8 10−3); rs139294; rs139283; rs139271; rs2413570). Three SNPs located nearby had p-values close to 0.05 (rs139284; rs139279; rs6519165). Several of these SNPs have been described by OhAinle (rs139302) [8], and Harari et al [24] (rs139294 and rs139302) and Zhen et al. [1] with the rs139302/D178E position. For these, an association signal is replicated by the present study. The 178 position is part of the Harari et al. ‘RDD’/‘GKE’ haplotype, or the Zhen et al ‘RED’ haplotype. These relationships are detailed in Figure 1.
Table 2. APOEBC3H SNP Associations with APOBEC3F/H (GA->AA) and APOBEC3G (GG->AG) signature mutated fraction.
APOBEC3H region SNPs associated with A3F/H or A3G mutated fraction, ranked by p-value of the association to the A3F/H mutated fraction.
| SNP number | SNP | Sample Size | APOBEC3F/H Mutated Fraction (GA->AA) | APOBEC3G Mutated Fraction (GG->AG) | ||
|---|---|---|---|---|---|---|
| Uncorrected P-value | Rank | Uncorrected P-value | Rank | |||
| 23 | rs139339 | 92 | 0.01006 | 2* | 0.1168 | 4 |
| 22 | rs139336 | 92 | 0.01006 | 2* | 0.1168 | 4 |
| 21 | rs139323 | 92 | 0.01006 | 2* | 0.1168 | 4 |
| 20 | rs139317 | 92 | 0.01177 | 6* | 0.1648 | 11 |
| 19 | rs139316 | 92 | 0.01006 | 2* | 0.1168 | 4 |
| 18 | rs139314 | 92 | 0.2082 | 33 | 0.2698 | 22 |
| 17 | rs139302 | 87 | 0.004802 | 1** | 0.2935 | 27 |
| 16 | rs139296 | 91 | 0.2108 | 34 | 0.2563 | 20 |
| 15 | rs139294 | 92 | 0.01401 | 7* | 0.1105 | 3 |
| 14 | rs139285 | 92 | 0.09264 | 22 | 0.201 | 16 |
| 13 | rs139284 | 92 | 0.05855 | 15 | 0.257 | 21 |
| 12 | rs139283 | 92 | 0.01616 | 8* | 0.1564 | 10 |
| 11 | rs139279 | 92 | 0.06322 | 17 | 0.3194 | 28 |
| 10 | rs139271 | 92 | 0.02301 | 10* | 0.1989 | 15 |
| 9 | rs11705514 | 92 | 0.944 | 66 | 0.9954 | 68 |
| 8 | rs5995668 | 92 | 0.4026 | 43 | 0.4089 | 31 |
| 7 | rs6001423 | 92 | 0.399 | 42 | 0.5164 | 40 |
| 6 | rs5757472 | 92 | 0.1802 | 30 | 0.2458 | 19 |
| 5 | rs17537581 | 92 | 0.247 | 35 | 0.4431 | 33 |
| 4 | rs2413570 | 92 | 0.01828 | 9* | 0.1932 | 14 |
| 3 | rs5757465 | 92 | 0.1747 | 29 | 0.07682 | 1 |
| 2 | rs6519165 | 92 | 0.0542 | 14 | 0.1691 | 12 |
| 1 | rs4820365 | 92 | 0.08952 | 20 | 0.5384 | 42 |
Figure 1. Relationship of A3H Core Risk Haplotype (CTGCT) to Previously Described SNPs within APOBEC3H.
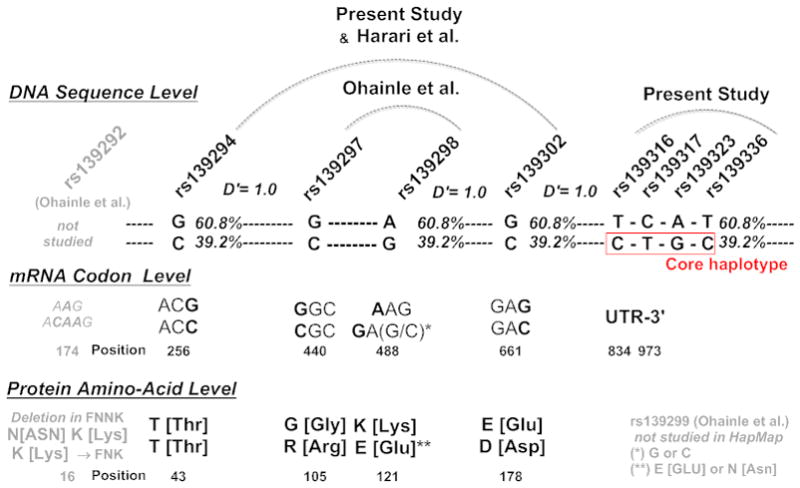
The figure displays A3H genetic patterns in the present study, prior studies (Harari et al.), and HapMap, at the DNA sequence level, mRNA codon level, protein amino-acid level. Linkage studies within HapMap reveal that the ‘CTGCT’ A3H core haplotype defined in the current study is in strong linkage to a ‘RED’ haplotype pattern within A3H.
3.5 Haplotype Estimation
In order to refine the observed association, we estimated haplotype frequencies in our dataset using the most associated 10 markers listed above, plus an additional 3 markers found in the telomeric region. Working from this set of SNPs, we identified a risk haplotype within the A3H region of 5 non-coding SNPS (rs139339*C; rs139336*T; rs139323*G; rs139317*C; rs139316*T, also referred to here as ‘CTGCT’ or the A3H “core” risk haplotype (A3Hrh), for convenience). The frequency of this haplotype was estimated to be 35.3 % in our cohort.
3.6 Haplotype Analyses
When a haplotype association test was run after phase inference with the A3Hrh 5 core SNPs (CTGCT), the association remain significant (ANOVA, p=0.01), and suggested a recessive model (p=0.05). Four of five members of the A3Hrh were found in A3H haplotypes found in HapMap (rs139336; rs139323; rs139317; rs139316). We examined our A3H core risk haplotype (4 of 5 members, rs139316/C; rs139317T; rs139323G; rs139336C) for association with previously reported SNPs (rs139302, rs139298, rs139297, and rs139294 [8, 10, 11]). We found a 4 member version (CTGC) of our A3H ‘CTGCT’ core haplotype was in complete linkage to rs139302C, rs139298G, rs139297C and rs139294C, with a D′ of 1.0 in all cases within HapMap. This indicates our core haplotype recovered an extended A3H haplotype coding for previously described A3H positions R105, E121 and D178 (“RED”) [1, 8, 10, 11].
3.7 SNP Typing Methods, HapMap Codon Inference and the A3H 121 Position
The A3H 121 codon, present as the second position our A3H risk haplotype (A3H 105/121/178) contains two SNPs, at positions 1 and 3 of the codon. Each of these nucleotides are coding in position 121 (E, K, N or D are possible). However, closely spaced SNPs cannot be simultaneously detected in modern high-throughput SNP assays due to high overlap (loss of specificity) in the binding of probe or primer to the target DNA region. Hence, HapMap only tracks the first position and not the third. We were able to link our A3H risk haplotype to only one SNP within codon 121, position 1 (rs139298) but not codon position 3 (rs139299, not present in HapMap). Based on available information, we inferred a G (GAG) coding for E, or Glutamine. Alternatively, the K121 is coded by AAG (K or Lysine), whereas an N121 (N or Asparagine) is coded by an AAC. Hence, it remains possible a GAC and not GAG were present in some study subjects. This change would lead to expression of D or Aspartic Acid that could yield an RDD haplotype and not the RED haplotype in some subjects.
3.8 Clinical Associations with APOBEC3H Risk Haplotype
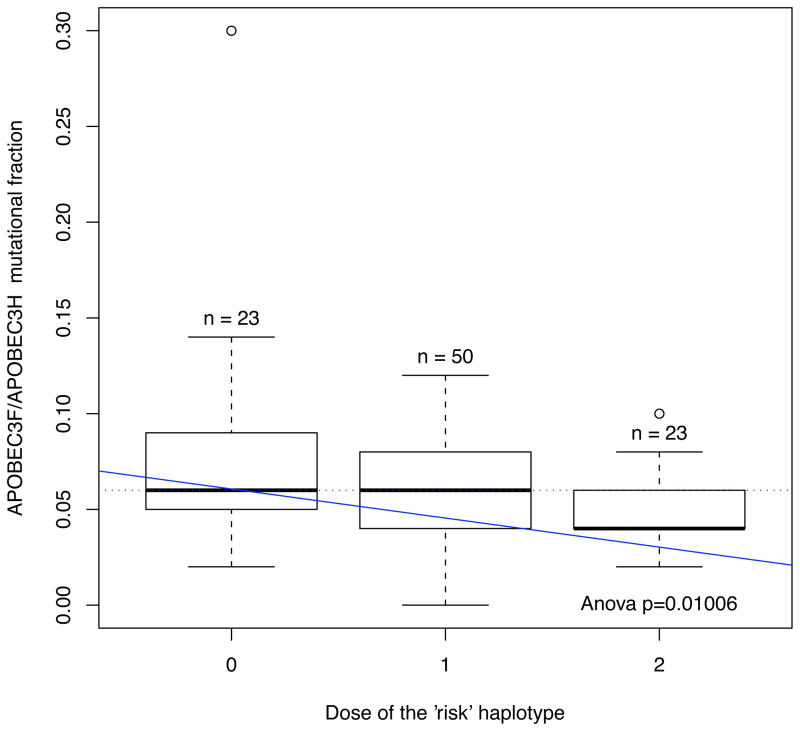
Homozygote carriers of the A3H risk haplotype had lower A3F associated mutational activity, than heterozygote of homozygote nulls (Figure 2). In a longitudinal mixed effects models, homozygote carriers of the A3H risk haplotype ‘CTGCT’ had lower HIV-1 RNA levels during 1 year or more of early untreated HIV-1 infection, compared to heterozygote carriers (+0.17 log10 copies/mL, p = 0.015) or homozygote nulls (+0.20 log10 copies/mL, p = 0.018). We did not observe an association of the A3H risk haplotype with CD4+ T cell counts over time.
Figure 2. APOBEC3H Risk Haplotype and APOBEC3F (GA->AA) Signature Mutation Fraction (Proportion of Motifs Converted Relative to HXB2).
A haplotype association test run after phase inference with the 5 core SNPs reveals a significant association is found between carriage of the core ‘RED’ haplotype and A3F signature upon HIV-1 Vif (ANOVA, p=0.01). The results suggest a recessive model (p= 0.05446).
3.9 Confirmation of Prior Findings
Our aim was confirmation and extension of previous findings on the existence of a strong LD block within A3H that associates with sequence variation in HIV-1 vif DNA [1, 8, 10, 11]. Therefore, we did not perform multiple comparison adjustments.
4. Discussion
We surveyed genetic variation in the APOBEC3 gene family locus with a panel of single nucleotide polymorphisms (SNPs), and paired this data to the sequence of proviral HIV-1 vif from peripheral blood mononuclear cells of recently HIV-1, anti-retroviral naïve, infected adults. We observed significant associations between a set of SNPs in the APOBEC3H region, and a resultant A3H risk haplotype, to reduced GA->AA sequence editing pattern typically associated with A3F activity [10], and to lower HIV-1 RNA levels over time.
We found our A3H risk haplotype was in strong linkage disequilibrium with several coding positions within A3H (105/121/178). The second of these, A3H 121, falls in a region of A3H with high structural homology to the closely related A3G protein. This loop region of A3G is bound by the HIV-1 Vif protein that routes A3G for proteolytic degradation and hence suppression of activity. A3H proteins bearing an E121 variant in this region are susceptible to HIV-1 Vif mediated suppression [1] despite demonstrating robust anti-retroviral activity in vitro. In contrast, the A3H D121 and K121 variants show reduced susceptibility to HIV-1 Vif in vitro, and would be predicted to be expressed and retain anti-HIV-1 sequence editing activity during infection in vivo. In our analysis we found evidence that our A3H risk haplotype is in strong linkage with the previously described ‘RED’ haplotype (R105/E121/D178) [10][1]. Hence, the association of our risk haplotype with reduced APOBEC3 signature upon HIV-1 DNA sequence may be due to the carriage of the A3H E121 variant, rendering that A3H protein susceptible to Vif targeting and proteolytic degradation, blocking A3H activity against HIV-1. That said, it is possible that our A3H haplotype does not contain an E at position 121 in all carriers in this study, with a D (or Aspartic acid) carried instead. Few human A3H genes have been re-sequenced in persons with HIV-1 infection, and the true frequency of an E121, K121, D121 and N121, and their linkages, are not yet known. In future studies, we plan to re-sequence A3H and other A3 members in cohorts of HIV-1 infected to determine frequencies of these distinct genetic forms.
We observed that homozygote carriers of the A3H risk haplotype, linked to the previously described RED or RDD haplotypes, appear to have lower A3F signature (GA->AA) nucleotide sequence editing activity, and also lower HIV-1 RNA levels over time early, untreated HIV-1 infection. Lower A3H activity against HIV-1 may slow viral sequence diversification and escape from immune responses, leading to the lowered viral loads observed here. The A3H risk haplotype may bear the E amino acid at A3H position 121 that has been recently reported to confer susceptibility of A3H proteins to targeting by HIV-1 Vif [1]. Further study will be required to determine the true frequency of the E121 variant in humans, and the expression level of an E121 bearing variant in the CD4+ T-cells which support HIV-1 replication. That A3H, or some of its variants, may cause a detectable mutational signature on HIV-1 genomes in vivo suggests A3H may play an independent role in HIV-1 sequence diversification and evolution [25].
Supplementary Material
Table S1 summarizes all original candidate SNPs
Table S2 summarizes associates of QC’d SNP set with A3F and A3G signature fractions.
Acknowledgments
We would like to acknowledge grant support from the University of California San Francisco Academic Senate Individual Investigators Research Grant (JDB), and the NIAID (AI 066917/JDB) and the UCSF-GIVI CFAR (P30 AI027763). We would like to thank the participants of the OPTIONS Cohort at San Francisco General Hospital. We would like to thank Sophie Stephenson and Robert Atchison for technical assistance with DNA extractions and amplification of HIV-1 vif DNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J Virol. 84(4):1902. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9(11):1404. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 3.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 4.Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol Mol Biol Rev. 2009;73(2):211. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11(5):435. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 6.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78(11):6073. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai SK, Wong JK, Barbour JD. Turning up the volume on mutational pressure: is more of a good thing always better? (A case study of HIV-1 Vif and APOBEC3) Retrovirology. 2008;5:26. doi: 10.1186/1742-4690-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80(8):3853. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourara K, Liegler TJ, Grant RM. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 2007;3(10):1477. doi: 10.1371/journal.ppat.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83(1):295. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4(3):249. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armitage AE, Katzourakis A, de Oliveira T, Welch JJ, Belshaw R, Bishop KN, Kramer B, McMichael AJ, Rambaut A, Iversen AK. Conserved footprints of APOBEC3G on Hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. J Virol. 2008;82(17):8743. doi: 10.1128/JVI.00584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kijak GH, Janini LM, Tovanabutra S, Sanders-Buell E, Arroyo MA, Robb ML, Michael NL, Birx DL, McCutchan FE. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology. 2008;376(1):101. doi: 10.1016/j.virol.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Reddy K, Winkler CA, Werner L, Mlisana K, Abdool Karim SS, Ndung’u T. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS. 24(2):195. doi: 10.1097/QAD.0b013e3283353bba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbour JD, Hecht FM, Wrin T, Segal MR, Ramstead CA, Liegler TJ, Busch MP, Petropoulos CJ, Hellmann NS, Kahn JO, Grant RM. Higher CD4+ T cell counts associated with low viral pol replication capacity among treatment-naive adults in early HIV-1 infection. J Infect Dis. 2004;190(2):251. doi: 10.1086/422036. [DOI] [PubMed] [Google Scholar]
- 16.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourraud PA, Feolo M. The Babel Tower revisited: SNPs - Indels - CNVs. Confusion in naming sequence variant always rises from ashes. Tissue Antigens. 75(3):199. doi: 10.1111/j.1399-0039.2009.01424.x. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC. Haploview. Visualization and analysis of SNP genotype data. CSH Protoc. 2009;(10) doi: 10.1101/pdb.ip71. pdb ip71, 2009. [DOI] [PubMed] [Google Scholar]
- 20.Gourraud PA, Genin E, Cambon-Thomsen A. Handling missing values in population data: consequences for maximum likelihood estimation of haplotype frequencies. Eur J Hum Genet. 2004;12(10):805. doi: 10.1038/sj.ejhg.5201233. [DOI] [PubMed] [Google Scholar]
- 21.Yu Z, Schaid DJ. Methods to impute missing genotypes for population data. Hum Genet. 2007;122(5):495. doi: 10.1007/s00439-007-0427-y. [DOI] [PubMed] [Google Scholar]
- 22.Kothe D, Byers RH, Caudill SP, Satten GA, Janssen RS, Hannon WH, Mei JV. Performance characteristics of a new less sensitive HIV-1 enzyme immunoassay for use in estimating HIV seroincidence. J Acquir Immune Defic Syndr. 2003;33(5):625. doi: 10.1097/00126334-200308150-00012. [DOI] [PubMed] [Google Scholar]
- 23.Barbour JD, Hecht FM, Wrin T, Liegler TJ, Ramstead CA, Busch MP, Segal MR, Petropoulos CJ, Grant RM. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS. 2004;18(12):1683. doi: 10.1097/01.aids.0000131391.91468.ff. [DOI] [PubMed] [Google Scholar]
- 24.Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105(14):5501. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li MM, Wu LI, Emerman M. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J Virol. 84(1):88. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 summarizes all original candidate SNPs
Table S2 summarizes associates of QC’d SNP set with A3F and A3G signature fractions.