Abstract
Three Indian rhesus macaques, Ad-SIV primed/protein boosted and exposed twice to high-dose mucosal SIVmac251 challenges, exhibited elite control of viremia over 6½ years. They were negative for host factors associated with control of SIV infection. After a third intrarectal challenge with SIVsmE660, all controlled viremia, with one (macaque #5) maintaining undetectable viremia in blood. Acquisition was not blocked, but virus was contained in the jejunum and draining lymph nodes. Polyfunctional memory T cell responses and high-titered neutralizing and non-neutralizing serum and mucosal antibodies were present before and maintained post-challenge. The level of protection seen for animal #5 was predicted from analyses of gene transcription in jejunum 2 weeks post-challenge. Macaques #7 and #9, exhibiting lower pre-challenge cellular and humoral immunity, partially controlled the SIVsmE660 challenge. Initial vaccine-induced control by macaque #5 extended to the SIVsmE660 challenge due to multiple immune mechanisms that were boosted and augmented by cryptic SIV exposure.
Keywords: SIV, elite-control, virus sequestration, multifaceted immunity, vaccine, memory
Introduction
Identification of immune correlates of protection from HIV infection has proven to be very difficult given the multifaceted nature of the host immune response and factors associated with viral control (Deeks and Walker 2007), as well as the sequence variability of HIV and its complex life cycle. Natural, elite control of HIV infection is rare. Initially defined as control of viremia to undetectable levels without drug therapy, the extent of elite control using highly sensitive PCR methods has been recently determined as median plasma RNA levels of 2 copies/mL (Pereyra et al., 2009). Elite controllers are invaluable for deciphering mechanisms of host immune control of viremia. Multiple factors have been implicated, including HLA class I alleles B*27 and B*57 associated with protective CTL responses; the presence of polyfunctional, mucosal CTLs; enrichment of certain killer cell Ig-like receptors (KIR) and/or ligands; the presence of viruses with attenuated replication; a general decrease in levels of immune activation; and the presence of intracellular host factors that limit virus replication, such as APOBEC3G and TRIM5α (Baker et al., 2009; Deeks and Walker, 2007; Ferre et al., 2009; O’Connell et al., 2009; Prado et al., 2010). HIV-1 neutralizing antibody and antibody reactivity by Western blot have correlated with lower viral loads in one cohort of elite controllers (Pereyra et al., 2009), while higher titers of ADCC antibody have correlated in another (Lambotte et al., 2009). Despite their best efforts, researchers have been unable to point to a single definitive immune correlate shared among all elite controllers.
Elite control of SIV infection in rhesus macaques has also been demonstrated (Loffredo et al., 2007; Yant et al., 2006). Most attempts to understand immune control in macaques have focused on the role of the MHC class I allele Mamu-B*08 in protection of Indian rhesus macaques from pathogenic SIVmac239 infection. Fifty percent of all macaques possessing this allele establish elite control. In one study, peripheral CD8+ T cells were depleted in macaque elite controllers and the frequency and specificity of the CD8+ T cells reappearing upon control of viremia were evaluated (Friedrich et al., 2007). CD8+ T cells which recognized epitopes restricted by Mamu-B*08 were expanded. Upon mutation of associated epitopes and re-infection of Mamu-B*08 positive macaques, the majority of the mutated viruses were unable to establish elite control (Valentine et al., 2009). One cannot discount however the potential role of NK cells in control of viremia, since they express KIRs that recognize class I ligands. Evidence of KIR3DL polymorphisms and decline of NK function has been associated with higher SIV replication in rhesus macaques (Bostik et al., 2009). Other protective mechanisms may involve CD4+ T cells, as those isolated from SIV elite controller macaques are able to lyse infected macrophages in vitro (Sacha et al., 2009).
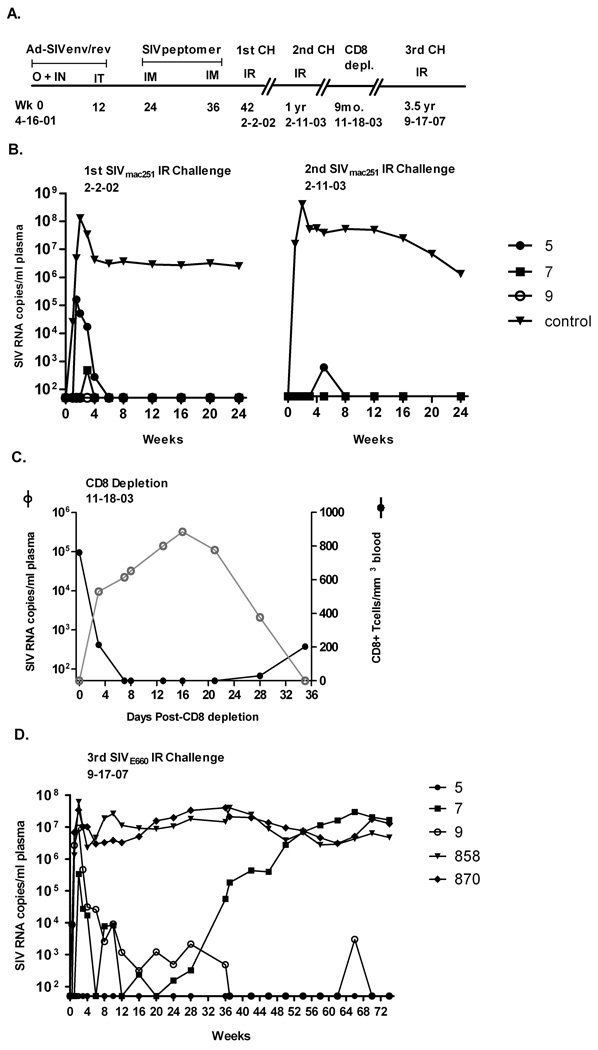
We have followed three Indian rhesus macaques, which over a 6½ year period maintained elite control of peak and chronic viremia after two separate high dose mucosal SIVmac251 challenges. These macaques are unique in that none of the reported MHC class I or II alleles associated with elite control or diminished chronic viremia (Mamu A*01, B*17, B*08, DRB1*0306 and DRB1*1003) (Giraldo-Vela et al., 2008; Loffredo et al., 2007; Pal et al., 2002; Yant et al., 2006; Zhang et al., 2002) have been detected. They were originally immunized with replicating adenovirus type 5 host range mutant (Ad5hr)-SIVsmH4env/rev and boosted with SIVmac251 peptomer (a conformational polypeptide composed of 18-mer units representing a partial sequence of the CD4 binding site) and exhibited elite control of a subsequent high dose SIVmac251 rectal challenge (Patterson et al., 2004) (Fig. 1A). Control was achieved not only during the chronic phase, but also during acute infection. One macaque (#9) showed complete lack of plasma viremia, one (#7) exhibited a single positive detection of 500 RNA copies/ml plasma, and the last (#5) displayed only low level acute viremia (peak of 105 RNA copies) and rapid clearance by week 6. All macaques were SIV proviral DNA positive at one or more time points, however. Twelve months later with no intervening immunization, a second rectal challenge with the same SIVmac251 virus stock resulted in no detectable acute or chronic viremia (macaques #7 and #9) or a single detection (macaque #5), and subsequent durable protection (Malkevitch et al., 2006) (Fig. 1B). In order to identify the mechanism of elite control, in vivo CD8 depletion experiments were conducted 9 months later. Whereas viremia rebounded with CD8 depletion and was controlled with reappearance of CD8 T cells in macaque #5, as shown in blood (Fig. 1C), lymph node and rectal tissue, viremia was undetectable throughout the time course for macaques #7 and #9 (Malkevitch et al., 2006).
Figure 1.
Immunization and challenge history of elite controller macaques (31, 40). (A) Macaques #5, #7, and #9 were originally immunized with an Ad-SIVsmH4 env/rev recombinant vector and boosted with a conformational CD4 binding site peptomer. Two priming immunizations given first orally and intranasally and second intratracheally were followed by two intramuscular boosts of peptomer in PBS. A high dose SIVmac251 rectal challenge was administered 6 weeks later. The times of second and third intrarectal challenges with SIVmac251 and SIVsmE660, respectively, and an intermediate in vivo CD8 depletion are indicated. (B) SIV RNA copies/ml plasma are shown for all three macaques, exhibiting elite control of the first high dose SIVmac251 rectal challenge and durable protection from a identical, second SIVmac251 rectal challenge one year later. Control group represents the geometric mean of 8 and 2 control macaques for the first and second challenges, respectively. (C) In vivo peripheral CD8+ T cell depletion resulted in reemergence of virus for macaque #5. Virus remained undetectable for #7 and #9 (not shown). (D) Outcome of a third heterologous rectal SIVE660 challenge, 3.5 years after the CD8 depletion study and 8 years since initial immunization. Macaques 858 and 870 are naïve controls.
A cellular component seemed to be involved in the durable protection of at least macaque #5. Due to productive virus exposure after the first SIV challenge, macaque #5 showed both Gag- and Env-specific cellular immune responses (Malkevitch et al., 2006). However, high titered SIV gp120 binding antibodies, neutralizing antibody to T-cell line adapted (TCLA)-SIVmac251 and antibody-dependent cellular cytotoxicity (ADCC) mediating antibodies were also present. Macaques #7 and #9 also exhibited cellular responses to Gag and other SIV antigens before and after the SIVmac251 rechallenge/CD8 depletion, in spite of their aviremic status, suggesting exposure to viremia below the detection limit of 50 copies or sequestration of virus at a secondary site. However, no anti-envelope binding antibody in serum or mucosal secretions was detected for either macaque #7 or #9.
Overall, no clear immune correlate of protection emerged from these earlier studies. Therefore, 3.5 years after CD8 depletion and 7 ½ years since initial immunization, we rechallenged the macaques using pathogenic, heterologous SIVsmE660. Our aims were to examine the extraordinary long-term viremia control exhibited by the macaques, evaluating the extent of virus in tissues as well as blood, and to comprehensively investigate durable immunity and protection using a spectrum of the newest methods to evaluate cellular, humoral and mucosal immunity. Extensive MHC class I plus TRIM5α genotyping, the latter recently implicated in modulation of SIV infection in rhesus macaques (Lim et al., 2010), as well as analysis of gene transcriptional profiles in jejunum before and after challenge were performed. Even though three macaques cannot provide statistical power, the opportunity to acquire valuable new information regarding elite control outweighed this concern.
Results
SIVsmE660 challenge results
Following SIVsmE660 challenge both naïve control macaques, #858 and #870, became rapidly infected, exhibiting peak viral loads between 107 and 108 copies/ml plasma. Subsequently over the entire monitoring period, the viral burdens for both macaques never dropped below 106 copies/ml. In evaluating viral loads of the elite controller macaques, amplification of the SIVmac251 gag gene by real-time PCR was not detected in any macaque plasma sample through week 12 post SIVsmE660 challenge (data not shown). Therefore, plasma viremia detected by NASBA during this time period is attributed solely to SIVsmE660. Following intrarectal SIVE660 challenge, macaque #5 continued to exhibit elite control of the heterologous, pathogenic SIV isolate (Fig. 1D). Viral RNA was undetectable in blood over the entire time course. Consistent with its lack of viremia, no significant drop in CD3+CD4+ T cells in blood was observed (supplementary Fig. S1A). Similarly, a high percentage of CD3+CD4+ T cells were present in jejunal tissue 2 weeks post-challenge (supplementary Fig. S1C). However, these cells declined in BAL implying that virus was present at a cryptic site (supplementary Fig. S1B). This was confirmed by the presence in lymph node of 17,512 and 29,798 viral RNA copies, at weeks 2 and 36 post-challenge, respectively, (Table 1) using NASBA, which does not discriminate SIVmac251 and SIVsmE660 gag sequences. A separate SIVmac251 PCR confirmed reactivated, low level RNA of 1650 and 349 viral copies at weeks 2 and 36 post-challenge, respectively for animal #5. Thus while undetectable in plasma, both viruses were detected in the lymph node of macaque #5 post-challenge. In jejunum, no SIVsmE660 was detected post-challenge, however pre-challenge, 108 copies of SIVmac239 were detected – 3 ½ years after CD8 depletion. Therefore, SIVmac251 originating from the first challenge was present and being controlled at very low levels.
Table 1.
Tissue viral loads
| Lymph node | ||||||
|---|---|---|---|---|---|---|
| Conserved SIV Gag (copies/µg tissue RNA) |
SIV251 (copies/µg tissue RNA) |
|||||
| Pre | Week 2 | Week 36 | Pre | Week 2 | Week 36 | |
| 5 | Neg (I) | 17,512 (M) | 29,798 (A) | Neg | 1650 | 349 |
| 7 | Neg (I) | 21,835 (M) | 85, 581(A) | Neg | Neg | Neg |
| 9 | Neg (A) | 745,813 (M) | 16,396 (A) | Neg | Neg | Neg |
| 858 | Neg (I) | 725,514 (M) | 1,216,698(I) | Neg | Neg | Neg |
| 870 | Neg (I) | 66,720 (M) | 586, 948(I) | Neg | Neg | Neg |
| Jejunum | ||||||
|---|---|---|---|---|---|---|
| SIV239 (copies/µg tissue RNA) |
SIVE660 (copies/µg tissue RNA) |
|||||
| Pre | Week 2 | Week 36 | Pre | Week 2 | Week 36 | |
| 5 | 108 | 6356 | 2400 | Neg | ND | Neg |
| 7 | Neg | Neg | Neg | Neg | ND | Neg |
| 9 | Neg | Neg | Neg | Neg | ND | Neg |
| 858 | Neg | Neg | Neg | Neg | ND | 223 |
| 870 | Neg | Neg | Neg | Neg | ND | 20,001 |
M = mesenteric; A = axillary; I = inguinal
For macaque #9, a 1 log blunting of acute plasma viremia compared to naïve control macaques was observed. Subsequently, viremia was controlled rapidly by week 10 to around 1000 copies and became undetectable after week 36 (Fig. 1D). A single viral detection occurred at week 66, but overall, this macaque exhibited sustained, strong control out to 74 weeks. The high acute plasma viremia was reflected in a lymph node biopsy, where the viremia level was similar to controls, but by week 36, the lymph node viral load was reduced (Table 1). No SIVmac251 virus was detected at any time point. In accordance with the high acute viremia levels, CD3+CD4+ T cells dropped in blood, BAL and jejunum (supplementary Figs. S1B, C), but thereafter remained stable in blood. Neither SIVmac239 nor SIVsmE660 was detected in jejunum before or after SIVsmE660 challenge (Table 1).
A blunted peak plasma viremia of 2 logs compared to control macaques was seen for macaque #7 with quick resolution of viremia by week 6 (Fig. 1D). Viral RNA loads thereafter vacillated broadly between ≤ 10,000 copies and undetectable levels until control was rapidly lost at week 36 (Fig. 1D). Afterwards, viremia continued to rise and reached control levels, suggesting viral escape by an as yet undetermined mechanism. Likewise, CD3+CD4+ T cell numbers were unchanged on average in blood until 28 weeks post-challenge, when a slow decline was observed (supplementary Fig. S1A). During the acute phase (week 2), no drop in CD3+CD4+ T cells was seen in either BAL or jejunum (supplementary, Figs. S1B, C), but viremia was detectable in lymph node at a level comparable to macaque #5 (Table 2). By week 36, virus (presumably SIVsmE660) spiked to approximately 85,000 copies, similar to the rising plasma levels. However, SIVsmE660 could not be amplified from week 36 jejunal biopsies, although it was readily detected in the control macaques at the same time point. Unlike macaque #5, SIVmac239 was not seen in jejunum prior to or after SIVsmE660 challenge.
Cellular immune responses
A battery of cellular immune responses was evaluated in blood and tissue of these macaques. Gag239- and Env239 (again, closely related to 251)-specific IFN-γ secreting cells were evaluated by ELISPOT in freshly prepared peripheral blood lymphocytes. Prior to challenge, animal #5 exhibited higher levels of Env-and Gag-specific responses than #9, whereas macaque #7 showed no SIV-specific IFN-γ-secreting cells (supplementary Fig. S2A). All three elite controllers exhibited strong T cell proliferative responses pre-challenge which declined immediately post challenge, but regained potency by week 12 (supplementary Fig. S2B). Gp120, p27 and Ald-SIV stimulations all elicited responses.
Multiparameter flow analysis of Env- and Gag-specific cytokine secreting T cells from blood, BAL and jejunal tissues was carried out using an 11-color antibody panel to distinguish T cell and memory subsets, as well as IFN-γ, IL-2, and TNF-α cytokines. No virus-specific responses were detected for either macaque #7 or #9 in PBMC prior to or following challenge in contrast to macaque #5 which exhibited broadly reactive cytokine positive responses that were especially potent among CD4 total memory T cells specific for both EnvsmH4 and Gag239 (Fig. 2A). Prior to SIVE660 challenge the Env-specific CD4 memory T cells recognized EnvsmH4 peptides, representing the env gene present in the Ad5hr-SIVsmH4env/rev vaccine and not the envelope matched to the first challenge virus, SIVmac251. After SIVsmE660 challenge however, the response of animal #5 became broader, with strong, sustained Env specific responses to Env239 peptides as well. Overall however, the highest frequency of response prior to and post challenge was to Gag239 peptides (Fig. 2A) reflecting prior challenge history rather than the vaccination regimen.
Figure 2.
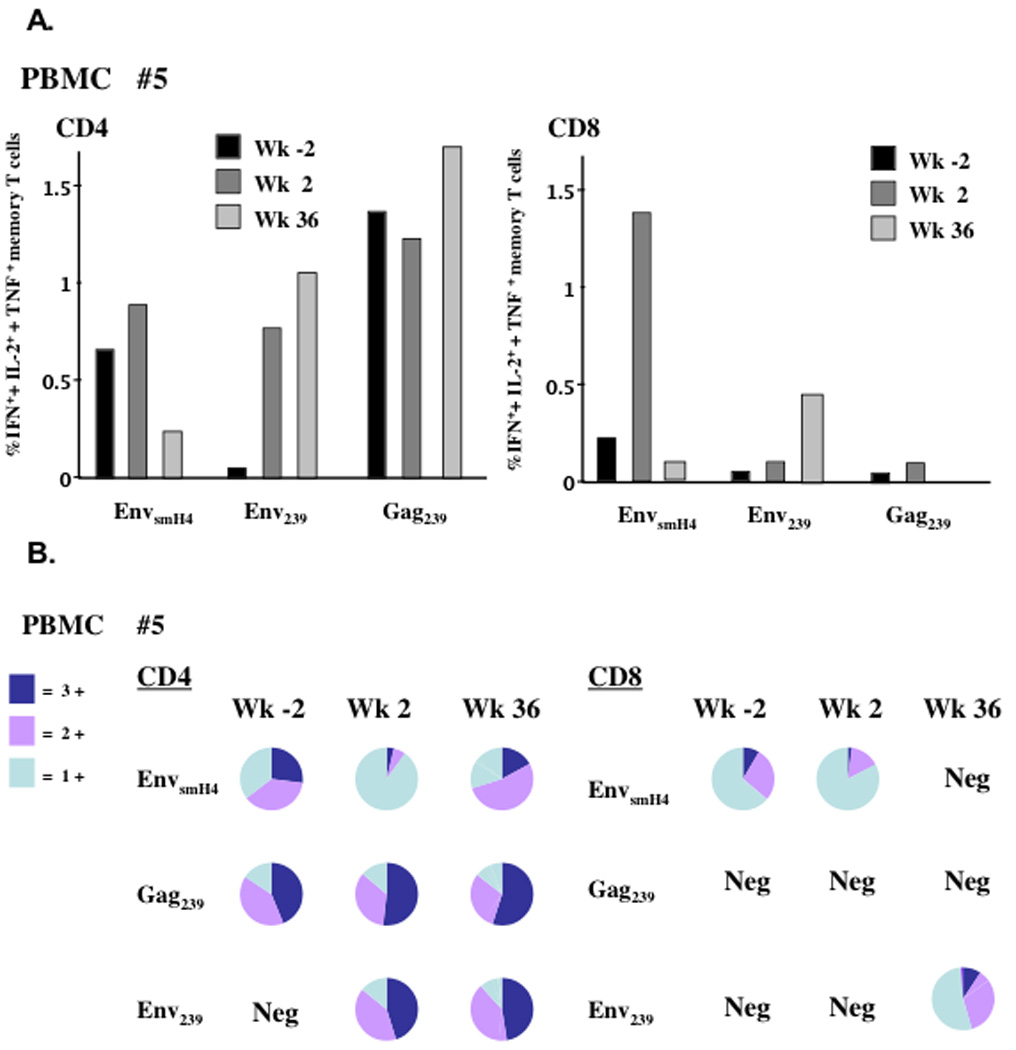
The sum percent of IFN-γ+, + IL-2+, + TNF-α+ total memory T cells in PBMCs for macaque #5. (A) Shown are the total cytokine positive cells among total memory CD4 or CD8 T cells at weeks −2, 2 and 36 post SIVsmE660 challenge after stimulation with three separate peptide pools. (B) Pie charts for each flow sample from panel A, representing the percent triple, double and single cytokine secreting cells. Neg = negative response.
The total memory cells in PBMC of macaque #5 were further examined for polyfunctionality, (cells secreting two or three cytokines), reported to correlate with protection (Darrah et al., 2007; Lindenstrom et al., 2009; Seder et al., 2008). Prior to challenge, up to 65% of EnvsmH4 and 80% Gag239 CD4 total memory cells were polyfunctional in nature (Fig. 2B) and predominately of a central memory phenotype (CD28+CD95+; not shown). For CD4 Gag239 memory cells, this polyfunctional response was maintained post challenge. Newly expanded CD4 memory cells in response to the SIVsmE660 challenge included Env239-specific polyfunctional cells that persisted to week 36. Interestingly, the CD4 EnvsmH4-specific memory cells seemed to evolve functionally post-challenge, and at week 2 during the acute phase displayed an effector-type profile with the majority secreting only IFN-γ (Fig. 2B).
The CD8 memory T cell response was of low frequency pre challenge for macaque #5 and only seen in response to EnvsmH4 stimulation (Fig. 2A). By week 2 however, these cells dramatically expanded up to 1.4% of total memory T cells, again with an IFN-γ secreting (Fig. 2B) effector memory phenotype (CD28−CD95+). Although not detected prior to challenge, CD8 total memory T cell responses for macaques #7 and #9 were also observed in PBMC post challenge. However, the frequency of the responses was low (0.1 – 0.2%), and no polyfunctionality was seen (data not shown).
For naïve controls, CD4 and CD8 Gag and Env specific total memory T cells were expanded post challenge with a peak CD4 Gag frequency of 1.5%. The cytokine profile of these memory cells was almost exclusively TNF-α (data not shown).
In contrast to responses only for animal #5 in peripheral blood, virus-specific cellular immune responses were detected for all three macaques in BAL. Figure 3A shows the frequency of total cytokine responses among CD4 and CD8 memory T cells for macaques #5, #7 and #9 pre- and post-challenge. As fewer lymphocytes were isolated from BAL at week 2, Env and Gag peptide stimulations had to be combined. Therefore, for ease in interpretation, stimulations shown in Fig. 3A are combined for all time points. CD4 total memory T cell responses were highest for animal #5 pre-challenge while responses for macaques #7 and #9 were detectable but low. None of the macaques exhibited post-challenge responses until week 36 when a very high total cytokine response was seen among CD4 total memory cells for animal #5 (2.68%) and lesser responses by macaques #7 (1.5%) and #9 (0.25%). For macaque #5, most of the pre-challenge CD4+ memory T cells were polyfunctional (Fig. 3B), although compared to blood where the majority were central memory, only 20–25% were triple positive central memory. The majority were double positive (IFN-γ+, TNF-α+) effector cells. These T cells disappeared completely at week 2, but by week 36, they were detected at high levels, were polyfunctional (Fig. 3B), and both central and effector memory cells were expanded (data not shown).
Figure 3.
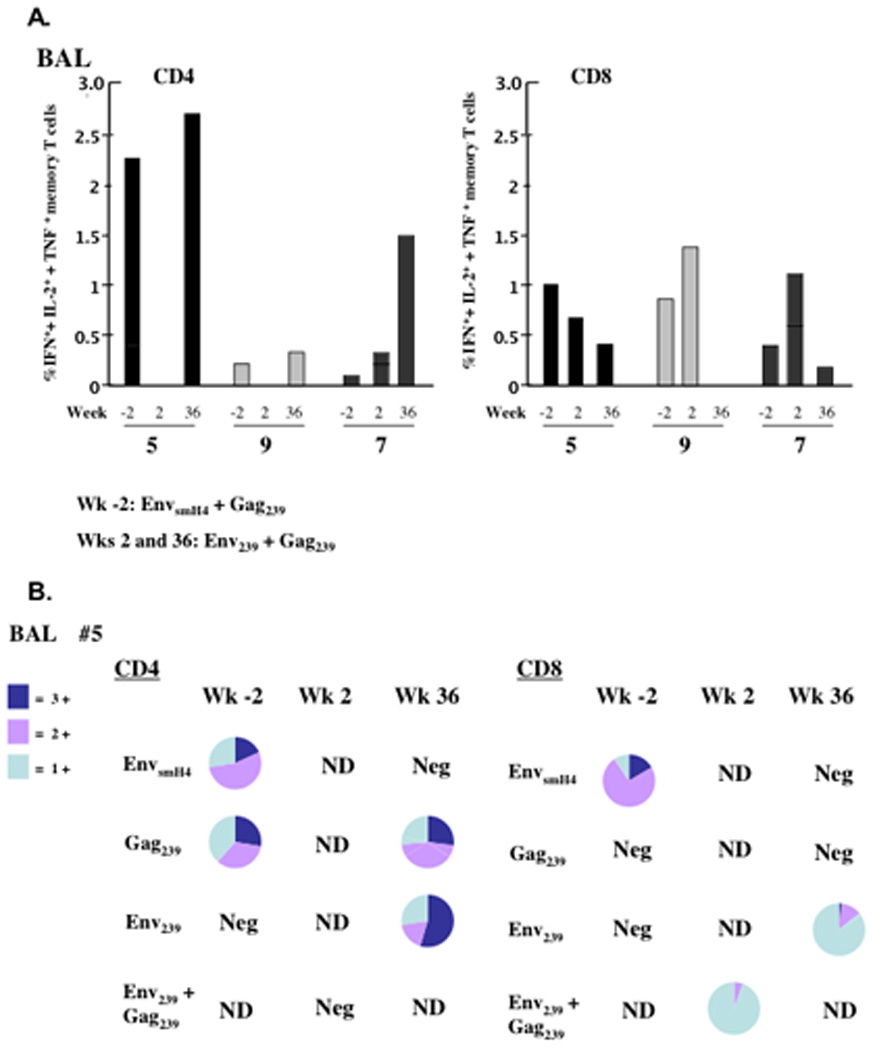
The sum percent of IFN-γ+, + IL-2+, + TNF-α+ total memory T cells in BAL samples for macaques #5, #7 and #9 over time. (A) Shown are the total cytokine positive cells among total memory CD4 or CD8 T cells at weeks -2, 2 and 36 post SIVsmE660 challenge after stimulation with peptide pools noted. Cell numbers were lower at week 2 post-challenge, so peptide pools were combined for stimulations. For ease in interpretation, responses were summed at weeks -2 and 36 as well. (B) Pie charts for each flow sample from panel A, representing the percent triple, double and single cytokine secreting cells. Neg = negative response. ND = denotes not done.
CD8 memory cell responses in BAL for all three macaques displayed a different pattern (Fig. 3A). A relatively high frequency of cytokine secreting memory cells was exhibited by all animals pre-challenge (0.4 to 1%). The cells were generally of the expected effector memory phenotype (not shown). The responses extended to week 2 post-challenge for all three macaques (Fig. 3A) but by week 36 there was a noticeable decrease in frequency of cytokine secreting cells. At this time point, most CD8 memory T cells of macaque #5 secreted only IFN-γ (Fig. 3B). Cytokine responses among CD8+ T cells of macaques #7 and #9 were exclusively IFN-γ+ (not shown). Of note was the lack of cytokine secreting T cells in the jejunum for all macaques. Even post challenge, negligible responses were seen (data not shown).
Humoral immune responses
Binding antibodies in serum to SIVmac251 gp120 were continually monitored over the immunization and challenge history of these macaques. Prior to the first SIVmac251 challenge, cross-reactive binding antibodies to SIVmac251 envelope were elicited by Ad5hr-SIVsmH4env priming, but were not boosted by the peptomer immunizations (Fig. 4A). Post challenge, only macaque #5 showed a dramatic spike in binding antibody, coinciding with the presence of detectable viremia in blood. Subsequently, before, during and after the second and third challenges, high levels of antibody were maintained in animal #5 (Fig. 4B, C). For macaques #7 and #9, only low titers of 100 were seen post 2nd challenge, but clearly macaque #9 exhibited an anamnestic response within the first two weeks after the third challenge. We believe the apparent drop in binding titer for animal #7 at week 2 may have resulted from an inadvertent switching of sample aliquots for macaques #7 and #9 at week 1. The binding results were reproducible, however, for all other humoral assays (ADCC, neutralization, ADCVI, see below) the antibody activity of macaque #9 appeared earlier and at higher titer than for macaque #7, consistent with a possible switch of the binding assay aliquots. The binding titers for macaque #7 peaked at week 4 post-challenge, whereas binding titers of controls were not detectable until this time point.
Figure 4.
SIVmac251 serum binding antibody titers (log10) throughout the three SIV challenges.
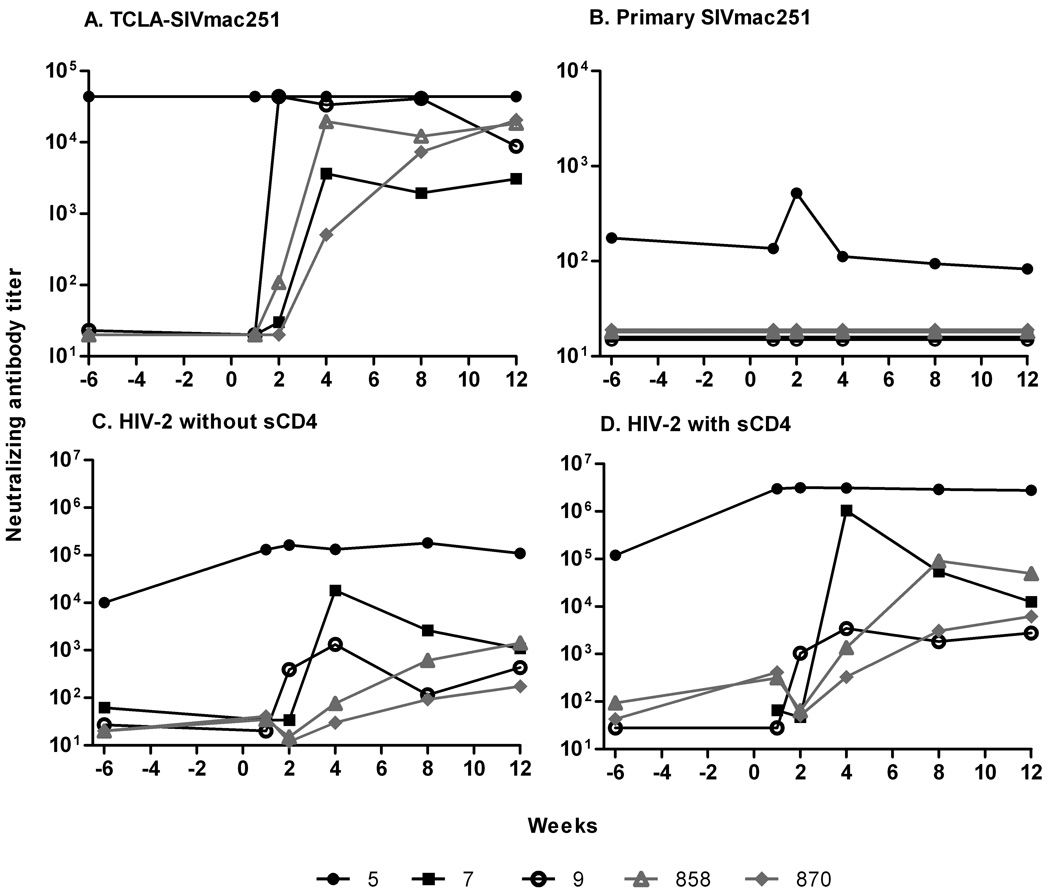
The original immunization regimen did not elicit neutralizing antibody against primary SIVmac251 in the three macaques pre-challenge (Patterson et al., 2004), however all three macaques exhibited low titer neutralization of TCLA-SIVmac251 before the 1st challenge which was strongly boosted in macaque #5 but not macaques #7 and #9 post-SIVmac251 challenge. Macaques #5 and #9 also developed antibodies able to neutralize SIVsmH4 pre-challenge. This activity was strongly boosted in animal #5 post-challenge in conjunction with its transient plasma viremia, but not in macaque #9 which remained aviremic (Patterson et al., 2004). Here, we examined whether exposure to the second and/or third challenge viruses elicited additional neutralizing antibody activities. The virus panel was expanded to include a primary SIVmac251 pseudovirus and an uncloned SIVsmE660 virus grown in CEMX174 cells which exhibits tier 2 neutralization in M7-Luc cells. None of the macaques was able to neutralize SIVsmE660 over the course of immunization and the three challenges (data not shown). Additionally, the three macaques were originally boosted with a peptomer mimicking the CD4 binding site of the virus. Development of anti-CD4 binding site antibodies and their binding to this site could have caused a conformational change in the envelope leading to antibody development against a CD4-induced site on the virus. Although not definitive, we included HIV-2 virus with and without the addition of soluble CD4 as described (Decker et al., 2005) to assess this possibility.
Only macaque #5 exhibited neutralizing antibody to all viruses tested except SIVsmE660 prior to the heterologous challenge (Fig. 5). In keeping with its subsequent aviremic status, the titers observed were maintained at constant levels post-challenge. High titers were seen against TCLA-SIVmac251 (Fig. 5A) and HIV-2 without sCD4 (Fig. 5C) and even higher titers against HIV-2 in the presence of sCD4 (Fig. 5D). Albeit at low level, difficult to neutralize primary SIVmac251 was neutralized by sera of macaque #5 and an anamnestic response was exhibited post-challenge (Fig.5B). Although macaques #7 and #9 were negative for neutralizing antibody activity prior to the SIVsmE660 challenge, both exhibited anamnestic neutralizing responses post-challenge against TCLA-SIVmac251 (Fig. 5A) and HIV-2, with and without sCD4 (Fig. 5C,D), but remained negative against primary SIVmac251 pseudovirus (Fig. 5B) and SIVsmE660 (data not shown).
Figure 5.
Neutralizing antibody titers in plasma after SIVsmE660 challenge. Neutralization titers to (A) TCLA-SIV mac251 and (B) primary SIVmac251. CD4-induced antibody neutralization of HIV-2 without sCD4 added (C) and with sCD4 added (D). In (A), neutralizing titers for macaque #5 were >4.4×104 at all time points.
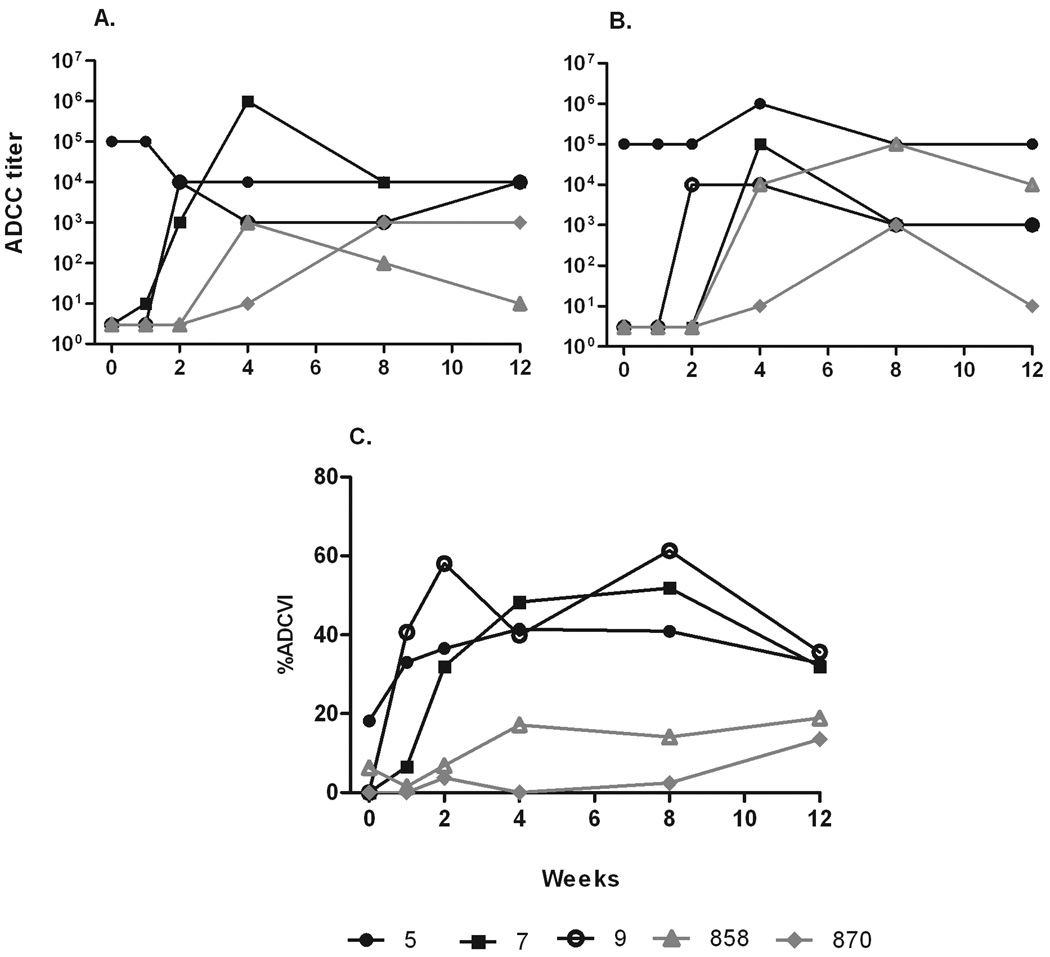
Non-neutralizing antibodies have been shown repeatedly in our laboratory to be correlated with control of SIV and SHIV infection in the rhesus macaque model (Florese et al., 2009; Gomez-Roman et al., 2005; Hidajat et al., 2009; Xiao et al., 2010). We therefore evaluated sera of the 3 macaques for antibodies able to mediate ADCC, an activity requiring interaction between effector cells, SIV-specific serum, and target cells expressing SIV antigen. Macaque #5 displayed potent antibody mediating ADCC against CEM-NKR target cells coated with both SIVmac251 gp120 (Fig. 6A) and SIVsmE660 gp120 (Fig. 6B) prior to challenge. Similarly to its binding and neutralizing antibody activities, ADCC titers were maintained post-challenge. In contrast, sera of macaques #7 and #9 were negative before challenge, but exhibited anamnestic responses in ADCC activity against SIVmac251 or SIVsmE660 by week 2 post-SIVsmE660 challenge.
Figure 6.
Non-neutralizing antibody titers in plasma after SIVsmE660 challenge. ADCC titers of plasma using SIV251 gp120- (A) and SIVsmE660 gp120-coated target cells (B). (C) % ADCVI of SIVsmE660 infection of rhesus PBMC using plasma diluted 1:200.
ADCVI has also been associated with control of viremia (Florese et al., 2009; Forthal et al., 2006; Hidajat et al., 2009; Xiao et al., 2010). Here, the three macaques exhibited a pattern of ADCVI activity similar to that seen for ADCC (Fig. 6C). Serum of macaque #5 was positive for ADCVI pre-SIVE660 challenge and the activity was sustained post-challenge. In contrast, macaques #7 and#9 were negative prior to challenge but developed strong anamnestic responses by week 2 which peaked at 60% and 50% inhibition respectively. Only low level ADCVI activity developed in controls over weeks 4 to 12.
Given the macaques’ history of vaccination with Env immunogens, multiple challenges, and their years of elite control, we expected they would have developed memory B cells associated with their sustained antibody and/or anamnestic responses. Therefore, bone marrow cells obtained at week 25 post challenge were evaluated for memory B cells. Prior to polyclonal stimulation, animal #5 exhibited the highest level of Env-specific IgG memory B cells compared to the other macaques (supplementary Fig. S3). The day 0 values represent primarily plasma cells in bone marrow which secrete low levels of antibody. Following 3 days of polyclonal stimulation, Env-specific ASC increased in all macaques. Again, the highest level was seen for macaque #5. This increase is attributed to memory B cells which differentiated in vitro into short-term plasma blasts or plasma cells. The increase in ASC in control macaque #858 indicates a cross-reactivty between the SIVmac251 gp120 used in the ELISPOT assay and the Env of the SIVsmE660 challenge stock.
Mucosal immune responses
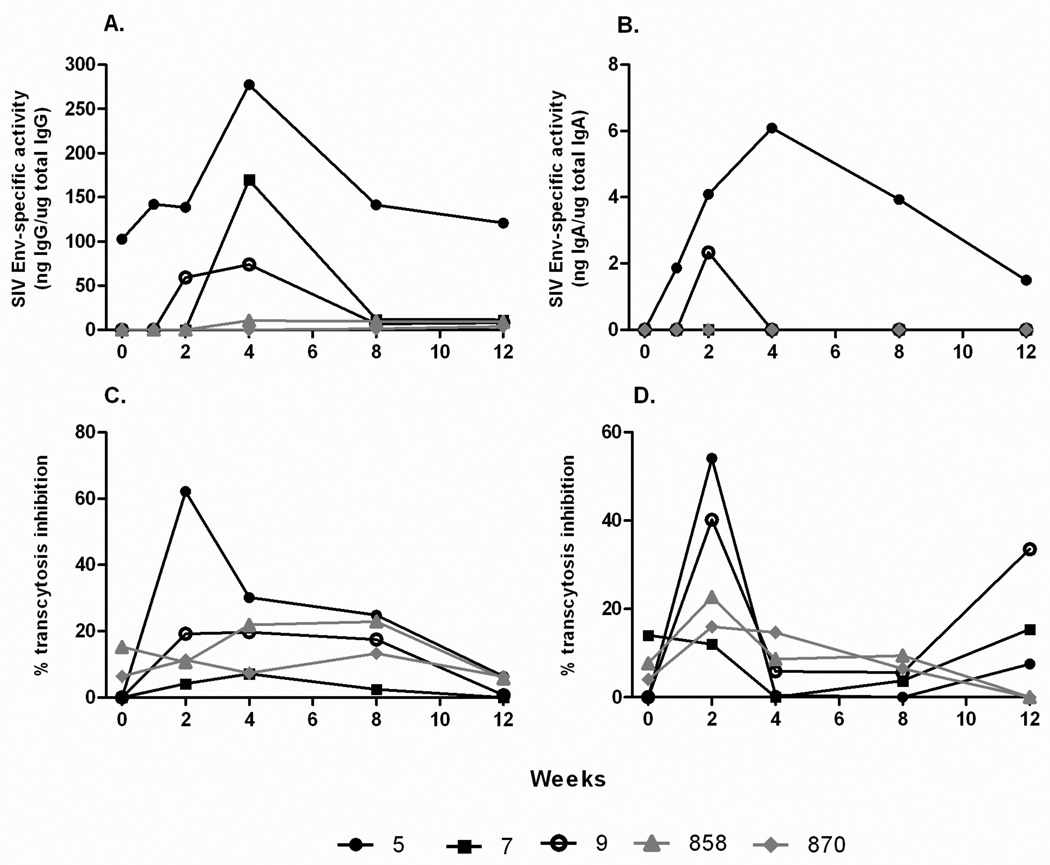
The three macaques were originally immunized with replicating Ad-recombinants which target mucosal epithelium, and subsequent challenges were mucosally administered. Therefore, it was prudent to assess induction of SIV-specific binding antibodies in rectal secretions (Fig. 7 A, B). Only macaque #5 showed measurable anti-Env IgG specific activity before challenge. While all 3 macaques displayed anamnestic responses in rectal IgG and/or IgA binding antibodies, the IgA response was first apparent in macaque #5. Macaque #9 exhibited both IgG and IgA anamnestic binding antibody responses, whereas macaque #7 lacked an IgA response.
Figure 7.
Mucosal antibody binding titers and transcytosis inhibition by rectal secretions post SIVsmE660 challenge. SIVmac251 Env specific IgG (A) and IgA (B) titers shown as specific activity values (ratio of specific divided by total IgG or IgA in each sample). Transcytosis inhibition of SIVmac251 (C) and SIVsmE660 (D) across a tight epithelial cell barrier.
To assess functional activity of the rectal antibodies, their ability to inhibit transcytosis of SIVsmE660 and SIVmac251 across tight intestinal epithelial cell barriers was evaluated. Rectal secretion of macaque #5 was able to inhibit transcytosis of both SIVmac251 and SIVsmE660 at 2 weeks post challenge (Fig. 7 C, D). Rectal antibody of macaque #9 also inhibited SIVsmE660 transcytosis at week 2 post-challenge, whereas rectal antibodies of #7 did not exhibit inhibition above levels seen in control animals.
Rhesus macaque microarray
To obtain a more comprehensive view of responses associated with elite control, we examined the transcriptional profile of genes in the jejunum 2 weeks after SIVsmE660 challenge. Comparisons were made two separate ways. First, a “fold-change” in gene expression in comparison to a pre challenge baseline was performed. Baseline samples included the two naïve macaques #858 and #870, in addition to macaques #7 and #9, since statistically they were not different from naives, and unlike macaque #5, remained essentially aviremic for approximately 7 years before the 3rd challenge. Finally, the greater number of animals included in the baseline allowed for greater statistical power for analyses. Macaque #5 was excluded from the baseline group since it exhibited significant differences in gene expression at the pre-challenge time point compared to samples from the other macaques. Figure 8A summarizes significant upregulation in all three macaques of genes involved in barrier integrity and epithelial repair and regeneration, a known hallmark for protection from SIV infection in rhesus macaques (George et al., 2003; George et al., 2005). Only macaque #5 displayed significant upregulation of immune response genes, however, including those associated with innate immunity, antigen presentation, chemotaxis and interferon induction (Fig. 8B). The latter upregulation was lower compared to that for the control macaques, where levels were 30X higher. Most importantly, genes associated with humoral immunity, including Ig lamba chain, were highly upregulated in macaque #5, hinting at a mechanism of protection seen at the gene, protein, and functional level.
Figure 8.
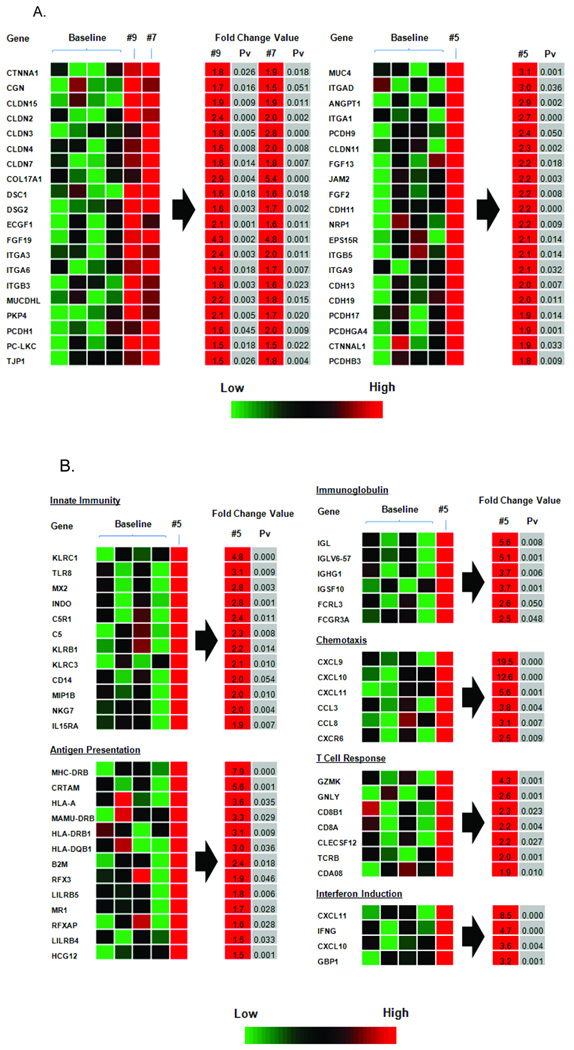
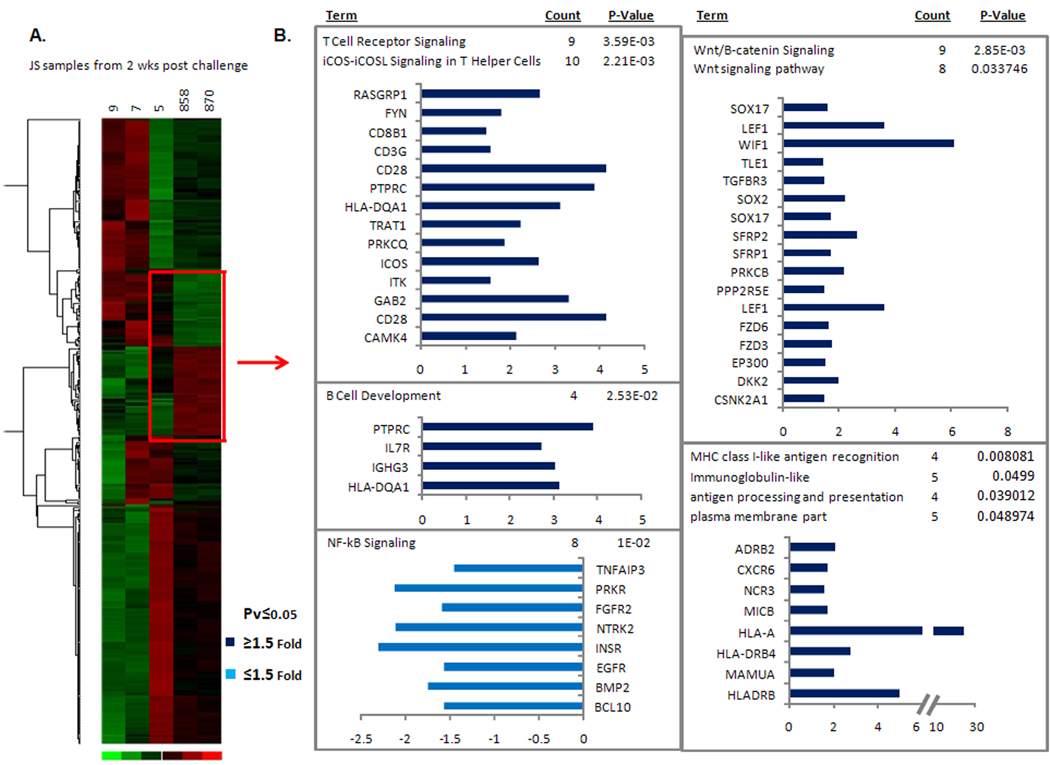
Transcriptional profile of jejunal biopsies week 2 post challenge. (A) Up-regulated genes involved in barrier integrity, epithelial repair and regeneration. (B) Up-regulated immune response genes for macaque #5.
Secondly, gene expression in jejunal tissues of all five macaques 2 weeks post-challenge was also examined. A comparison of macaque #5 to both naïve animals is shown in Figure 9 with genes of interest highlighted in red. Within this box, 1410 genes were significantly different, with relevant pathways listed to the right. Again, genes involved in immune responses such as T cell receptor signaling, B cell development and MHC Class I antigen presentation were all upregulated in macaque #5, as well as Wnt/B-catenin signaling, which includes genes that downstream can be responsible for epithelial regeneration, cell development and repair. In turn, innate immune responses such as NF-kB signaling were comparatively downregulated for macaque #5. Finally, a preliminary comparison of macaque #5 with macaques #7 and #9 revealed significant differences for 6852 genes at week 2 post challenge, none of which however, included immune response genes (data not shown). While beyond the scope of this study, detailed data analyses will need to be performed in order to tease out relevant pathways.
Figure 9.
Changes in gene expression levels in jejunal tissue in vaccinated and control macaques 2 weeks post SIVsmE660 challenge. (A) Heat diagram of changes in gene expression in each vaccinated animal as compared to control animals (#858 and #870) following SIVsmE660 challenge. Relative increase (red) or decrease (green) of mRNA levels are shown. (B) Following hierarchical clustering, gene expression levels for the elite controller macaque #5 were then compared to the two control macaques (genes within red box). Pathways statistically over represented in the gene lists are shown to the right of the red arrow with the genes and P value indicated.
MHC class I and TRIM5α genotyping
A thorough examination of rhesus macaque MHC class I genotyping using 454 deep sequencing (Wiseman et al., 2009) was performed for all macaques in this study (supplementary Fig. S4). None were Mamu A*01 positive and results confirmed the lack of Mamu B alleles (B*008 or B*017) known to be associated with control of SIV replication (Loffredo et al., 2007; Yant et al., 2006).
The existence of specific TRIM5α allele polymorphisms were shown recently to have an attenuating affect on SIVmac251 replication in rhesus macaques (Lim et al., 2010). More directly applicable to this study, three TRIM5 alleles, TFP, Q and CypA were recently identified as correlating with viral outcome from a clonal SIVsmE543 challenge in rhesus macaques (Kirmaier et al., 2010). While still unproven for the closely related SIVsmE660 swarm used here (a single rhesus #E660 passage from original RM #E543), animals that were homozygous Q/Q were most permissive for SIVsmE543 infection, while TFP/TFP, TFP/CypA, and CypA/CypA macaques were more resistant. Retrospective typing of macaques used in this study showed that they were heterozygous for TRIM5 alleles TFP and Q, except for the control naïve macaque #870 which was homozygous Q/Q (data not shown). Higher viral loads were not seen for #870 in comparison to the heterozygous TFP/Q control macaque #858 (Fig. 1C), however. The strong control of viremia observed for the elite controllers (7-log reduction compared to controls) argues against TRIM5 alleles being the sole mediator of control here. Previously, viremia differences of approximately 3 logs between TFP/TFP and Q/Q macaques following SIVsm543 infection were seen, while heterozygotes displayed lesser differences. However, we can’t discount that the TRIM5 haplotype exerted some effect.
Discussion
The goal of this study was to comprehensively assess the level of viral control and virus induced immunity after a third, heterologous high dose mucosal SIV challenge in three rhesus macaque elite controllers. In contrast to other elite controller macaques studied, our macaques were unique in possessing no MHC haplotypes and TRIM5α polymorphisms known to be associated with elite control. Even given the complicated immunization/challenge history of these macaques, they provide an opportunity to undercover alternative, non-MHC-restricted immune responses associated with protection. The myriad array of positive immune responses we observed in these macaques points to multiple immune protective mechanisms, especially potent humoral immunity. Coupled with comprehensive assessments of viremia throughout the body and mucosal gene transcription, this study has implicated a broad range of factors involved in maintenance of strong viremia control.
Of the three original macaques, only macaque #5 continued to maintain strict, elite control, as defined by undetectable plasma viremia, of the heterologous SIVsmE660 mucosal challenge. However, examination of viral loads in mesenteric and axillary lymph nodes showed that this macaque, while negative pre-challenge, became infected. Both SIVsmE660 and SIVmac251, reactivated at low level from previous exposures, were undetected in blood, but present and contained locally in the lymph node (Table 1). When SIV is administered across a mucosal barrier, virus establishes an initial localized infection in a small number of cells, the founder population (Li et al., 2005; Miller et al., 2005). Dissemination to draining lymph nodes can occur within 24 hours, but until viremia hits a threshold level in the mucosa – usually 5–6 days – viremia in the distal sites and periphery is unsustainable. We did not assess the level of viremia at the rectal mucosa, the site of mucosal challenge, but SIVsmE660 was disseminated to draining lymph nodes and remarkably, subsequently contained. This control of viremia was mediated at the local site of infection, in the lymph node itself, or at both sites.
It was surprising that before rechallenge, a low level SIVmac251 infection (108 copies) was detected in the jejunum more than 3 years after CD8 depletion and re-appearance of viremia in macaque #5 (Table 1). Interestingly, SIVsmE660 was never detected in the jejunum, although a critical 2-week timepoint could not be evaluated due to lack of sufficient sample. However, the original SIVmac251 infection was reactivated in the jejunum following SIVE660 challenge, increased 10-fold, and thereafter remained stable. To our knowledge, this is the first detection of long term viremia sequestration and control at a mucosal site. Whatever the mechanism(s) of mucosal immune control, it did not prevent SIVE660 acquisition. Nevertheless, SIVE660 replication was subsequently controlled in the lymph node to a level that precluded spread of viremia to the blood over the entire post challenge period. It was recently shown that in human elite suppressors, low level, ongoing viral replication did not reseed the latent reservoir, and most likely originated from an as yet unknown cryptic reservoir (O’Connell et al., 2010). It is known that upon removal of HAART therapy and necropsy of SIV-infected rhesus macaques, virus can be detected in numerous tissue sanctuaries, in particular lymph nodes, spleen, and gastrointestinal sites (North et al., 2010), similar to sites where viremia was detected here. While we have learned much regarding initial viral infection at mucosal sites and its eventual systemic spread, greater understanding of the immune processes that contain it mucosally and control its dissemination is needed.
In spite of the strong control of SIVmac251 in jejunal tissue and the inability to detect SIVE660 at that site, we were not able to associate local cellular immune responses with viremia control. Virus-specific responses in the jejunum were consistently negative by intracellular cytokine staining. Continued stimulation by the low level viremia at this mucosal effector site might have resulted in already highly activated cells becoming anergic and unresponsive to further in vitro stimulation. However, in a study in which a large amount of jejunal tissue was obtained for analysis by necropsy of macaques immunized with live attenuated virus, cytokine secreting cells were correlated with protection (Genesca et al., 2008). Furthermore, a recent study in which virus-specific effector cells were visualized in situ (Li et al., 2009) supports the notion that these cells exist at this site. We may have missed detection of cytokine-positive cells due to sampling issues. Alternatively, other mechanisms may have played a role in the protection observed.
In contrast to results obtained with jejunal tissue, macaque #5 exhibited outstanding cellular immune responses in terms of breadth and frequency, particularly in blood and BAL. SIV-specific IFN-γ secreting PBMC in addition to broad, potent T cell proliferative responses existed prior to SIVE660 challenge and were maintained throughout the post-challenge phase. Polyfunctional, CD4 and CD8 central and effector memory cells specific for both Env and Gag were present at extremely high frequency pre-challenge and were boosted post-challenge in both peripheral blood and BAL (Figs. 2, 3). The expansion post-challenge of vaccine-specific EnvsmH4 CD4 memory cells in PBMC to an effector phenotype during the acute phase of infection followed by contraction during set point to polyfunctional cells (Figs. 2B) is of note. The complex nature of viremia control in macaque #5 makes definitive conclusions difficult, but these effector cells may have been contributing to control of SIVE660 infection, especially if they homed back to the rectal mucosa. The concomitant disappearance of CD4 memory cells from the lung at week 2 (Fig. 3A) could also have provided for a transient influx of effector cells to the rectal mucosa. Vaccine elicited effector memory T cell responses have recently been shown to be associated with protection from SIV mucosal challenge (Hansen et al., 2009).
Macaques #9 and #7 exhibited lesser control of SIVsmE660 viremia in comparison to macaque #5, although both showed blunted acute viremia and subsequent control to undetectable levels during chronic infection. Macaque #7 rapidly controlled viremia by week 6 and maintained this control until week 36 when a sharp increase in viral load indicated immune escape. We plan to sequence viruses around this time point after single genome amplification in order to determine if a recombination event occurred and shed light on a possible mechanism of escape. The kinetics of viremia control for animal #9 differed. This macaque exhibited undetectable viremia beginning at week 36, however, after week 75 viral loads became positive and increased slowly (data not shown). Therefore, escape from immune control may be occurring for this macaque as well. SIVmac251 was never detected before or after SIVsmE660 challenge in plasma or tissues of either of these macaques. Unlike animal #5, both macaques controlled the early SIVmac251 challenges to undetectable levels and therefore were not exposed to transient SIVmac251 viremia. We speculate that the low-level SIVmac251 infection of macaque #5, kept under control by potent immune responses, may be playing a significant role in its maintenance of stronger viremia control by providing a persistent immunologic stimulus.
Although macaques #7 and #9 lacked SIV-specific cells in PBMC pre- or post-SIVsmE660 challenge, both macaques displayed pre-existing virus specific memory cells in BAL which were boosted following SIVsmE660 exposure. Since we consistently detected SIV-specific CD8+ effector cells at this site for all three macaques, we speculate that long term memory cells might reside in the upper respiratory tract (URT), possibly in the lymph nodes draining the lungs. Our Ad5hr-SIV recombinant vaccines replicate in the URT after intranasal/intratracheal immunization in the rhesus macaque model. Therefore, it is likely that durable vaccine-induced immune cells would reside there.
Functionally diverse, mucosal and plasma antibody responses were uniquely observed here for all macaques, but especially for #5. Macaque #5 exhibited serum antibodies able to mediate multiple activities prior to SIVE660 challenge. The antibodies, both binding and neutralizing, were generally of high titer and were maintained post-challenge. In addition to neutralization of primary and TCLA-SIV, sera of macaque #5 neutralized HIV-2, with enhanced titers seen with addition of sCD4, implying recognition of CD4-induced epitopes. As SIVsmE660 was not neutralized, activities mediated by non-neutralizing antibodies such as ADCC and ADCVI were assessed. Both activities were mediated by sera of macaque #5, and both have been correlated with reduced viremia (Gomez-Roman et al., 2005; Xiao et al., 2010). Most of these functional antibody activities were detected post-challenge for both macaques #9 and #7 too, albeit at lower levels. Macaque #9 showed more potent SIVsmE660-specific ADCVI activity and a more rapid SIVE660-specific ADCC anamnestic response than #7, which may have contributed to the better viremia control of this macaque during chronic infection.
IgG memory B cells specific for SIV Env were present in bone marrow of all three macaques, indicative of their long vaccination and challenge history. Macaque #5 again exhibited the highest levels. With regard to mucosal antibody, anamnestic SIV Env-specific IgG responses were seen for all three macaques, and IgA responses for macaques #5 and #9. Again, macaque #5 had the highest rectal antibody levels. The mucosal antibodies elicited mediated transcytosis inhibition against SIVmac251 (macaque #5) and SIVsmE660 (macaques #5 and #9). Inhibition of transcytosis has been associated with protection both in macaques (Hidajat et al., 2009; Xiao et al., 2010) and in humans (Devito et al., 2000; Shen et al., 2010). We have observed that transcytosis inhibitory activity is significantly correlated with reduced chronic viremia in the SHIV89.6P rhesus macaque model (Xiao et al., 2010) suggesting that it might not only be important in protecting against acquisition, but might also play a role in protection against cell to cell transmission. This would be relevant here regarding control of viremia at tissue sites.
The evaluation by microarray of intestinal gene expression over time has provided a number of novel observations. Firstly, humoral immune response genes at this site were upregulated post virus exposure for macaque #5. Although we detected mucosal antibodies in rectal secretions, we can now link these functional responses to gene expression directly after virus exposure since transcriptional analysis was performed at week 2 post-challenge.
Secondly, numerous genes involved in induction of immune responses were upregulated in macaque #5 immediately following challenge. These were related to processes such as innate immunity, antigen presentation, chemotaxis and T cell responses. A lesser upregulation of inflammatory response genes, in particular interferon known to be induced to high levels in SIV unprotected macaques (Bosinger et al., 2009), was observed for macaque #5. In addition, tight junction and epithelial repair and regeneration genes associated with protection from SIV infection (George et al., 2003; George et al., 2005) were upregulated in all three macaques, but more so for macaque #5, reflecting greater control of viremia. Further post challenge analysis of clear differences seen between the elite controller macaque #5 and macaques #7 and #9, which did not control viremia to undetectable levels, might in the future, yield new clues to SIV protection in the rhesus macaque model.
It is likely that the stronger protection seen in macaque #5 resulted in part from the cryptic exposure to SIVmac251. In this animal, vaccine-elicited anti-envelope antibody was boosted by exposure to SIVmac251 and also augmented by development of anti-gag cellular responses. The resultant local containment of virus by potent immunity had the effect of creating a “live-attenuated virus” from the pathogenic SIVmac251 challenges, leading to subsequent complete protection against the heterologous SIVsmE660. In contrast, macaques #7 and #9, originally better protected and not subjected to transient SIVmac251 viremia, initially controlled, but later were less able to sustain SIVsmE660 control. A key difference was their lack of long term maintenance of a potent anti-envelope antibody response. The peptomer, administered without an adjuvant to all three, was not a good booster immunogen in comparison to SIV envelope itself, and induced only low-titer antibodies at the time of the first SIVmac251 challenge. Whether a vaccine regimen that provides better initial anti-envelope boosting and maintenance of high-titer, functional antibodies will preclude the necessity for continuous, cryptic antigen exposure will require further investigation. The mechanism by which macaque #7 lost viremia control is also worthy of pursuit, and should elucidate whether the initial control was primarily cellular or humoral.
Overall, study of the three elite controller macaques has attributed the potent viremia control to multifunctional cellular, humoral, and mucosal immune responses. This extraordinary level of control appropriately matches the complex nature of SIV infection and pathology.
Materials and methods
Animals, viruses and reagents
In addition to the 3 elite controller macaques, 2 naïve rhesus macaques, #858 and #870, negative for prior exposure to SIV, simian retrovirus type D, and simian T-cell leukemia virus, served as controls for the SIVsmE660 challenge. All macaques were housed and maintained according to NIH guidelines.
The SIVsmE660 challenge stock was used undiluted (Hirsch and Johnson 1994). Intrarectal administration of undiluted stock previously infected 100% (4/4) of naive macaques, whereas a 1:10 dilution was not infectious by this route in 4 macaques.
Expression and purification of SIVsmE660 gp120 protein was accomplished as follows. The gp120 gene was optimized using human codon bias and synthesized by Geneart AG (Regensburg, Germany). The gp120 gene was cloned into the mammalian expression plasmid pJWIRES Puro in frame with the tissue plasminogen activator (tPa) signal peptide which replaced the relatively weak native signal peptide, residues M1-C22, in order to improve secretion efficiency of the mature gp120 protein. A termination signal was included after R533, at the native cleavage site. Production of the gp120 was driven by the cytomegalovirus (CMV) immediate early promoter. The pJWIRES Puro vector contains the puromycin acetyl-transferase (PAC) gene for selection of stable transfectants in the presence of puromycin. The PAC gene is linked to the gp120 gene on the bicistronic RNA via an internal ribosomal entry site (IRES). 293H cells were transfected with pJWIRES using Lipofectamine 2000 and the cells were plated and grown under selection with puromycin. High gp120 expressing cell lines were expanded and gp120 was purified from culture supernatants by loading onto a GNL column and eluting with 0.4M methyl alpha-D-mannopyranoside in PBS. The UV peak was concentrated in Vivaspin 30K MWCO tubes, acidified to pH 4 and passed through a C18 column. Fractions were collected over a 0–100% acetonitrile in 0.1% TFA gradient. The purified gp120 was lyophilized, resuspended in PBS, and frozen at −70°C.
Sample collection
Lymphocytes were isolated from fresh blood using Ficoll-Paque Plus (GE), and used fresh or else resuspended in FBS + 7% DMSO and stored in liquid nitrogen for later use. Serum and rectal secretions were serially collected and stored at −70°C. Before use in functional assays, serum samples were thawed at room temperature, diluted 10-fold with R-10 medium (RPMI 1640 containing 10% FCS, 2mM L-glutamine, and pen-strep), and heat inactivated at 56°C for 30 min. Rectal secretions were obtained using cotton-tipped swabs and placed in 1 ml of PBS containing 0.1% bovine serum albumin, 0.01% thimerosal, and 750 Kallikrein inhibitor units of aprotinin. The samples were tested for blood contamination using Chemstrips 4 (Boehringer-Mannheim); samples testing positive were not analyzed. For assay, rectal secretions were thawed, pelleted, and the supernatant passed through a 0.45 µm SPIN-X tube (Corning Inc.). Bronchoalveolar lavage (BAL), lymph node (mesenteric, axillary, and/or inguinal), and jejunal strip biopsy samples were collected and processed as previously described (Hidajat et al., 2009; Malkevitch et al., 2006). Lymphocytes were isolated from BAL and jejunal biopsies using a percoll gradient of 35% and 65% layered solutions. Lymphocytes from lymph nodes were used directly in assays. Separate frozen lymph node (mesenteric, axillary, inguinal) and jejunal samples were used for viral RNA determination and for microarray analysis.
Viral load measurement by NASBA
Nucleic acid sequence based amplification (N ASBA) sensitive to <50 SIV copies/ml, was utilized to quantitate viral loads in blood and lymph node samples (Lee et al., 2010; Romano et al., 2000). Primers were specific for a conserved sequence in the Gag gene, therefore discrimination of SIVmac251 and SIVsmE660 was not possible with this assay.
Quantitative real time RT-PCR
In order to discriminate between SIVmac251 and SIVsmE660 viral burdens in rhesus plasma, lymph nodes and jejunal tissue, two Gag strain-specific quantitative real-time RT-PCR (qRT-PCR) assays were adapted from previously published methods (Leutenegger et al., 2001; Yeh et al., 2009). Viral RNA was extracted and purified from plasma and/or tissue using the QIAmp viral RNA minikit (Qiagen, Valencia, CA). 0.5 µg RNA was then reverse transcribed using a high capacity cDNA reverse transcriptase kit (Applied Biosystems, Foster City, CA) using a gag RT primer 5’GGTGCAGCAAATCCTCT 3’ to generate a cDNA product suitable for future quantitative PCR. The PCR reaction mixture included (1X) TaqMan®Universal PCR master mix (Applied Biosystems, Foster City, CA), 200nM forward/reverse primers, 250nM probe and 25 µl cDNA template. cDNAs were subsequently amplified using either SIVsmE660 or SIVmac251 gag primer-probe sets as follows: SIVsmE660 gag forward primer; 5’ CAAGGGTCTGGGTATGAATCC 3’, reverse primer; 5’ TCAATGCTTCTGCCATTAATCTAG 3’, probe; 5’ (FAM) TCCTGGCCCTCCTATTCCCTGACA (BHQ-1) 3’. SIVmac251 gag forward primer; 5’ GCCAGGATTTCAGGCACTGT 3’, reverse primer; 5’ GCTTGATGGTCTCCCACACAA 3’, probe; 5’ (FAM) AAGGTTGCACCCCCTATGACATTAATCAGATGT (BHQ-1) 3’. Note, for jejunal tissue only, primers specific for SIVmac239 gag (closely related sequence to 251), were used as follows: SIV239 gag forward primer; 5’ GCCAGGATTTCAGGCACTGT 3’; reverse primer; 5’ GCTTGATGGTCTCCCACACAA 3’; probe; 5’ (FAM) AAGGTTGCACCCCCTATGACATTAATCAGATGTTA (BHQ-1) 3’. Probes were labeled with a 6-carboxyfluorescein (FAM) and quencher dye BHQ-1 (Integrated DNA technologies Coralville, IA). Fluorescent signal was detected by the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). Cycling conditions for SIVsmE660 were 2 min at 50°C, 10 min at 90°C, followed by 40 cycles of 30 sec at 90°C and 1 min at 63°C. For SIVmac251 and SIV mac239, cycling conditions were: 2 min at 50°C, 10 min at 90°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Duplicate or triplicate test reactions were performed for each sample. Dilutions of RNA standards were used to extrapolate SIV viral RNA copies/ml. The sensitivity for detection of SIVmac251 gag was 222 copies, SIVmac239 gag was 90 copies, while for SIVsmE660 gag it was 115 copy eq/ml plasma.
ELISpot and T cell Proliferation assays
SIV-specific interferon-gamma (IFN-γ) secreting cells were measured using freshly isolated PBMC after stimulation with SIV Gag239, Env239, or EnvsmH4 overlapping peptide pools using an ELISpot assay as described (Malkevitch et al., 2006; Patterson et al., 2003). Mean spot forming cells (SFC) from triplicate determinations are reported as SFC/106 PBMC following subtraction of background spots.
T cell proliferative responses to purified proteins including SIV251 gp120, SIV239 p27 and aldrithiol-2-inactivated SIVmac239 (Ald-SIV) were assessed in fresh PBMC by 3H-thymidine incorporation as described previously (Patterson et al., 2008). Samples were assayed in triplicate, and the stimulation index (SI) was calculated as the mean counts per minute (cpm) for the stimulated wells divided by the mean cpm for the R10 or microvesicle control wells.
Intracellular Cytokine Staining
Intracellular cytokine staining was utilized to measure SIV-specific IFN-γ, IL-2, and TNF-α cytokine secreting cells among freshly isolated PBMC and lymphocytes from BAL and jejunal strip biopsies. A total of 1–2 × 106 freshly isolated lymphocytes in 400 µl R10 from each tissue were stimulated with peptides at a 2 µg/ml final concentration. SIV peptide pools were made up of 15-mers overlapping by 11 amino acids and included EnvsmH4 (Advanced BioScience Laboratories, Inc., Kensington, MD), Env239, and Gag239 (AIDS Research Reference and Reagent Program, NIAID). Control tubes included a non-stimulated and an SEB (1 µg/ml) positive control. Anti-CD28-Alexa 680 (custom conjugate), and anti CD49d (BD Biosciences), were also added during stimulation along with brefeldin A inhibitor (BD Biosciences), all at 10 µg/ml final concentration. Tubes were incubated at 37°C for 6 hours. Thirty minutes before the 6 hour incubation was over, anti-CCR7-Pacific Blue (R&D Systems) was added. Cells were washed twice with PBS, resuspended in 95 µl of PBS plus 5 µl of Aqua Blue viability dye (Invitrogen, 1:40 dilution of DMSO stock) and incubated at room temperature (RT) for 10 min. Cells were washed once with PBS, and surface stained for 20 min at RT with the following antibodies: CD4-Qdot605 (Invitrogen), CD8-Qdot655 (Invitrogen), CD95-APC (BD Pharmingen) and CD45RA-ECD (Immunotech) at concentrations determined by dilution of stock or product insert. Cells were washed twice, resuspended in 250 µl fix/perm solution (BD Biosciences) for 20 min at 4°C, washed twice with BD wash buffer and resuspended in 100 µl wash buffer plus the following antibodies: CD3-APC-Cy7, IFN-γ-FITC, IL-2-PE and TNF-α-PE Cy7 (BD Pharmingen). After 20 min incubation at 4°C in the dark, the cells were washed twice with BD wash buffer and pellets were resuspended in 1% formaldehyde solution (Tousimis) for acquisition on an LSR II. Approximately one million lymphocytes were collected for PBMC samples, and 30,000 – 100,000 for tissue. Compensation was performed using the same antibodies bound to BD CompBeads. A singlet, followed by live/dead and then lymphocytic gates were first applied. CD3+ T cells were divided into CD4 and CD8 populations and for each, CD45RA+CD95+ and CD45RA−CD95+ cells were further subdivided into CD28+CD95+ central and CD28−CD95+ effector memory cells. Percent cytokine secreting cells among each memory subset was then determined. Data were analyzed using FloJo software (TreeStar Inc., Ashland, OR) and Pestle and Spice (version 4.2.2) programs (Mario Roederer, VRC, NIAID).
Binding and neutralizing antibody assays
Serum binding antibodies reactive with SIVmac251 gp120 Env protein were measured in an ELISA assay as previously described (Buge et al., 1997). The reciprocal of the dilution at which the absorbance was two times greater than a 1:50 dilution of the control serum was defined as the antibody titer.
Neutralizing antibody titers for the primary SIVmac251 CS.41 pseudovirus were determined using the TZM-bl Luc cell line as previously described (Montefiori 2004). For TCLA-SIVmac251 and -SIVsmE660 (H9 and CEM × 174-grown respectively), neutralizing titers were obtained using M7-Luc cells (Montefiori 2004). Titers are defined as the reciprocal plasma dilution at which there was a 50% reduction in relative luminescence units (RLUs) compared to virus control wells which contained no test sample.
CD4-induced neutralizing antibodies (CD4i Nabs) were assayed using the HIV-2 strain 7312A as described (Decker et al., 2005). Briefly, the HIV-2 proviral clone pJK7312A (GeneBank/EMBL/DDBJ accession no. L36874) was transfected into 293T cells to obtain HIV-2 provirus 7312A. 7×103 TZM-bl cells were plated in 96-well tissue culture plates and cultured overnight in DMEM supplemented with 10%FBS. 3,000 infectious units of HIV-2 were combined in a total volume of 60ul with or without a 2X concentration of sCD4 (R&D Systems) in DMEM with 6% FBS and 80µg/ml DEAE-dextran. The concentration of sCD4 was chosen so that the final 1X concentration after the addition of test serum/plasma corresponded to the IC50 of sCD4 specific for each virus. After 1 hr incubation at 37°C, an equal volume of serially diluted test serum/plasma in DMEM containing 6% FBS and 10% normal human plasma (NHP) was added. The virus ± sCD4 + test serum/plasma mixture was incubated for 1hr at 37°C, then transferred to the overnight-cultured TZM-bl cells. Media was removed completely from the adherent TZM-bl cells just before the addition of the virus ± sCD4 + test serum/plasma mixture. Controls included wells without virus (uninfected) and those without serum (virus control). RLU were calculated for duplicate samples, and 50 % neutralizing titers are reported.
ADCC and antibody-dependent cell-mediated viral inhibition (ADCVI)
Measurement of ADCC activity was performed as previously described (Gomez-Roman et al., 2005). Human PBMC served as effectors, whereas SIVmac251 and SIVsmE660 gp120 protein coated CEM-NKR cells separately served as targets, used at an E:T ratio of 50:1. ADCC titers are defined as the reciprocal dilution at which the percent ADCC killing was greater than the mean percent killing of the negative controls plus three standard deviations.
ADCVI activity was assessed as described (Xiao et al., 2010), using rhesus PBMC as effectors and targets. The latter were infected with SIVE660 and used at an E:T ratio of 20:1. Serum samples were tested at a 1:200 dilution.
B cell memory ELISpot
Bone marrow (BM) lymphocytes were isolated for enumeration of total and SIV251 gp120-specific (IgG secreting memory B cells with or without prior polyclonal stimulation as described (Brocca-Cofano et al., submitted). Briefly, lymphocytes were cultured for 3 days in R10 medium alone or with R10 medium supplemented with 1 µg/ml CpG(ODN-2006), 0.5 µg/ml recombinant human sCD40L (Peprotech), and 50 ng/ml recombinant human IL-21 (Peprotech). The cells were washed in R10 and antibody secreting cells (ASC) were enumerated by ELISPOT. Multiscreen 96-well filter plates (MSIPS4510, Millipore, Bedford, MA) were activated with 70% ethanol, washed three times with PBS, and coated overnight at 4°C with 100µl of 10µg/ml goat anti-monkey IgG (KPL) to measure total IgG or with 300ng/well of gp120 to detect virus-specific ASC. After washing with R-10, total IgG plates were blocked with 200µl/well of R-10 plus a 1/20 dilution of milk (KPL), while plates for specific IgG were blocked with R-10 alone and incubated for 2 h at 37°C. Uncultured or stimulated BM cells (2 × 105 for specific or 104 for total) were added to wells and incubated at 37°C overnight. Specific IgG plates were washed once with R-10 and blocked with 200µl/well of R-10 plus 3% bovine serum albumin (BSA, fraction V; Sigma) for 2 h at 37°C. Then, all plates were washed 4X with PBS containing 0.05% Tween 20 (Sigma) (PBST) followed by addition of biotinylated goat anti monkey IgG (Rockland) (1µg/ml) in PBST + 1% FBS for 2 h at 37°C. After four washes with PBST, the plates were incubated for 1 h at RT with 5 µg/ml HRP-conjugated avidin-D (Vector Laboratories, Burlingame, CA) in PBST + 1% FBS. Plates were washed 3X with PBST and 3X with PBS, and developed using 3 amino-9 ethyl-carbazole (AEC, Sigma). Reactions were stopped by washing twice with water. Spots were counted using an automated Elispot reader (Axioplan 2 imaging; Zeiss, Munchen, Germany). Assays were carried out in triplicate, and data are represented as the percentage of gp120-specific ASC relative to the number of total IgG ASC.
Inhibition of SIV transcytosis
SIV transcytosis across a monolayer of HT-29 intestinal epithelial cells was performed according to the method of Bomsel (Bomsel 1997), modified as previously described (Hidajat et al., 2009). Rectal secretions diluted 1:10 were evaluated, and inhibition of SIV transcytosis was expressed as the percentage of p27 antigen recovered in the basal chamber in the presence of rectal sample, relative to the amount of p27 antigen recovered in the absence of rectal sample.
Mucosal IgA and IgG binding antibodies
Anti-SIVmac251 gp120 IgA and IgG antibodies in rectal secretions were measured by ELISA as described (Bertley et al., 2004). Briefly, Nunc MaxiSorp plates were coated with 100 ng of SIVmac251 gp120 (ABL). After blocking, plates were loaded with serial dilutions of secretions as well as dilutions of Env-specific IgA and IgG standards derived from IgG-depleted pooled serum or purified serum IgG, respectively, obtained from SIVmac251- infected macaques and calibrated as described (Manrique et al., 2009). Plates were developed with 0.1µg/ml HRP-conjugated goat anti-monkey IgA or IgG (Alpha Diagnostic International, San Antonio, TX) and developed with TMB substrate, followed by measurement of absorbance at 450nm. To account for the variable immunoglobulin concentration in secretions, the concentration of antigen-specific IgA or IgG in each secretion was divided by the total IgA or IgG concentration to obtain specific activity (ng of specific IgA or IgG/ ug total IgA or IgG). Total IgA/IgG was measured by ELISA using plates coated with 1µg/ml affinity-purified goat anti-monkey IgA/IgG antibody (AlphaDiagnostic). The standards for total IgA/IgG were a dilution series of a normal rhesus macaque serum containing a known amount of IgA and IgG.
Rhesus macaque microarrays
Intestinal gene expression in rhesus macaques was evaluated using the rhesus macaque genome GeneChips® (Affymetrix, Santa Clara, CA) comparable to previously published methods (George et al., 2003; George et al., 2005). Briefly, total RNA was extracted from flash-frozen biopsies using the RNeasy isolation kit (Qiagen). cDNA was synthesized by reverse transcription, which was then used as a template for synthesis of cRNA in an in vitro transcription reaction using biotin-labeled nucleotides. The biotin labeled cRNA was hybridized overnight (16hr) to the rhesus genome specific high density oligonucleotide microarray (Affymetrix) and subsequently stained with a strepavidin-phycoerythrin conjugate and scanned with an Affymetrix 3000 scanner. Fluorescence emitted was measured and used to calculate expression levels of each gene. Expression levels of the experimental samples were then compared to baseline samples to generate fold changes. A mean fluorescent intensity ≥1.5-fold above or below mean baseline was considered significantly different (P ≤ 0.05). Background correction, normalization, and statistical analyses were performed using dChip (http://biosun1.harvard.edu/complab/dchip) algorithms. Assignment of genes to functional categories was performed through annotation of gene lists using the Affymetrix NetAFFX web interface, the DAVID (http://apps1.niaid.nih.gov/david) annotation tool, Ingenuity Pathway Analysis (IPA), and through literature-based classification by hand. The entire microarray data set is deposited at the Gene Expression Omnibus in the National Center for Biotechnology (accession number GSE23722).
Supplementary Material
Supplementary Figure S1. % CD3+CD4+ T cells in peripheral blood and tissue biopsies following SIVE660 challenge. (A) The percent CD3+CD4+ T cells in peripheral blood over the course of this study. The frequency of these same cells prior to and during acute phase infection (Wk 2) in bronchoalveolar lavage samples (B) and jejunal biopsies (C). The jejunal biopsy from Wk 0 for macaque #5 was not of sufficient quality (NS) to measure.
Supplementary Figure S2. IFN-γ ELISpot and T cell proliferative responses in PBMCs collected prior to and after challenge. (A) Stacked responses are shown after stimulation with SIV239 gag and SIV239 env peptide pools. Values for media alone or media + DMSO control wells were subtracted from peptide stimulated wells. No spots were detected after stimulation with SIVsmH4 env peptide pools. (B). Stacked T cell proliferative responses after stimulation with SIV239 Gag and SIV251 Env proteins and inactivated SIV (Ald-SIV).
Supplementary Figure S3. SIV Env-specific IgG memory B cells in bone marrow 25 weeks post SIVsmE660 challenge. Shown is the specific activity of SIVmac251 Env-specific ASC, defined as the % of Env-specific IgG ASC relative to the total IgG ASC. The memory B cells were assessed on day 0 (fresh, unstimulated) and after 3 days of polyclonal stimulation.
Supplementary Figure S4. MHC class I genotypes for elite-controller macaques. Heat map showing normalized data as a % of the total class I sequence reads identified/animal with scarlet (>5%), rose (1–5%) and pink (<1%).
Acknowledgements
We gratefully acknowledge the animal caretakers at Advanced BioScience Laboratories and Bioqual, Inc. for their expertise in caring for our macaques and collection of serial samples, and Steve Perfetto at the VRC, NIAID, for help in troubleshooting the 11-color flow panel and useful discussion. We thank Julian Bess and Jeffrey D. Lifson for the aldrithiol-inactivated SIV and control microvesicles; George Shaw for the HIV-2 proviral clone, pJK7312A; Wendy Yeh for SIV plasmid standards; and Wendy Yeh and Sam Sankaran for consultation on the real time PCR assay. The following reagents were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: CEM-NKR cells from Dr. Peter Cresswell; complete sets of SIVmac239 Env peptides and SIVmac239 Gag peptides. We thank Welkin Johnson, C. Audin, L. Hall, J. Morgan and the NEPRC Genetics Core, funded by NIH/NCRR grant RR00168, for TRIM5α genotyping. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker BM, Block BL, Rothchild AC, Walker BD. Elite control of HIV infection: implications for vaccine design. Expert Opin. Biol. Ther. 2009;9:55–69. doi: 10.1517/14712590802571928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertley FM, Kozlowski PA, Wang S-W, Chappelle J, Patel J, Sonuyi O, Mazzara G, Montefiori D, Carville A, Mansfield KG, Aldovini A. Control of simian/human immunodeficiency virus viremia and disease progression after IL-2-augmented DNA-modified vaccinia virus Ankara nasal vaccination in nonhuman primates. J. Immunol. 2004;172:3745–3757. doi: 10.4049/jimmunol.172.6.3745. [DOI] [PubMed] [Google Scholar]
- Bomsel M. Transcytosis of infectious human immunodeficiency virus across a tight human epithelial cell line barrier. Nat. Med. 1997;3:42–47. doi: 10.1038/nm0197-42. [DOI] [PubMed] [Google Scholar]
- Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, Zeng M, Masopust D, Carlis JV, Ran L, Vanderford TH, Paiardini M, Isett RB, Baldwin DA, Else JG, Staprans SI, Silvestri G, Haase AT, Kelvin DJ. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostik P, Kobkitjaroen J, Tang W, Villinger F, Pereira LE, Little DM, Stephenson ST, Bouzyk B, Ansari AA. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J. Immunol. 2009;182:3638–3649. doi: 10.4049/jimmunol.0803580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buge SL, Richardson E, Alipanah S, Markham P, Cheng S, Kalyan N, Miller CJ, Lubeck M, Udem S, Eldridge J, Robert-Guroff M. An adenovirus-simian immunodeficiency virus env vaccine elicits humoral, cellular, and mucosal immune responses in rhesus macaques and decreases viral burden following vaginal challenge. J. Virol. 1997;71:8531–8541. doi: 10.1128/jvi.71.11.8531-8541.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah PA, Patel DT, De Luca PM, Lindsay RWB, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Dwong PD, Robinson JE, Shaw GM. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Devito C, Broliden K, Kaul R, Svensson L, Johansen K, Kiama P, Kimani J, Lopalco L, Piconi S, Bwayo JJ, Plummer F, Clerici M, Hinkula J. Mucosal and plasma IgA from HIV-1-exposed uninfected individuals inhibit HIV-1 transcytosis across human epithelial cells. J. Immunol. 2000;165:5170–5176. doi: 10.4049/jimmunol.165.9.5170. [DOI] [PubMed] [Google Scholar]
- Ferre AL, Hunt PW, Critchfield JW, Young DH, Morris MM, Garcia JC, Pollard RB, Yee HF, Jr, Martin JN, Deeks SG, Shacklett BL. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood. 2009;113:3978–3989. doi: 10.1182/blood-2008-10-182709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florese RH, Demberg T, Xiao P, Kuller L, Larsen K, Summers LE, Venzon D, Cafaro A, Ensoli B, Robert-Guroff M. Contribution of nonneutralizing vaccine-elicited antibody activities to improved protective efficacy in rhesus macaques immunized with Tat/Env compared with multigenic vaccines. J. Immunol. 2009;182:3718–3727. doi: 10.4049/jimmunol.0803115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthal DN, Landucci G, Cole KS, Marthas M, Becerra JC, Van Rompay K. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J. Virol. 2006;80:9217–9225. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, Leon EJ, Soma T, Napoe G, Capuano SV, III, Wilson NA, Watkins DI. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genesca M, Skinner PJ, Bost KM, Lu D, Wang Y, Rourke TL, Haase AT, McChesney MB, Miller CJ. Protective attenuated lentivirus immunization induces SIV-specific T cells in the genital tract of rhesus monkeys. Mucosal Immunol. 2008;1:219–228. doi: 10.1038/mi.2008.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Reay E, Sankaran S, Dandekar S. Early antiretrovial therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T-cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J. Virol. 2005;79:2709–2719. doi: 10.1128/JVI.79.5.2709-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Sankaran S, Reay E, Gelli AC, Dandekar S. High-throughput gene expression profiling indicates dysregulation of intestinal cell cycle mediators and growth factors during primary simian immunodeficiency virus infection. Virology. 2003;312:84–94. doi: 10.1016/s0042-6822(03)00207-1. [DOI] [PubMed] [Google Scholar]
- Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, Borchardt GJ, Fisk DL, Glidden CE, Loffredo JT, Piaskowski SM, Furlott JR, Morales-Martinez JP, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and –DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J. Virol. 2008;82:859–870. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman VR, Patterson LJ, Venzon D, Liewehr D, Aldrich K, Florese R, Robert-Guroff M. Vaccine-elicited antibodies mediate antibody-dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. J. Immunol. 2005;174:2185–2189. doi: 10.4049/jimmunol.174.4.2185. [DOI] [PubMed] [Google Scholar]
- Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M, Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidajat R, Xiao P, Zhou Q, Venzon D, Summers LE, Kalyanaraman VS, Montefiori DC, Robert-Guroff M. Correlation of vaccine-elicited systemic and mucosal nonneutralizing antibody activities with reduced acute viremia following intrarectal simian immunodeficiency virus SIVmac251 challenge of rhesus macaques. J. Virol. 2009;83:791–801. doi: 10.1128/JVI.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Johnson PR. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O’Conner S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000462. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambotte O, Ferrari G, Moog C, Yates NL, Liao H-X, Parks RJ, Hicks CB, Owzar K, Tomaras GD, Montefiori DC, Haynes BF, Delfraissy J-F. Heterologous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS. 2009;23:897–906. doi: 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EM, Chung HK, Livesay J, Suschak J, Fink L, Hudacik L, Galmin L, Bowen B, Markham P, Cristillo A, Pal R. Molecular methods for evaluation of virological status of nonhuman primates challenged with simian immunodeficiency or simian-human immunodeficiency viruses. J. Virol. Methods. 2010;163:287–294. doi: 10.1016/j.jviromet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Leutenegger CM, Higgins J, Matthews TB, Tarantal AF, Luciw PA, Pedersen NC, North TW. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res. and Hum. Retro. 2001;17:243–251. doi: 10.1089/088922201750063160. [DOI] [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Ma Z-M, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- Li Q, Skinner PJ, Ha S-J, Duan L, Mattila TL, Hage A, White C, Barber DL, O’Mara L, Southern PJ, Reilly CS, Carlis JV, Miller CJ, Ahmed R, Haase AT. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 2009;323:1726–1729. doi: 10.1126/science.1168676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S-Y, Rogers T, Whitney JB, Kim J, Sodroski J, Letvin NL. TRIM5α modulates immunodeficiency virus control in rhesus monkeys. PLOS Pathogens. 2010;6(1):e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenstrom T, Agger EM, Korsholm KS, Darrah PA, Aagaard C, Seder RA, Rosenkrands I, Andersen P. Tuberculosis subunit vaccination provides long-term protective immunity characterized by multifunctional CD4 memory T cells. J. Immunol. 2009;182:8047–8055. doi: 10.4049/jimmunol.0801592. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkevitch NV, Patterson LJ, Aldrich MK, Wu Y, Venzon D, Florese RH, Kalyanaraman VS, Pal R, Lee EM, Zhao J, Cristillo A, Robert-Guroff M. Durable protection of rhesus macaques immunized with a replicating adenovirus SIV multigene prime/protein boost vaccine r egimen against a second SIVmac251 rectal challenge: role of SIV-specific CD8+ T cell responses. Virology. 2006;353:83–98. doi: 10.1016/j.virol.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Manrique M, Kozlowski PA, Wang S-W, Wilson RL, Micewicz E, Montefiori DC, Mansfield KG, Carville A, Aldovini A. Nasal DNA-MVA SIV vaccination provides more significant protection from progression to AIDS than a similar intramuscular vaccination. Muc. Immunol. 2009;2:536–550. doi: 10.1038/mi.2009.103. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Li Q, Abel K, Kim E-Y, Ma Z-M, Wietgrefe S, Franco-Scheuch LL, Compton L, Duan L, Shore MD, Zupancic M, Busch M, Carlis J, Wolinsky S, Haase AT. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J. Virol. 2005;79:9217–9227. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. Current Protocols in Immunology. John Wiley & Sons; 2004. pp. 12.11.1–12.11.15. [DOI] [PubMed] [Google Scholar]
- North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, Shacklett BL, Schinazi RF, Luciw PA. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J. Virol. 2010;84:2913–2922. doi: 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J. Virol. 2010;84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell KA, Han Y, Williams TM, Siliciano RF, Blankson JN. Role of natural killer cells in a cohort of elite suppressors: low frequency of the protective KIR3DS1 allele and limited inhibition of human immunodeficiency virus type 1 replication in vitro. J. Virol. 2009;83:5028–5034. doi: 10.1128/JVI.02551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, Tryniszewska E, Lewis MG, VanCott TC, Hirsch V, Woodward R, Gibson A, Grace M, Dobratz E, Markham PD, Hel Z, Nasca J, Klein M, Tartaglia J, Franchini G. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIV-mac-induced immunodeficiency. J. Virol. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Beal J, Demberg T, Florese RH, Marlkevich N, Venzon D, Aldrich K, Richardson E, Kalyanaraman VS, Kalisz I, Lee EM, Montefiori DC, Robey F, Robert-Guroff M. Replicating adenovirus HIV/SIV recombinant priming alone or in combination with a gp140 protein boost results in significant control of viremia following a SHIV89.6P challenge in Mamu-A*01 negative rhesus macaques. Virology. 2008;374:322–337. doi: 10.1016/j.virol.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Patterson LJ, Malkevitch N, Pinczweski J, Venzon D, Lou Y, Peng B, Munch C, Leonard M, Richardson E, Aldrich K, Kalyanaraman VS, Pavlakis GN, Robert-Guroff M. Potent, persistent induction and modulation of cellular immune responses in rhesus macaques primed with Ad5hr-simian immunodeficiency virus (SIV) env/rev, gag, and/or nef vaccines and boosted with SIV gp120. J. Virol. 2003;77:8607–8620. doi: 10.1128/JVI.77.16.8607-8620.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Malkevitch N, Venzon D, Pinczewski J, Gomez-Roman VR, Wang L, Kalyanaraman VS, Markham PD, Robey FA, Robert-Guroff M. Protection against mucosal simian immunodeficiency virus SIV(mac251) challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J. Virol. 2004;78:2212–2221. doi: 10.1128/JVI.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, Baker B, Rosenberg R, Cutrell E, Seaman MS, Coffin JM, Walker BD. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J. Infect. Diseases. 2009;200:984–990. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado JG, Prendergast A, Thobakgale C, Molina C, Tudor-Williams G, Ndung’u T, Walker BD, Goulder P. Replicative capacity of human immunodeficiency virus type 1 transmitted from mother to child is associated with pediatric disease progression rate. J. Virol. 2010;84:492–502. doi: 10.1128/JVI.01743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JW, Shurtliff RN, Dobratz E, Gibson A, Hickman K, Markham PD, Pal R. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J. Virol. Methods. 2000;86:61–70. doi: 10.1016/s0166-0934(99)00184-6. [DOI] [PubMed] [Google Scholar]
- Sacha JB, Giraldo-Vela JP, Buechler MB, Martins MA, Maness NJ, Chung C, Wallace LT, Leon EJ, Friedrich TC, Wilson NA, Hiraoka A, Watkins DI. Gag- and Nef-specific CD4+ T cells recognize and inhibit SIV replication in infected macrophages early after infection. PNAS. 2009;106:9791–9796. doi: 10.1073/pnas.0813106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- Shen R, Drelichman ER, Bimczok D, Ochsenbauer C, Kappes JC, Cannon JA, Tudor D, Bomsel M, Smythies LE, Smith PD. Gp41-specific antibody blocks cell-free HIV type 1 transcytosis through human rectal mucosa and model colonic epithelium. J. Immunol. 2010;184:3648–3655. doi: 10.4049/jimmunol.0903346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine LE, Loffredo JT, Bean AT, Leon EJ, MacNair CE, Beal DR, Piaskowski SM, Klimentidis YC, Lank SM, Wiseman RW, Weinfurter JT, May GE, Rakasz EG, Wilson NA, Friedrich TC, O’Conner DH, Allison DA, Watkins DI. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J. Virol. 2009;83:11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Wright C, Harkins T, O’Conner DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat. Med. 2009;15:1322–1327. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P, Zhao J, Patterson LJ, Brocca-Cofano E, Venzon D, Kozlowski PA, Hidajat R, Demberg T, Robert-Guroff M. Multiple vaccine-elicited non-neutralizing anti-envelope antibody activities contribute to protective efficacy against SHIV89.6P challenge in rhesus macaques. J. Virol. 2010;84:7161–7173. doi: 10.1128/JVI.00410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O’Conner DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WW, Jaru-ampornpan P, Nevidomskyte D, Asmal M, Rao SS, Buzby AP, Montefiori DC, Korber BT, Letvin NL. Partial protection of simian immunodeficiency virus (SIV)-infected rhesus monkeys against superinfection with a heterologous SIV isolate. J. Virol. 2009;83:2686–2696. doi: 10.1128/JVI.02237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-Q, Fu T-M, Casimiro DR, Davies M-E, Liang X, Schleif WA, Handt L, Tussey L, Chen M, Tang A, Wilson KAn, Trigona WL, Freed DC, Tan CY, Horton M, Emini EA, Shiver JW. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J. Virol. 2002;76:12845–12854. doi: 10.1128/JVI.76.24.12845-12854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. % CD3+CD4+ T cells in peripheral blood and tissue biopsies following SIVE660 challenge. (A) The percent CD3+CD4+ T cells in peripheral blood over the course of this study. The frequency of these same cells prior to and during acute phase infection (Wk 2) in bronchoalveolar lavage samples (B) and jejunal biopsies (C). The jejunal biopsy from Wk 0 for macaque #5 was not of sufficient quality (NS) to measure.
Supplementary Figure S2. IFN-γ ELISpot and T cell proliferative responses in PBMCs collected prior to and after challenge. (A) Stacked responses are shown after stimulation with SIV239 gag and SIV239 env peptide pools. Values for media alone or media + DMSO control wells were subtracted from peptide stimulated wells. No spots were detected after stimulation with SIVsmH4 env peptide pools. (B). Stacked T cell proliferative responses after stimulation with SIV239 Gag and SIV251 Env proteins and inactivated SIV (Ald-SIV).
Supplementary Figure S3. SIV Env-specific IgG memory B cells in bone marrow 25 weeks post SIVsmE660 challenge. Shown is the specific activity of SIVmac251 Env-specific ASC, defined as the % of Env-specific IgG ASC relative to the total IgG ASC. The memory B cells were assessed on day 0 (fresh, unstimulated) and after 3 days of polyclonal stimulation.
Supplementary Figure S4. MHC class I genotypes for elite-controller macaques. Heat map showing normalized data as a % of the total class I sequence reads identified/animal with scarlet (>5%), rose (1–5%) and pink (<1%).