Abstract
The maintenance and differentiation of embryonic stem cells (ES cells) depends on the regulation of gene expression through the coordinated binding of transcription factors to regulatory promoter elements. One of the genes involved in embryonic development is the chromatin factor CP27. Previously, we have shown that NF-Y interacted with the CP27 proximal promoter CCAAT-box. Here we report that CP27 gene expression in mouse ES cells is controlled by CCAAT and E-box cis-acting regulatory elements and their corresponding transcription factors NF-Y and USF1. Specifically, USF1 interacts with the E-box of the CP27 proximal promoter and NF-Y interacts with the CCAAT box. NF-Y and USF1 also interacted with each other and activated the CP27 promoter in a synergistic fashion. Together, these studies demonstrate that gene expression of the chromatin factor CP27 is regulated through the interaction of the transcription factors NF-Y and USF1 with the CP27 proximal promoter.
Keywords: Promoter, ES cell, NF-Y, USF1, CP27
1. Introduction
The eukaryotic transcriptional machinery controls gene expression through its interaction with the promoters of target genes, most commonly through specific promoter elements such as TATA boxes. Frequently, eukaryotic promoter elements act in concert with other regulatory elements such as enhancers or silencers (Griffiths et al., 2000). For example, the CCAAT box acts in conjunction with the NF-Y transcription factor as a crucial promoter organizer, facilitating the recruitment of polymerase II and of neighboring transcription factors (Frontini et al., 2002; Kabe et al., 2005; Donati et al., 2006). The combinatorial binding of transcription factors to promoter elements plays a key role during eukaryotic development, when unique combinations of transcription factor binding sites establish specific regulatory codes (Narlikar and Ovcharenko, 2009). The quintessential role of a finely tuned transcriptional machinery is illustrated by the regulatory circuitry involved in the maintenance of embryonic stem cell (ES cell) pluripotency and progression into differentiated lineages, which is controlled by the promoters of chromatin factors (Boyer et al., 2005).
One of the chromatin factors required for ES cell pluripotency maintenance is the CP27 gene, a highly-conserved transcriptional co-regulator characterized by a TATA-less proximal promoter (Luan et al., 2010). Previously we have reported that CP27 function elimination caused early embryonic lethality and failure of epiblast expansion in CP27-null mouse embryos (Ito et al., 2005). The CP27 promoter contains several CCAAT-boxes, common regulatory elements in eukaryotic promoters usually found in the vicinity of other promoter elements such as E-box and GC-rich elements (Luan et al., 2010; Bucher and Trifonov, 1988; Magan et al., 2003; Schuettengruber et al., 2003; Zhu et al., 2003; Zhu et al., 2005). Most prominent among CCAAT-box binding factors is the ubiquitous transcription factor NF-Y, a trimeric transcriptional activator with histone-like subunits (Mantovani, 1999; Guerra et al., 2007). NF-Y binding to the CCAAT-box is required for ES cell proliferation (Grskovic et al., 2007).
To understand the mechanisms that govern CP27 regulation in ES cell maintenance and differentiation, we turned to the CP27 promoter to identify specific regulatory elements and to test their function in mouse ES cells. We found two major cis-acting elements in the CP27 proximal promoter, a CCAAT-box and an E-box. Here we report how these two key CP27 proximal promoter elements are regulated by the transcription factors NF-Y and USF1 and provide an explanation for the role of CP27 in ES cell growth.
2. Materials and Methods
2.1. Mouse embryonic stem (ES) cell culture
Mouse ES cells were cultured in DMEM supplemented with 15% FCS, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 0.1 mM 2-mercaptoethanol and 1000 U/ml leukemia inhibitory factor (LIF) (Chemicon, Temecula, CA). The culture medium was changed everyday and the cells were passed onto gelatin coated cell culture dish at subconfluence every 3 days.
2.2. Nuclear Extract Preparation
ES cells (5×107) in 100 mm dishes were harvested and cell lysates were centrifuged. The pelleted nuclei were resuspended in nuclear extraction buffer with a final concentration of 300 mM NaCl, and centrifuged at 24,000 g for 20 min at 4°C. Aliquots of the supernatant were frozen at −70° C. Protein concentrations were determined using a protein assay reagent (Bio-Rad, Philadelphia, PA).
2.3. Electrophoretic Mobility Shift Assay (EMSA)
Double-stranded oligonucleotides (Table 1) for EMSAs were labeled with (gamma-32P) ATP using T4 kinase. EMSAs were performed by incubating 5µg of nuclear extract with labeled double-stranded oligonucleotide at room temperature for 20 min. For competition analysis, 25- or 50-fold molar excess of unlabeled double-stranded oligonucleotide was added to the nuclear extracts prior to the addition of the labeled probe. For supershift assays, polyclonal antibodies against NF-YA, NF1, USF1 and USF2 (Santa Cruz Biotechnology, Santa Cruz, CA) were incubated with the nuclear extracts for 15 min followed by the addition of radio-labeled probe. NF1 antibody was used as antibody control. DNA-protein complexes were resolved on a 5 % non-denaturing polyacrylamide gel in 1× TBE.
TABLE 1.
Oligonucleotide sequences used for CP27 promoter analysis
| Oligo Name | Sequence | Experiment |
|---|---|---|
| −207/−186 | TATTAGCTTGTGAGCAAATT | Construct |
| −93/−73 | TGAGTGTAGACTGACCAATCGC | Construct/ChIP |
| −55/−36 | GTCTCTGACCACGTGGCACT | Construct |
| −17/+4 | CCTCTAGGGCGGCCCTAGCT | Construct |
| +48/+25 | GCGAAGCTAGATATAGGGCGAGAC | Construct/ChIP |
| CP−93/−56 | TGAGTGTAGACTGACCAATCGCAGCAGCCGGAAGTGTC | EMSA |
| CP−93/−56 CAATmut | TGAGTGTAGACTGACacATCGCAGCAGCCGGAAGTGTC | EMSA |
| CP−55/−27 | TCTCTGACCACGTGGCACTGCCTGCGCA | EMSA |
| CP−55/−27 Emut | TCTCTGACCAttTGGCACTGCCTGCGCA | EMSA |
| CP sense | CGTCCAGACTTCTCCACATCGGA(FAM)G | PCR |
| CP antisense | CTCCACCGGACGGCATAGTAGT | PCR |
| NF-Ya sense | ATGGAGCAGTATACGACAAACAGCA | PCR |
| NF-Ya antisense | TTAGGACACTCGGATGATCTG | PCR |
| USF1 sense | ACCCCAACGTCAAGTACGTC | PCR |
| USF1 antisense | TATGTTGAGCCCTCCGTTTC | PCR |
| beta actin sense | TTCCTGACAGGATGCAGAAG | PCR |
| beta actin antisense | GTACTTGCGCTCAGGAGGAG | PCR |
Oligonucleotide sequences are listed in 5’ to 3’ orientation.
2.4. DNase I footprinting analysis
This assay was performed using the Core Footprinting System (Promega, Madison, WI). An end-labeled probe CP −207/+48 was generated from the plasmid pGL −207/+48 by first digesting the DNA with Sac I, end-labeling with T4 polynucleotide using (gamma-32P) ATP, and then digesting the DNA with Hind III. DNA-protein complexes were formed by incubating the end-labeled DNA with 100 µg of nuclear extracts in binding buffer. BSA was used as negative control. RQ1 RNase-free DNase was added to the reaction containing Ca2+/Mg2+. DNA samples were then separated by denaturing polyacrylamide gel-electrophoresis.
2.5. DNA constructs
For reporter constructs, fragments of the mouse CP27 promoter were amplified by PCR with a screened genomic clone as a template using a common 3’ primer and selected 5’ primers (Table 1). These primers also introduced a Sac I site at the 5’ end and a Hind III site at 3’ end of the amplified fragments. The PCR fragments were subcloned into the pGL3-basic vector (Promega). Correct sequence and orientation of all inserts in respect to the pGL3 vector was verified by DNA sequencing. Four reporter constructs (pGL-207, pGL-93, pGL-55, and pGL-17) were generated and each construct contained part of the exon 1 noncoding region. For the generation of expression constructs, the full length of NF-YA (long isoform) cDNA was generously provided by Dr. Sanker N. Maity (M.D. Anderson Cancer Center, University of Texas, Huston, TX), and subcloned into the Sal I and Sac I sites of the pIRES-GFP vector (Clontech, San Francisco, CA). The USF1 expression vector pCMV-SPORT6.1-USF1 was obtained from Open Biosystems (Huntsville, AL). The position of probes, constructs, the E-box, and the CCAAT box on the proximal CP27 promoter are annotated in Table 2.
Table 2.
Annotated Sequence of the CP27 Proximal Promoter
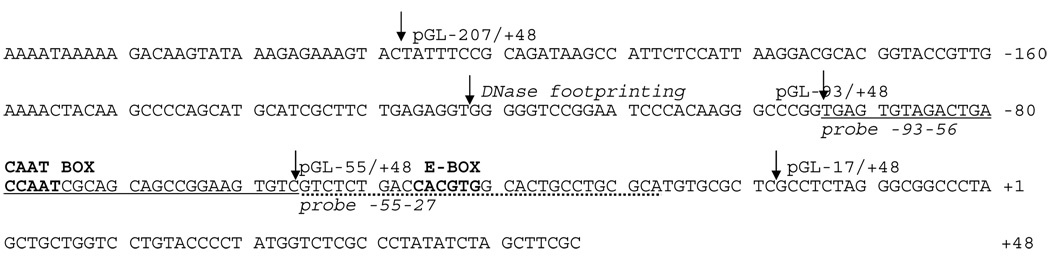 |
2.6. Transient Transfection and Dual Luciferase Assay
Mouse ES cells were used as the recipient cells for transient transfection assays. Cells (106 cells/transfection) were transfected with 5 µg of each promoter-reporter plasmid only or with expression vectors pIRES-NF-YA and/or pCMV-SPORT6.1-USF1 using an electroporator (Nucleofector, Amaxa, Gaithersburg, MD). 0.01 µg of pRL-TK (Promega) was co-transfected with promoter-reporter plasmid as internal control for transfection efficiency. Cells were incubated in 3 ml complemented medium for 48 h and then subjected to a dual luciferase assay (Promega) or RNA extraction. Overexpression of NF-Ya and USF1 was monitored by RT-PCR and compared with β-actin.
2.7. Mutagenesis
Site-directed mutagenesis was performed using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega). The sense oligonucleotides (Table 1) used in the mutation analysis of competition EMSA served as mutagenesis primers. The template for the mutagenesis of the CCAAT-box and the E box was construct pGL −93/+48. Mutant strands were synthesized and ligated in synthesis buffer using T4 DNA polymerase and T4 DNA ligase (Promega). To confirm the fidelity of mutations, plasmids were analyzed by DNA sequencing.
2.8. Immunofluorescence
Cells were cultured on glass slides and fixed in 0.1% paraformaldehyde. For detection of NF-Ya and USF1 expression in ES cells, fluorescent immunohistochemistry was performed using goat anti-NF-Ya antibody or rabbit anti-USF1 antibody (Santa Cruz). Samples were incubated overnight at 4° C with primary antibodies and then with secondary FITC-conjugated anti-goat IgG antibody or Texas Red-conjugated anti-rabit antibody (Invitrogen, Carlsbad, CA) and then analyzed using fluorescence microscopy. The same fluorescence settings and intensities were used for all experiments.
2.9. Chromatin immunoprecipitation (ChIP)
ChIP was performed using Chromatin-Immunoprecipitation Assay Kit (Millipore, Billerica, MA). ES cells were fixed in 1 % formaldehyde. Nuclei were isolated, sonicated and pre-cleaned with protein A Agarose/Salmon Sperm DNA. For ChIP, the pre-cleaned chromatin solution was set as input or incubated with 5 µg of anti-NFYa, anti-USF1 (Santa Cruz) or anti-Flag antibody (Sigma, St. Louis, MO) on a rotation platform at 4° C overnight. After reversal of the cross-links, the DNA was purified from the immune complex and amplified using PCR primers (Table 1) specific for the CP27 promoter region.
2.10. Coimmunoprecipitation and Western blotting
Whole cell lysates were centrifuged at 10,000 g to pellet cellular debris. The supernatants were precleared with protein A-agarose beads and then incubated with NF-YA or USF1 antibodies (Santa Cruz) at 4° C overnight and with protein A-agarose beads for 1 hour. The bead-antibody pellets were washed and fragmented by SDS-PAGE on 15 % gels. The separated proteins were transferred to a nitrocellular membrane. The membranes were blocked with 5 % non-fat dried milk in PBS overnight at 4° C and incubated with 1:2000 diluted USF1 or NF-YA (Santa Cruz), and then with a 1:2000 diluted HRP-conjugated secondary antibody (Invitrogen, Carlsbad, CA). The immune complexes were detected with SuperSignal West Pico Chemilumnescent Substrate (Thermo Scientific, Rockford, IL).
2.11. Quantitative real time RT-PCR
Total RNA was extracted from mES cells using Trizol (Invitrogen). Total RNA quality and quantity were tested by spectrophotometry and agarose gel electrophoresis. 2 µg RNA was reverse transcribed and cDNA amplified using selected primers. Real time quantitative PCR was conducted using SuperScript III Platinum two step qPCR kit with LUX fluorogenic primer (Invitrogen). Primers were designed using the LUX™ Designer software (FAM labeled LUX primer, Invitrogen) and listed in Table 1. Reaction conditions were as follows: 2 min at 50° C (one cycle), 10 min at 95° C (one cycle), and 15 second at 95° C and 1 min at 60° C (40 cycles). PCR products were continuously monitored with an ABI PRISM 7900 detection system (RRC-Core at UIC). Samples were normalized using ribosome 18 RNA (JOE labeled LUX primer set, Invitrogen). CP27 expression levels were calculated in relationship to the β-actin internal control using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.12. Statistical Analysis
Statistical significance was determined by Student’s T-test or 1-way analysis of variance (ANOVA).
3. Results
3.1. Two regulatory elements, a CCAAT-box and an E-box, are located in the mouse CP27 proximal promoter region
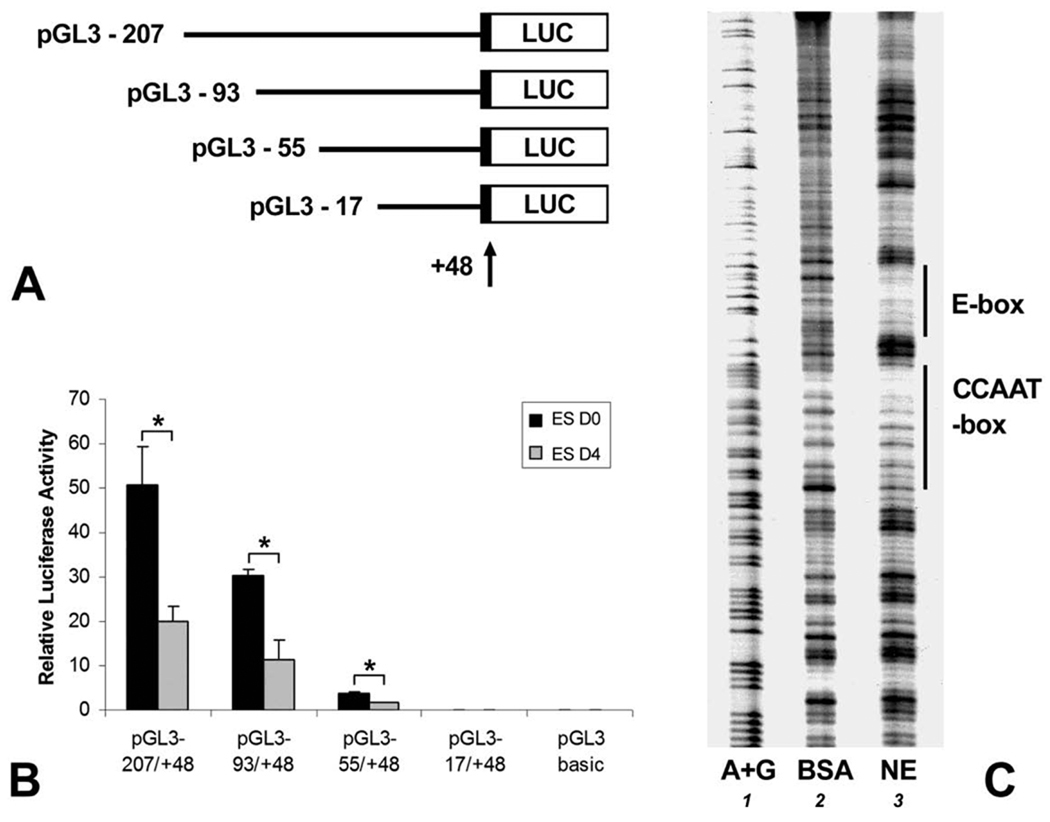
To understand how CP27 expression is regulated in ES cells, putative cis-acting elements within the CP27 proximal promoter region were localized by progressive 5’ deletion mutations using reporter gene constructs in mouse ES cells with or without LIF (Fig. 1A). Luciferase activity assays demonstrated that the region between −93 and −56 contained a strong enhancer activity because the 5’ deletion mutation from nucleotides − 93 to −56 resulted in a 9.6 times decrease in luciferase activity. The construct pGL −55/+48 had a 60 times higher luciferase activity than the pGL3-basic control. The activity of the pGL −17/+48 construct was on the same level with the pGL3-basic vector (Fig. 1B).
Fig. 1. Characterization of the mouse CP27 proximal promoter activity in ES cells.
A, schematic representation of reporter plasmids. The arrow indicates the position of the transcription start site. Reporter constructs descriptors indicate the 5’-end nucleotide number of the inserted promoter sequence. B, luciferase activity resulting from the expression of the CP27 promoter-reporter gene constructs. D0 represents cell culture in the presence of LIF and D4 represents LIF-withdrawal for 4 days. The results were normalized by co-transfection with a pRL-TK reporter plasmid. Error bars represent the standard error for three samples in five independent experiments. C, DNase I footpriting analysis. The DNA fragment CP−207/+48 was 3’-end labeled and incubated with BSA (lane 2) or nuclear extract from ES cells (lane 3). Lane 1 represents a G+A Maxam-Gilbert sequencing ladder. Two DNase I protected regions are recognized, and their sequences match the E-box and the CCAAT-box consensus sequences of the proximal CP27 promoter. The asterisks in Fig. B indicate significant differences (P < 0.05).
The transient transfection experiments described above indicate the presence of two positive regulatory elements within 93 bp of the mouse CP27 proximal promoter region. In order to elucidate the nuclear factor binding sites, DNase I footprint assays were performed. Using end-labeled probe CP −207/+48 and nuclear extracts from ES cells, two major protected DNA regions were identified between nucleotides −83 and −61, and from −52 to −37 (Fig. 1C, lane 3). These protected sequences were identified as a CCAAT-box and an inverted E-box by comparison with a DNA ladder.
3.2. Transcription factors binding to corresponding cis-regulatory elements on the CP27 proximal promoter in vitro and in vivo: NF-Y binds to the CCAAT-box and USF1 binds to an E-box
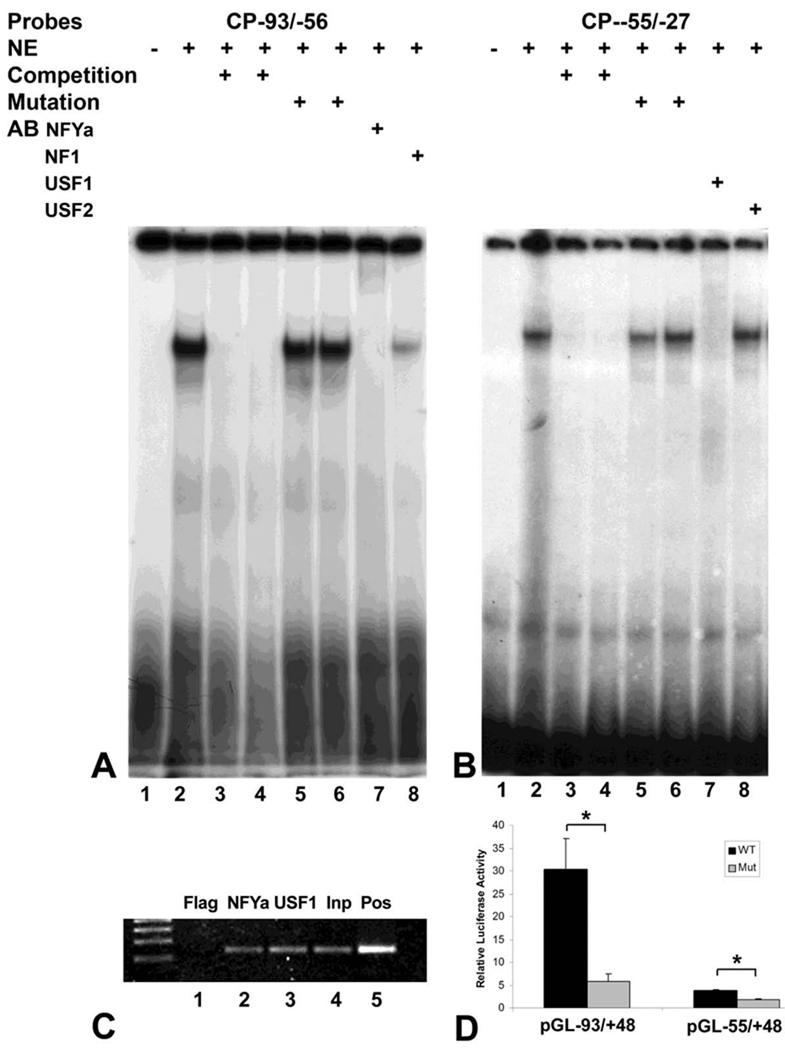
To confirm that the CCAAT-box and the E-box are regulatory elements in the mouse CP27 proximal promoter, oligonucleotides containing the CCAAT-box (CP−93/−56) or the E box (CP−55/−27) were subjected to EMSA. The EMSA revealed formation of protein-DNA complexes using nuclear extracts from ES cells and the radioactively labeled oligonucleotide CP−93/−56 or oligonucleotide CP−55/−27. Competition experiments were performed to determine the sequence specificity of the protein-DNA complex. 25- or 50-fold molar excess of unlabeled CP−93/−56 or CP−55/−27 eliminated the formation of the complexes (Figs. 2A and 2B, lane 3 and 4). However, when a mutation in the CCAAT-box was introduced by mutating CCAAT to CACAT or a mutation in E box was generated by mutating CGTG to AATG, the mutated oligonucleotides did not affect protein/DNA bindings (Figs. 2A and 2B, lanes 5 and 6).
Fig. 2. Identification of two active cis-regulatory elements and corresponding transcription factors on the CP27 proximal promoter: NF-Y binds to the CCAAT-box and USF1 binds to an E-box.
A and B, electrophoretic mobility shift analysis. Nuclear extracts from ES cells were incubated with 32P-labeled oligonucleotide probes CP−93/−56 (A) or CP−55/−27 (B) in the presence of various competitors, mutants and antibodies. The labels above individual lanes read NE = nuclear extract, AB = antibody. C, chromatin immunoprecipitation. NF-YA (lane 2) or USF1 associated DNA fragments (lane 3) were immunoprecipitated using anti-NF-YA or USF1 antibodies. Flag antibody served as negative control. D, transient transfection experiments. The plasmid pGL−93/+48 CCAATm is a modified construct of pGL−93/+48 in which the CCAAT-box had been mutated; the plasmid pGL−55/+48 Em is a mutated construct of pGL−55/+48 in which the E box was modified. The asterisks in Fig. D indicate significant differences (P < 0.05).
To determine which transcription factors contributed to the complexes formed with the CCAAT-box or the E-box, the proximal CP27 promoter region was analyzed using MatInspector (www.genomatrix.de) and Signal Scan (www.bimas.dcrt.nih.gov). This analysis identified a NF-Y binding site and a USF1 binding site. To further confirm that NF-Y and USF1 bind to the CCAAT-box and the E-box in the CP27 proximal promoter respectively, super-gel shift assays were performed. Incubation of anti-NF-YA antibody together with the CP27−93/−56 DNA-protein complex led to the formation of slower migrating supershifted bands (Fig. 2A, Lane 7). The anti-NF1 antibody did not form a supershifted band with CP27 proximal promoter sequences (Figs. 2A, Lane 8). The antibody to USF1 completely obliterated the CP27−55/−27 DNA/protein complex (Fig. 2B, Lane 7), whereas the antibody to USF2 did not recognize this band (Fig. 2B, lane 8), indicating that USF1 binds to the E-box within the CP27 proximal promoter. Note that the USF1 antibody did not supershift but caused the disappearance of the cognate ESMA band. These results further confirmed that the CP27 −93/−56 DNA-protein complex contained the NF-Y transcription factor (Fig. 2A, lane 7) while the CP27−55/−27 DNA-protein complex contained the USF1 transcription factor (Fig. 2B, lane 7).
Chromatin immunoprecipitation was performed in order to verify the binding of NFY and USF1 to the CP27 promoter in ES cells in vivo. The cross-linked chromatin of ES cells was immunoprecipitated with anti-NF-YA or anti-USF1 antibodies and subjected to PCR amplification using primers spanning the CP27 promoter region (Table 1), resulting in the amplification of a 141 bp fragment corresponding in size to the CP27 proximal promoter fragment (Fig. 2C, lane 2 and 3). In contrast, a control anti-flag antibody failed to precipitate a chromatin fragment containing the endogenous CP27 promoter (Fig. 3C, lane 1). These results confirmed the presence of an active CCAAT-box and an active E-box in the CP27 proximal promoter.
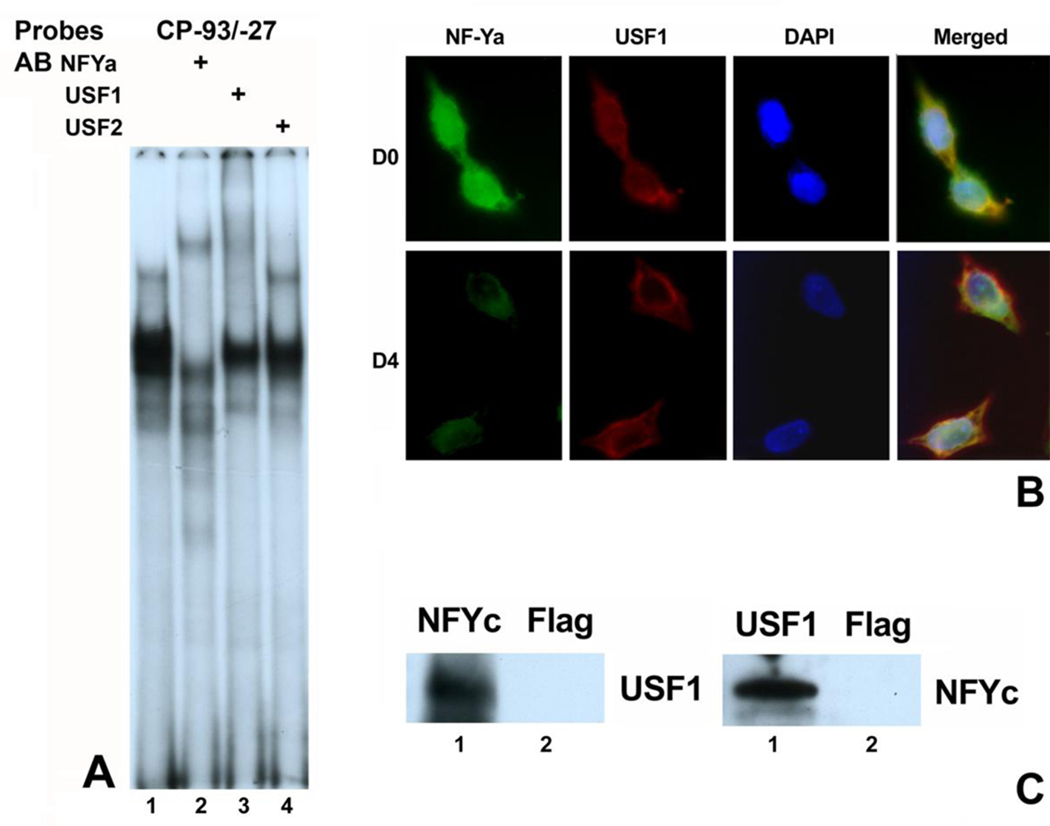
Fig. 3. Interaction between NF-Y and USF1 on the CP27 proximal promoter.
A, supershift assay. Oligonucleotide CP−93/−27 containing both the CCAAT-box and the E box was 32P-labeled and incubated first with NE from ES cells and then with various antibodies. AB = antibody. B, Immunofluorescent assay for NF-YA and USF1 expression. ES cells were cultured in the presence of LIF or without LIF for four days, fixed and subjected to double immunofluorescence staining using NF-YA (green) and USF1 (red) antibodies. The nucleus was stained with DAPI (blue). D0 represents cell culture in the presence of LIF and D4 represents LIF-withdrawal for 4 days. C, co-immunoprecipitation. ES cell lysates were precipitation with either NF-YC antibody or USF1 antibody. Flag antibody was used as negative control.
To determine whether the CCAAT-box and the E-box are involved in the regulation of the CP27 promoter, the CCAAT-box and the E-box of the intact CP27 promoter were mutated (Table 1). Specifically, the activity of the mutated plasmid pGL−93/+48 CCAAT-m was compared with that of the wild-type pGL−93/+48 and the mutated plasmid pGL−55/+48 E-m was compared with that of the wild-type pGL−55/+48. The mutations had significant effects on the activity of the CP27 promoter. Mutation in the CCAAT-box reduced CP27 promoter activity by 80.74% (Fig. 2D), while mutation of the E box decreased CP27 promoter activity by 53.12% (Fig. 2D). These results demonstrate that the CCAAT-box and the E-box are functionally significant regulatory elements located in the mouse CP27 proximal promoter.
3.3. NF-Y interacts with USF1 on the CP27 proximal promoter
DNase I footprinting and functional studies described earlier have identified two active cis-regulatory elements in the proximal CP27 promoter, an E-box and an adjacent CCAAT-box. Previous studies have suggested that a combined CCAAT-box-E box motif regulates gene expression directly and co-operatively (Zhu et al., 2003; Grskovic et al., 2007). To explore the possibility of an interaction between the CCAAT-box and the E-box in the CP27 proximal promoter, we subjected oligonucleotide (CP−93/−27) containing both the CCAAT-box and the E-box to EMSA. The CP−93/−27 probe generated three EMSA bands from ES cell nuclear extracts (Fig. 3A, lane 1). The fastest migration band was generated by USF1 because the USF1 antibody completely abolished the band (Fig. 3A, lane 3). The second fast migrating band was generated by NFY because the NF-YA antibody obliterated the band and formed a supershifted band (Fig. 3A, Lane 2). The slowest band was generated by both NF-Y and USF1 since inclusion of antibody against either USF1 or NF-YA into the promoter/NE reaction disrupted the formation of this band (Fig. 3A, Lanes 1 to 3). The USF2 antibody did not interact with any protein/DNA complexes (Fig. 3A, Lane 4).
To determine whether NF-Y and USF1 were co-localized in ES cells, NF-YA and USF1 expression were examined by immunohistochemistry. Immunofluorescent assays illustrated that NF-YA and USF1 proteins were expressed and overlapped in the nucleus with and without addition of the growth factor LIF (Fig. 3B).
To confirm that NF-Y interacts with USF1 in vivo, we performed coimmunoprecipitation experiments. ES Cell lysates were used for immunoprecipitation with antibodies against NF-YC or USF1. Flag antibody was served as a negative control. As shown in Fig. 3C, USF1 was detected in anti-NF-YC immunoprecipitates but not in immunoprecipitates with Flag antibody. Conversely, NF-YC was detected in the USF1 immunoprecipitates. These results provide additional evidence that NF-Y and USF1 physically interact in vivo.
3.4. Synergistic activation of CP27 expression by NF-Y and USF1
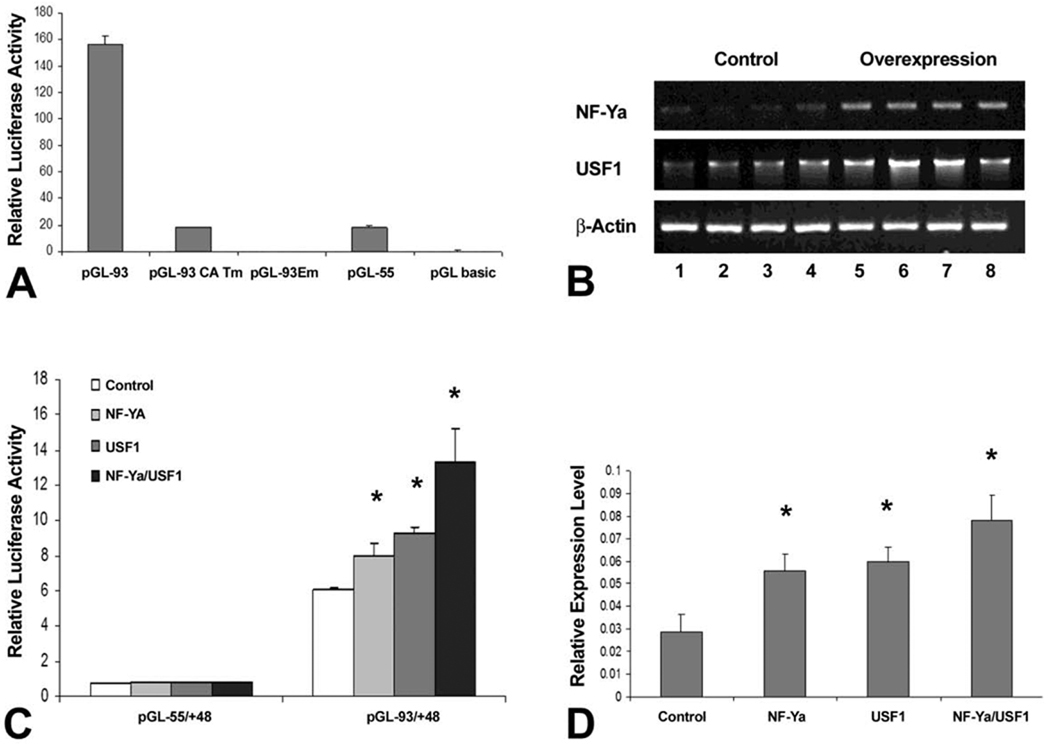
To investigate the involvement of NFY-USF interactions in the regulation of the CP27 promoter, mutated CCAAT-box and E-box constructs of the intact CP27 promoter were generated (Table 1). Specifically, the activity of the mutated plasmid pGL−93/+48 CAT-m or pGL−93/+48 E-m was compared with that of the wild-type pGL−93/+48. The mutation in the CCAAT-box reduced luciferase activity by 90.49% (Fig. 4A). Surprisingly, the mutation in the E-box completely abolished the activity of the CP27 proximal promoter even though the CCAAT-box remained intact (Fig. 4A). These results suggest that the function of the CCAAT-box is dependent on the E-box as an essential regulatory element located in the mouse CP27 proximal promoter.
Fig. 4. Effect of the interaction between NF-Y and USF1 on CP27 gene expression.
A, function study to determine whether the CCAAT-box and the E box are essential regulatory elements of the CP27 promoter. The wild type promoter-reporter construct pGL−93/+48 and its mutated homologues pGL−93/+48CCAATm or pGL−93/+48Em were transfected into ES cells. B, RT-PCR analysis of NF-YA or USF1 overexpression efficiency. ES cells were transfected with expression vectors pIRES-NF-YA or pCMV-SPORT6.1-USF1. Each lane (1–8) represents RNA extracts from individually cultured wells. Lanes 1–4 are from controls and 5–8 are from overexpressing mouse ES cells. C, Regulation of CP27 promoter activity by overexpression of NF-YA or USF1 or a combination of both. The construct pGL−93/+48 was co-transfected with the expression vectors pIRES-NF-YA or pCMV-SPORT6.1-USF1 or a combination of both into ES cells. Transfection with pIRES-GFP or pCMV-SPORT6.1 was used as controls. D, qRT-PCR analysis of CP27 mRNA expression regulated by gene manipulation of NF-YA or USF1 or a combination of both. The asterisk indicates a statistically significant increase in luciferase activity and gene expression levels. CP27 expression levels were calculated in relationship to the β-actin internal control using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
To determine whether NF-Y and USF1 can function synergistically in activation of the CP27 promoter, expression vectors for NF-YA (pIRES-NF-YA) or USF1 (pCMV-SPORT6.1-USF1) were introduced into ES cells and expression levels in transfected cells were significantly higher than in control cells as demonstrated by RT-PCR (lanes 5–8 compared to lanes 1–4, determined by densitometry)(Fig. 4B). Vectors pIRES-NF-YA, pCMV-SPORT6.1-USF1 or a combination were then co-transfected with the promoter-reporter construct pGL-CP−93/+48 containing both the CCAAT-box and the E-box and a high degree of synergy (2.19 folds) between NF-YA and USF1 was observed (Fig. 4C).
To confirm that the functional collaboration between NFY and USF1 influences endogenous CP27 gene expression, ES cells were transfected with pIRES-NF-YA or pCMV-SPORT6.1-USF1 or a combination of both, and CP27 mRNA was quantified by real time RT-PCR. The endogenous CP27 mRNA expression was increased by 1.93-fold following NF-YA overexpression, 2.06-fold following USF1 overexpression, and 2.69-fold following NF-YA-USF1 overexpression, respectively (Fig. 4D).
4. Discussion
In the present study we have identified a CCAAT-box and an E-box as cis-acting promoter elements of the TATA box-less CP27 promoter and characterized their regulation in ES cells through the transcription factors NF-Y and USF1. The CP27 gene encodes a novel chromatin factor required for mouse development and germ layer differentiation, and CP27 is highly expressed in inner cell mass (ICM) of developing blastocysts. Loss of CP27 caused severely disturbed epiblast development (not published). The distinct expression of CP27 in the ICM and the effect of CP27 on epiblast development were the reason for selecting ES cells as a model system to study the regulation of CP27 in ES cells.
Two cis-regulatory elements, a CCAAT-box and an E-box, were identified in the CP27 proximal promoter by deletion mutation and footprint analysis. Among several transcription factor binding candidates, NF-Y was identified as the CCAAT-box binding protein and USF1 as the E-box binding protein. Previous studies have reported combined CCAAT-box/E-box structures in the HOXB4 proximal promoter (Zhu et al., 2003) and in HOXC4 and HOXD4 promoters of in both human and mouse (Zhu et al., 2005). The close relationship between the CCAAT-box and the E-box and the interaction between NF-Y and USF1 suggests that NF-Y and USF1 might cooperate in the regulation of CP27 gene expression. In our studies, overexpression of exogenous NF-YA and USF1 upregulated CP27 promoter activity and endogenous CP27 gene expression, suggesting a synergistic interaction between NF-Y and USF1. Explaining the function of a combined CCAAT-box/E-box structure, previous studies have indicated that NF-Y is not a powerful transcription activator but rather a promoter organizer which co-operates with neighboring transcription factors to modulate the transcriptional activity of target genes (Testa et al., 2005; Ceribelli et al., 2008; Nicolas et al., 2003). In the case of the CP27 proximal promoter, this would imply that NF-Y might cooperate with USF1 to regulate CP27 transcription. In support of a functional interaction between NF-Y and USF1 we have shown that CCAAT-box mutation reduced the activity of the CP27 proximal promoter by 80.74%, while maintaining the CP27 basal promoter function. In contrast, mutation of CP27 E-box completely abolished the function of the CP27 proximal promoter even though the CCAAT-box structure remained intact. These results indicate that the regulatory role of NF-Y may depend on the transcriptional regulatory partners to which it binds in a given genomic and cellular context (Zhu et al., 2003; Matuoka et al., 1999).
On a biological level, these data indicate that the effect of the ES cell proliferation transcription factor NF-Y on the CP27 promoter is synergistically modulated by a second transcription factor, USF-1. Here we have shown that in tandem with NF-Ya downregulation, USF-1 shuttles from the center of the nucleus to the nuclear envelope, a potential transcription factor resting place (Heessen and Fornerod, 2007). Thus, regulation of CP27 gene expression to facilitate ES cell proliferation may require the physical interaction between USF-1 and NF-Ya in ES cell nuclei.
We conclude that through CCAAT-box and E-box promoter elements, the CP27 proximal promoter is jointly and interactively regulated by two important transcriptional factors, NF-Y and USF1. NF-Y is a ubiquitous transcription factor that promotes proliferation while USF1 has been linked to the inhibition of cell differentiation (Zhu et al., 2003; Grskovic et al., 2007; Jiang and Mendelson, 2003). Our studies provide a functional link between the transcriptional regulators NF-Y and USF1 and ES cell growth through the regulation of CP27 gene expression.
Acknowledgments
Studies were generously supported by the National Institute of Dental and Craniofacial Research, National Institute of Health Grant R01 DE013095 to TGHD.
Glossary
- CP27
Craniofacial protein 27
- NF-Y
Nuclear transcription factor Y
- USF1
Upstream stimulatory factor 1
- USF2
Upstream stimulatory factor 2
- ES
Embryonic stem cell
- DMEM
Dulbecco’s modified eagle medium
- FCS
Fetal calf serum
- LIF
Leukemia inhibitory factor
- EMSA
Electrophoretic mobility shift assay
- NF-YA
Nuclear transcription factor Y subunit alpha
- NF1
Nuclear transcription factor 1
- BSA
Bovine serum albumin
- PCR
Polymerase chain reaction
- CBF-B
Core binding transcription factor-subunit B
- ChIP
Chromatin immunoprecipitation
- ICM
Inner cell mass
- HOX
Homeobox protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P, Trifonov EN. CCAAT box revisited: bidirectionality, location and context. J Biomol Struct Dyn. 1988;5:1231–1236. doi: 10.1080/07391102.1988.10506466. [DOI] [PubMed] [Google Scholar]
- Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008;28:2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Imbriano C, Mantovani R. Dynamic recruitment of transcription factors and epigenetic changes on the ER stress response gene promoters. Nucleic Acids Res. 2006;34:3116–3127. doi: 10.1093/nar/gkl304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontini M, Imbriano C, diSilvio A, Bell B, Bogni A, Romier C, Moras D, Tora L, Davidson I, Mantovani R. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J Biol Chem. 2002;277:5841–5848. doi: 10.1074/jbc.M103651200. [DOI] [PubMed] [Google Scholar]
- Griffiths JFA, Miller HJ, Suzuki TD, Lewontin CR, 3, Gelbart MW. An Introduction to Genetic Analysis. 7th edition. New York: 2000. [Google Scholar]
- Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra RF, Imperadori L, Mantovani R, Dunlap DD, Finzi L. DNA compaction by the nuclear factor-Y. Biophys J. 2007;93:176–182. doi: 10.1529/biophysj.106.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO reports. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Luan X, Fan J, Diekwisch T. CP27 function related to early inner cell mass differentiation and pluripotent network maintenance. ASCB. 2005:L349. [Google Scholar]
- Jiang B, Mendelson CR. USF1 and USF2 mediate inhibition of human trophoblast differentiation and CYP19 gene expression by Mash-2 and hypoxia. Mol Cell Biol. 2003;23:6117–6128. doi: 10.1128/MCB.23.17.6117-6128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luan X, Ito Y, Zhang Y, Diekwisch TG. Characterization of the mouse CP27 promoter and NF-Y mediated gene regulation. Gene. 460:8–19. doi: 10.1016/j.gene.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magan N, Szremska AP, Isaacs RJ, Stowell KM. Modulation of DNA topoisomerase II alpha promoter activity by members of the Sp (specificity protein) and NF-Y (nuclear factor Y) families of transcription factors. Biochem J. 2003;374:723–729. doi: 10.1042/BJ20030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Matuoka K, Yu Chen K. Nuclear factor Y (NF-Y) and cellular senescence. Exp Cell Res. 1999;253:365–371. doi: 10.1006/excr.1999.4605. [DOI] [PubMed] [Google Scholar]
- Narlikar L, Ovcharenko I. Identifying regulatory elements in eukaryotic genomes. Brief Funct Genomic Proteomic. 2009;8:215–230. doi: 10.1093/bfgp/elp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Noe V, Ciudad CJ. Transcriptional regulation of the human Sp1 gene promoter by the specificity protein (Sp) family members nuclear factor Y (NF-Y) and E2F. Biochem J. 2003;371:265–275. doi: 10.1042/BJ20021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Simboeck E, Khier H, Seiser C. Autoregulation of mouse histone deacetylase 1 expression. Mol Cell Biol. 2003;23:6993–7004. doi: 10.1128/MCB.23.19.6993-7004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A, Donati G, Yan P, Romani F, Huang TH, Vigano MA, Mantovani R. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J Biol Chem. 2005;280:13606–13615. doi: 10.1074/jbc.M414039200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Giannola DM, Zhang Y, Rivera AJ, Emerson SG. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood. 2003;102:2420–2427. doi: 10.1182/blood-2003-01-0251. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci U S A. 2005;102:11728–11733. doi: 10.1073/pnas.0503405102. [DOI] [PMC free article] [PubMed] [Google Scholar]