Abstract
The bodies of most animals are populated by highly complex and genetically diverse communities of microorganisms. The majority of these microbes reside within the intestines in largely stable but dynamically interactive climax communities that positively interact with their host. Studies from this laboratory have shown that stressor exposure impacts the stability of the microbiota and leads to bacterial translocation. The biological importance of these alterations, however, is not well understood. To determine whether the microbiome contributes to stressor-induced immunoenhancement, mice were exposed to a social stressor called social disruption (SDR), that increases circulating cytokines and primes the innate immune system for enhanced reactivity. Bacterial populations in the cecum were characterized using bacterial tag-encoded FLX amplicon pyrosequencing. Stressor exposure significantly changed the community structure of the microbiota, particularly when the microbiota were assessed immediately after stressor exposure. Most notably, stressor exposure decreased the relative abundance of bacteria in the genus Bacteroides, while increasing the relative abundance of bacteria in the genus Clostridium. The stressor also increased circulating levels of IL-6 and MCP-1, which were significantly correlated with stressor-induced changes to three bacterial genera (i.e., Coprococcus, Pseudobutyrivibrio, and Dorea). In follow up experiments, mice were treated with an antibiotic cocktail to determine whether reducing the microbiota would abrogate the stressor-induced increases in circulating cytokines. Exposure to SDR failed to increase IL-6 and MCP-1 in the antibiotic treated mice. These data show that exposure to SDR significantly affects bacterial populations in the intestines, and remarkably also suggest that the microbiota are necessary for stressor-induced increases in circulating cytokines.
Keywords: Social Stress, Gastrointestinal Microbiota, Pyrosequencing, Inflammation, Immunomodulation
Introduction
The external surfaces of the body are colonized by vast arrays of microbes that outnumber cells of the body by a factor of 10 (i.e., 1014 bacterial cells to 1013 human cells). This means that 90% of the cells of our body are our commensal microbiome. The vast majority of these microbes reside in the intestines as part of the intestinal microbiota, with microbiota levels ranging from < 105 bacteria per gram of digesta in the upper parts of the small intestine, to > 1012 bacteria per gram of digesta in the large intestine (Sekirov et al., 2010). Many of these bacteria are simple opportunistic colonizers, while the majority are true symbiotic organisms in the sense that they have beneficial interactions with each other and the host. For example, many metabolic activities in the intestines are derived from the microbiota, such as the synthesis of vitamin K and vitamin B complex and the metabolism of carcinogens (O'Hara and Shanahan, 2006). In addition, studies utilizing germ-free mice have now linked changes in the intestinal microbiota to the development of obesity and diabetes (Backhed et al., 2004; Ley et al., 2005; Turnbaugh et al., 2006). Perhaps one of the most well studied effects of the microbiota on the host, however, is their impact on the immune system.
It has long been recognized that the intestinal microbiota affect the mucosal immune system. This has been well documented in germ-free animals that due to a lack of commensal microbiota have smaller Peyer’s patches, fewer intraepithelial lymphocytes, and lower levels of secretory IgA (Macpherson and Uhr, 2004). Importantly, colonizing the germ-free mice with commensal bacteria normalizes these immune parameters, indicating the importance of intestinal bacteria in shaping the mucosal immune response (Macpherson and Uhr, 2004). The impact that the microbiota can have on systemic immunity has not been as widely studied. But, alterations to the intestinal microbiota have been linked to inflammatory diseases, such as asthma, in animal models and also in humans, suggesting that the microbiota affect aspects of adaptive immune regulation (Huffnagle, 2010). Innate immunity has also been shown to be affected by the microbiota, with studies indicating that neutrophil activity is primed in vivo by microbiota-derived peptidoglycan acting on neutrophil nucleotide oligomerization domain-containing protein (Nod)1 receptors (Clarke et al., 2010). Moreover, translocation of intestinal bacteria from the lumen of the intestines to the interior of the body has been shown to result in increases in circulating cytokines like IL-6 (Ando et al., 2000). Thus, there is ample evidence linking the microbiota to innate and adaptive immune responses at mucosal, as well as systemic, sites.
The microbiota reside as a largely stable climax community as a result of a series of ecological successions involving the selection of species best adapted for the given niche (Huffnagle, 2010). This climax community is resistant and resilient to long-term disruptions in community structure (Allison and Martiny, 2008), but many factors, such as diet or antibiotic use, can cause more transient alterations in their community structure (Antonopoulos et al., 2009; Dethlefsen et al., 2008). Studies from our laboratories, and from others, demonstrate that stressor exposure, or exposure to neuroendocrine hormones, can significantly affect the microbiota (Bailey et al., 2010; Bailey and Coe, 1999; Knowles et al., 2008; Lizko, 1987; Lyte and Bailey, 1997; Tannock and Savage, 1974). A study using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) demonstrated that the community structure of microbiota from mice exposed to a prolonged restraint stressor was significantly different than the community structure found in non-stressed control mice (Bailey et al., 2010). The importance of these findings for the health of the host is not completely understood. While there is evidence to suggest that changes in the intestinal microbiota reduces resistance to infectious challenge with intestinal pathogens (Bailey et al., 2010), it is also known that altering the climax communities of microbiota is a predisposing factor for translocation of bacteria from the lumen of the intestines to the interior of the body (Berg, 1999) where they can stimulate the immune system (Clarke et al., 2010; Kim et al., 2009). Although we have reported that stressor exposure increases the translocation of gastrointestinal and cutaneous microbiota to secondary lymphoid organs (Bailey et al., 2006), the likelihood that stressor-induced effects on the microbiota also impact stressor-induced immunomodulation has not been tested.
Psychological stressors in humans, and in laboratory animals, often cause an increase in circulating cytokines. For example, prolonged stressors, such as the stress of caring for a spouse with dementia or chronic work stress, are associated with elevated cytokines like IL-6 and TNF-α (Hemingway et al., 2003; Kiecolt-Glaser et al., 2003). Acute laboratory stressors, like the Trier social stress test (TSST) and the Stroop task also cause elevated circulating IL-6 and TNF-α (Brydon et al., 2004; Brydon et al., 2005; Brydon and Steptoe, 2005; Edwards et al., 2006; von et al., 2006). In laboratory mice, repeated social defeat incurred during stressor paradigms including social conflict and social disruption (SDR) results in enhanced innate immune activity (Bailey et al., 2007; Bailey et al., 2009; Lyte et al., 1990; Powell et al., 2009). Cytokines, including IL-6, are increased in the circulation of mice exposed to the SDR paradigm (Stark et al., 2002), and splenic macrophages from the repeatedly defeated mice are primed for enhanced cytokine production and antimicrobial activity upon bacterial stimulation (Bailey et al., 2007; Bailey et al., 2009). The mechanisms linking the stress response to increases in immune function are not well understood, but evidence suggests that stressor-induced increases in sympathetic nervous system (SNS) activity can enhance innate immune activity (Bierhaus et al., 2003). Because stressor exposure changes the community structure of the microbiota (Bailey et al., 2010), and induces bacterial translocation (Bailey et al., 2006), and because microbiota products have been shown to be necessary for priming of innate immunity (Clarke et al., 2010), we hypothesized that microbiota are an important component of the stressor-induced enhancement of innate immune activity. To test this hypothesis, we first determined whether exposure to the SDR stressor changed the community structure of the microbiota. This was followed by experiments in which antibiotics were given to stressor-exposed, as well as non-stressed control, mice to determine whether reducing the microbiota would abrogate stressor-induced increases in immune activity.
Methods
Animals
Male CD-1 mice, 6–8 wks of age, were purchased from Charles River Laboratories (Wilmington, MA) and allowed to acclimate to the animal vivarium for 1 week prior to experimentation. The mice were housed in groups of 3–5 per cage and were kept on a 12 hr light:dark schedule with lights on at 0600. Food and water were available ad libitum. All experimental procedures were approved by The Ohio State University’s Animal Care and Use Committee.
Social Disruption
The social disruption (SDR) stressor occurred over a 2 hr period between 1630–1830, which is at the transition from the end of the light cycle to the beginning of the dark (i.e., active) cycle. SDR was initiated by placing an aggressive male mouse into the home cage of the resident mice as previously reported (Bailey et al., 2007). The aggressor was the same strain as the residents and was originally isolated from the rest of the colony due to observed aggressiveness towards cagemates. During SDR, agonistic interactions between the aggressor and the residents were observed for the first 20 min to be sure that the aggressor attacked and defeated all of the residents. If fighting did not begin within the first 5 min of the interactions, a different aggressor was placed in the cage. After fighting was initiated, the aggressors were left in the cages for 2 hrs. At the end of the 2 hr period, the aggressor was removed and the residents were left undisturbed until the following day when SDR was repeated. Thus, the residents were exposed to a total of 6, two-hour cycles of SDR. The subjects of the experiments were the residents that were repeatedly defeated by the aggressor. Mice were euthanized immediately after the last cycle of SDR (designated SDR + 0 hr) or the morning following the last cycle of SDR (designated SDR + 15 hr). Non-stressed home cage control mice were euthanized at the identical time points (designated HCC + 0 hr and HCC + 15 hr). When the mice were sacrificed, wounds that were incurred during SDR were assessed. In no case did wounding exceed superficial skin wounds from biting and scratching.
Microbiota Analysis
Mice were humanely euthanized via CO2 asphyxiation and the cecal contents were harvested using asceptic technique. The cecal contents were emptied into sterile 1.8 ml microcentrifuge tubes and snap frozen in liquid nitrogen. The samples were stored at −80°C until shipping overnight on dry ice for bTEFAP analysis.
Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP)
Because it is not possible to culture the vast majority of microbes residing in the intestines, we used bTEFAP to determine the relative percentages of the primary populations of microbes in the cecal contents.
DNA extraction
After thawing, the debridement samples were centrifuged at 14,000 rpm for 30 seconds and resuspended in 500µl RLT buffer (Qiagen, Valencia, CA) (with β-mercaptoethanol). A sterile 5mm steel bead (Qiagen, Valencia, CA) and a 500µl volume of 0.1mm glass beads (Scientific Industries, Inc., NY, USA) were added for complete bacterial lyses in a Qiagen TissueLyser (Qiagen, Valencia, CA), run at 30Hz for 5min. Samples were centrifuged briefly and 100µl of 100% ethanol added to a 100µl aliquot of the sample supernatant. This mixture was added to a DNA spin column, and DNA recovery protocols were followed as instructed in the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) starting at step 5 of the Tissue Protocol. DNA was eluted from the column with 30µl water and samples were diluted accordingly to a final concentration of 20 ng/µl. DNA samples were quantified using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France).
Pyrosequencing
bTEFAP was performed as described previously (Dowd et al., 2008b) at the Research and Testing Laboratory (Lubbock, TX). Based upon the number of sequences per sample (approximately 3000), the depth of sequencing was sufficient to evaluate primary gastrointestinal populations at > 0.1% of the total population.
Bacterial diversity data analysis
Following sequencing, all failed sequence reads, low quality sequence ends and tags were removed and sequences were depleted of any non-bacteria ribosome sequences and chimeras using custom software described previously (Dowd et al., 2008a) and the Black Box Chimera Check software (Gontcharova et al., 2010) B2C2 (freely available at http://www.researchandtesting.com/Bioinformatics.html). Sequences less than 250bp were removed. To determine the community compositions of bacteria based upon the remaining sequences, sequences were first queried using a distributed BLASTn .NET (www.krakenblast.com) algorithm (Dowd et al., 2005) against a database of high quality 16s bacterial sequences derived from NCBI. Database sequences were characterized as high quality based upon the criteria of RDP ver 9 (Cole et al., 2009). Using a .NET and C# analysis pipeline the resulting BLASTn outputs were compiled, validated using taxonomic distance methods, and data reduction analysis performed as described previously (Dowd et al., 2008b). Rarefaction, Ace, and Chao1 to estimate mathematically predicted diversity and richness in the treatments using of 250 bp trimmed, non-ribosomal sequence depleted, chimera depleted, high quality reads, was performed as described previously (Acosta-Martinez et al., 2008).
Bacterial identification
Sequences with identity scores, based upon best alignment to known or well characterized 16S sequences with greater than 96.5% identity (<3% divergence) were resolved at the species level, between 94.5% and 96.4% at the genus level, between 89.5% and 94.4% at the family and between 80% and 89.4% at the order level. After resolving based upon these parameters, the percentage of each bacterial ID was individually analyzed for each cecal sample providing relative abundance information within and among the samples based upon the relative numbers of sequences within a given sample.
Cytometric Bead Array
Blood was collected via cardiac puncture, and cytokine levels in the serum assessed using the mouse inflammation kit cytometric bead array per manufacturers instructions (BD, San Diego, CA). This allowed for the simultaneous assessment of IL-6, IFN-γ, IL-12p70, TNF-α, MCP-1 (i.e., CCL2), and IL-10.
Splenic iNOS mRNA
iNOS mRNA in the spleen was assessed using real-time PCR. RNA was isolated by placing the spleens into 1 ml of Trizol reagent (Gibco, Rockville, MD) and homogenizing in the Trizol reagent using a Tissue Tearor (Biospec Products, Bartlesville, OK). RNA was isolated according to the Trizol protocol provided by the manufacturer (Gibco). Total RNA was reverse transcribed using a commercially available kit (Promega, Madison, WI) according to manufacturer’s instructions. Briefly, 1 µg of total RNA was combined with 5 mM MgCl2, 1 mM of each dNTP, 1X RT buffer, 1 U/µl RNasin, 15 U/µg AMV reverse transcriptase, and primed with 0.5 µg/µg random hexamers and DEPC H2O to a volume of 50 µl. The reaction was first incubated for 10 min at room temperature, and then at 42°C for 1 h. This was followed by a 5 min incubation in boiling water and then a cooling period of 5 min on ice. The volume was adjusted to 100 µl by adding DEPC water.
Real time PCR
Sequences for the primers were previously published (Bailey et al., 2010) and were synthesized by Applied Biosystems. The 5’ – 3’ sequences are: iNOS forward, CAGCTGGGCTGTACAAACCTT, reverse TGAATGTGATGTTTGCTTCGG, probe CGGGCAGCCTGTGAGACCTTTGA and 18S forward CGGCTACCACATCCAAGGAA, reverse GCTGGAATTACCGCGGCT, probe TGCTGGCACCAGACTTGCCCTC. The PCR reaction mixture consisted of 2.5 µl of cDNA, 2.5 µl primer mix (900 nM), 2.5 µl of probe, 5 µl sterile dH2O, and 12.5 µl Taqman Universal Master Mix (PE Applied Biosystems, Foster City, CA) for a final volume of 25 µl. Following an initial 2-min cycle at 50 °C followed by 10 min at 95 °C, the reaction ran for an additional 40 total cycles which consisted of a 15-s denaturing phase (90 °C) and a 1-min anneal/extension phase (60 °C). The change in fluorescence was measured using an Applied Biosystems 7000 Prism Sequence Detector (PE Applied Biosystems) and analyzed using Sequence Detector version 1.0. The relative amount of transcript was determined using the comparative Ct method as described by the manufacturer. In these experiments, gene expression in the spleens of non-stressed HCC mice was used as the calibrator. Gene expression, therefore, is expressed as the fold increase in expression of these non-stressed control mice.
Antibiotic Administration
To determine whether indigenous microbiota were responsible for stressor-induced increases in immune activity, mice were treated with an antibiotic cocktail consisting of ampicillin (1 mg/ml), vancomycin (0.5 mg/ml), neomycin sulfate (1 mg/ml), and metronidazole (1 mg/ml) to reduce the intestinal microbiota as previously described (Ochoa-Reparaz et al., 2009; Rakoff-Nahoum et al., 2004). The antibiotics were administered by oral gavage in the morning and evening, beginning three days prior to initiating the stressor as well as on each day the stressor occurred. Vehicle Home Cage Control (Vehicle HCC) and Vehicle SDR mice were orally gavaged with sterile PBS as a control.
Statistics
Stressor-induced changes in the relative abundance of microbiota were assessed using a two-factor analysis of variance (ANOVA), with the groups (HCC vs. SDR) as one factor and the time of sample (0 hr vs. 15 hr) as the second factor. Cytokine levels were log (10) transformed prior to using ANOVA to determine statistically significant differences, and statistical analyses were performed on the ΔΔCt for the real-time PCR data. In the absence of antibiotics, a two factor ANOVA was used (HCC vs. SDR as factor 1 and 0 hr vs. 15 hr as factor 2). For the antibiotic studies, the + 0 hr and +15 hr time points were analyzed separately using a two factor ANOVA (HCC vs SDR; vehicle vs. antibiotic). When statistically significant interactions were found, protected t tests were used as post-hoc tests. Pearson’s correlation coefficient was calculated to determine statistical associations between bacterial species and log(10) transformed cytokine levels. In all cases, the level of statistical significant was set at p < .05 and were determined using SPSS for Windows, version 17.0 (SPSS, Chicago IL). Multivariate dual hierarchical clustering methods were performed using NCSS 2007 (NCSS, Kaysville, UT).
Results
Impact of Stress Exposure on the Intestinal Microbiota
Exposure to the SDR stressor significantly affected the intestinal microbiota. This was manifest as a reduction in microbial diversity and richness. Mathematical evaluation of diversity and richness estimates (at 3% and 5% divergence, which roughly correspond to species and genus levels) based upon Ace and Chao1, as well as curve fitting Richard's equations to predict maximum potential operational taxonomic units indicated a significant reduction in microbial diversity in the SDR + 15 hr group compared to all others (p < .05) (Table 1). Interestingly, this reduction in diversity was not evident immediately after the last exposure of SDR (i.e., no difference in the SDR + 0 hr group compared to the HCC groups) (Table 1). There were no differences in the HCC + 0 hr and HCC + 15 hr groups.
Table 1.
Microbial diversity and richness in the cecal contents
| OTU 3% | OTU 5% | ACE 3% | ACE 5% | Chao1 3% | Chao1 5% | |
|---|---|---|---|---|---|---|
| HCC + 0 hr | 746.8 43.1 | 462.4±24.9 | 1037.9±64.1 | 601.9±35.0 | 1027.1±66.3 | 599.4±35.8 |
| SDR + 0 hr | 809.2±45.0 | 507.6±27.4 | 1144.0±49.6 | 668.1±30.6 | 1117.9±54.6 | 664.8±32.6 |
| HCC + 15 hr | 847.4±50.5 | 541.6±29.8 | 1221.0±63.3 | 717.6±35.0 | 1186.3±68.4 | 700.2±35.2 |
| SDR + 15 hr | 656.0±33.7* | 416.2±20.0* | 956.7±48.3* | 529.8±25.7* | 934.0±50.2* | 523.2±23.5* |
Data are the mean±st. error at 3% and 5% divergence, which roughly corresponds to the species and genus level (respectively).
p <.05 vs. HCC + 15 hr
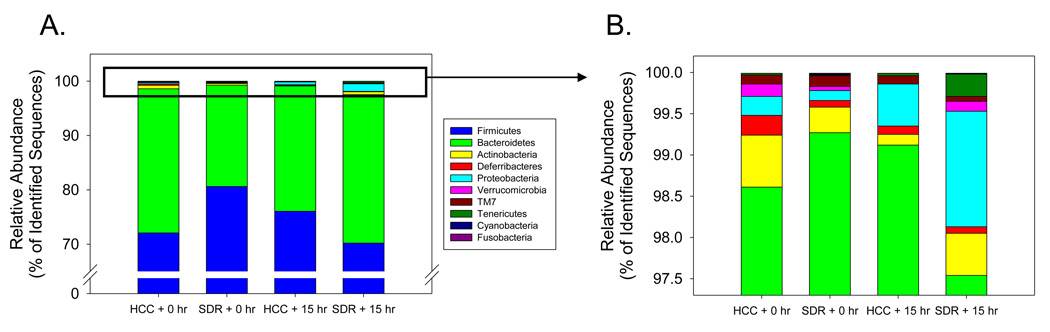
There were few differences between SDR and HCC samples at the phylum taxonomic level of analysis. There was a trend for the SDR + 0 hr mice to have fewer bacteria in the Firmicutes phylum and more bacteria in the Bacteroidetes phylum compared to HCC + 0 hr (Fig. 1A), but these differences were not quite large enough to be considered statistically significant (p = .09 and p = .12 respectively). Similarly, the mean levels of lower abundance phyla differed between the SDR + 0 hr and the HCC + 0, such as the relative abundance of Actinobacteria and Deferribacteria (Fig 1B), but these differences were too small to be considered statistically significant. Despite a higher mean proportion of Proteobacteria in the SDR + 15 hr mice compared to the HCC + 15 hr mice (i.e., 1.51±0.84 vs. 0.52±0.29, Fig. 1B), differences in the lower abundance phyla, including the Proteobacteria, did not approach statistical significance (p > .31 for all phyla).
Figure 1.
The cecal microbiota was comprised of bacteria from 10 divisions. A. The Firmicutes and Bacteroidetes phyla comprised approximately 98% of the identified sequences. There was a trend for SDR + 0 hr mice to have higher levels of Firmicutes (p = .08), but lower levels of Bacteroidetes (p = .11) compared to the HCC + 0 hr mice. None of the other groups were found to be significantly different. B. Eight bacterial divisions accounted for the remaining 2% of the identified sequences. There was a trend for SDR + 0 hr mice to have lower levels of Deferribacteres compared to the HCC + 0 hr mice (p = .08). None of the other groups were found to be significantly different. n = 5 per group
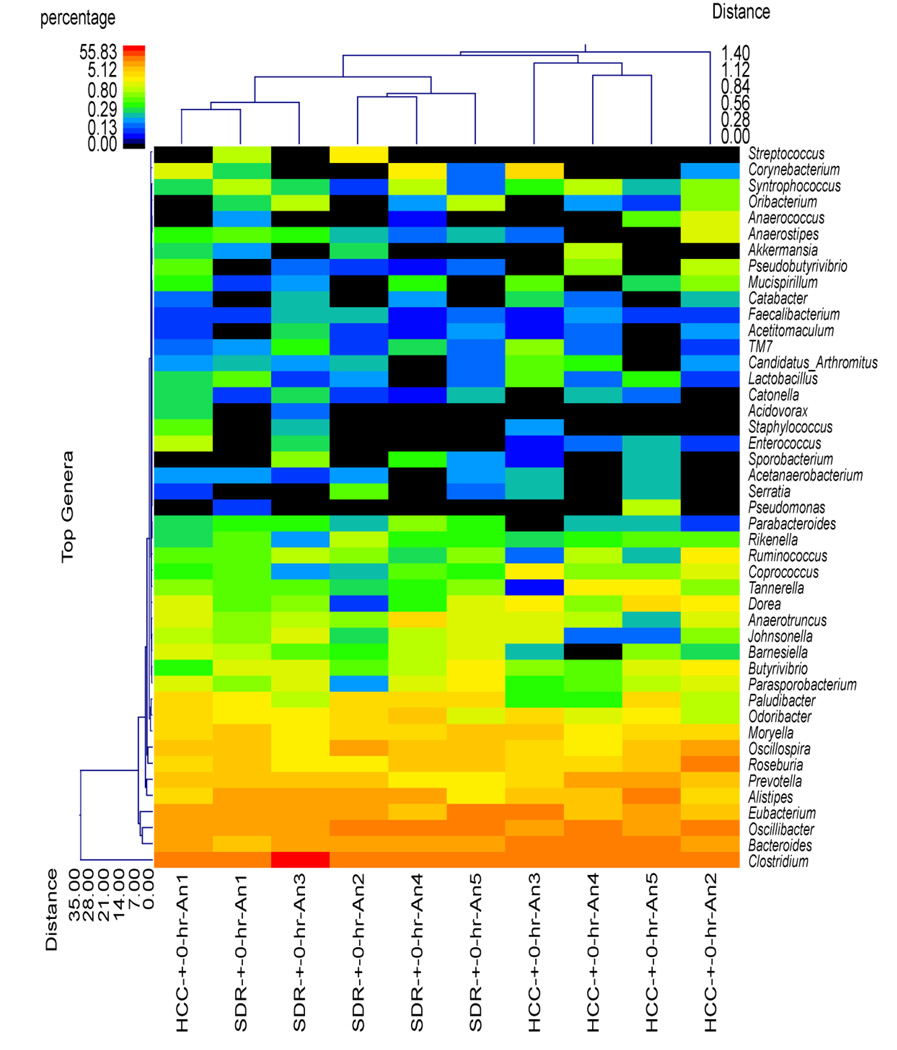
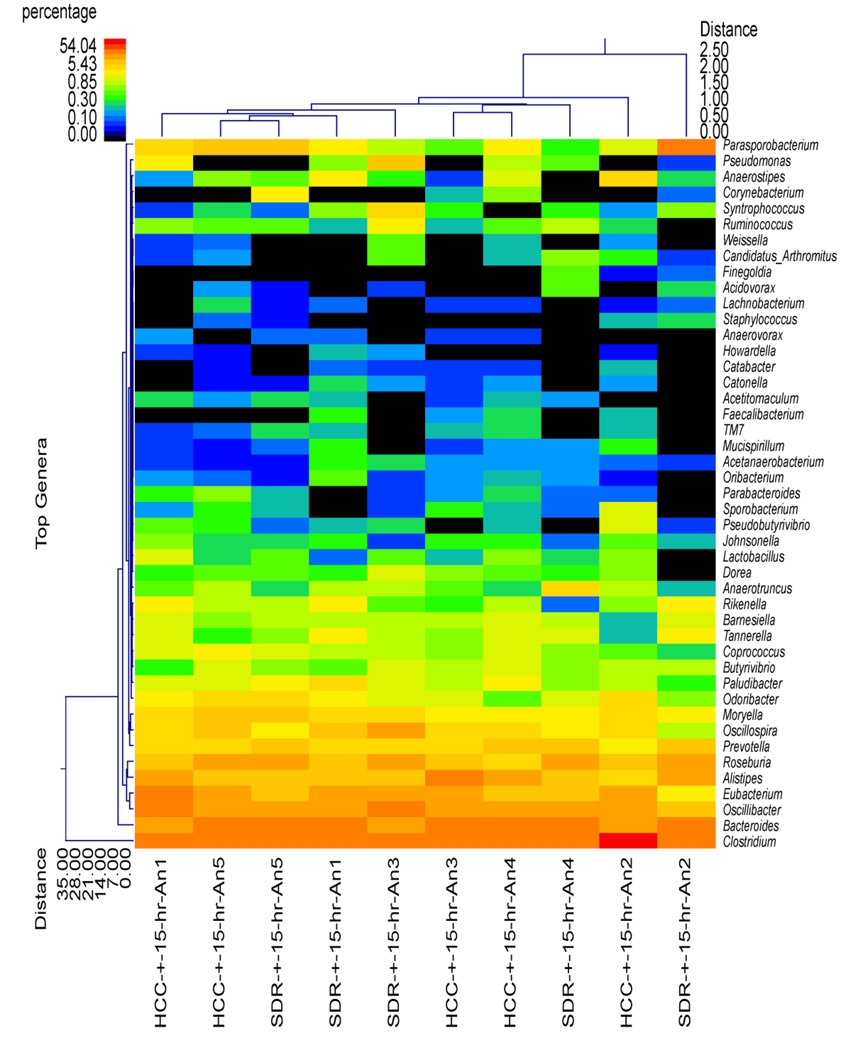
Although the SDR + 0 hr group did not significantly differ from HCC mice in estimates of diversity and richness or at the phylum level of taxonomic analysis, the use of hierarchical clustering indicated that the profile of bacterial genera in the SDR + 0 hr animals was significantly different than the profile in the HCC + 0 hr animals (Fig. 2). Overall, all of the SDR + 0 hr animals clustered together, with one of the HCC + 0 hr animals. The remaining HCC + 0 hr animals were in the same cluster, which was separate from the SDR + 0 hr animals (Fig. 2). The distinct clustering of SDR and HCC animals was not evident 15 hrs after the last cycle of SDR, where 3 of the 5 SDR + 15 hr animals clustered with 2 HCC + 15 hr animals, with the remaining 2 SDR + 15 hr animals clustering with the remaining 3 HCC + 15 hr animals (Fig. 3). The microbiome was more similar among the SDR + 0 hr animals compared to the HCC + 0 hr animals, the profiles of the SDR + 15 hr animals was not clearly distinguishable from the HCC + 15 hr animals.
Figure 2.
Cecal microbiota community structure was different in the SDR + 0 hr mice compared to the HCC + 0 hr mice. The dual hierarchal dendrogram describes the top 40 bacterial genera detected in the SDR + 0 hr and HCC + 0 hr samples. The heat map indicates the relative abundance of the given genus within each sample ID with a color legend and scale provided. The distance of the samples based upon weighted pair linkage and Manhattan distance methods with no scaling is provided at the top of the figure along with a distance score. The bacterial genus and the associated clustering are provided along the y axis, and their associated distance scores are indicated. The dual hierarchical clustering indicates that all 5 SDR + 0 hr samples cluster together, and thus have unique profiles compared to 4 of the 5 HCC + 0 hr samples that clustered together. n = 5 per group.
Figure 3.
Cecal microbiota community structure in the SDR + 15 hr and HCC + 15 hr mice. The dual hierarchal dendrogram describes the top 40 bacterial genera detected in the SDR + 15 hr and HCC + 15 hr samples. The heat map indicates the relative abundance of the given genus within each sample ID with a color legend and scale provided. The distance of the samples based upon weighted pair linkage and Manhattan distance methods with no scaling is provided at the top of the figure along with a distance score. The bacterial genus and the associated clustering are provided along the y axis, and their associated distance scores are indicated. The dual hierarchical clustering indicates that 3 of the 5 SDR + 15 hr samples clustered together with, and thus are similar to, 2 of the 5 HCC + 15 hr samples. One SDR + 15 hr sample clustered with the remaining 3 HCC + 15 hr samples, and one SDR + 15 hr sample was found to be unique from the other HCC + 15 hr and SDR + 15 hr samples. n = 5 per group
Differences in the intestinal microbiota were further evident when the relative abundance (or percentage of total sequences) of the top 30 genera were investigated. The most abundant genus was Clostridium, and there was a trend for the clostridia to be higher in the SDR mice compared to HCC mice at the + 0 hr time point (p = .07), but lower than HCC mice at the + 15 hr time point (p = .07)(Table 2). This was a mirror image of the second most abundant genus, i.e., Bacteroides, which was lower in SDR + 0 hr compared to the HCC + 0 hr mice (Table 2)(p < .05). Although the mean abundance of Bacteroides spp. in the SDR + 15 hr mice was higher than in the HCC + 15 hr mice, this difference did not reach statistical significance (Table 2). In the SDR + 15 hr mice, there was a significant increase in the abundance of bacteria in the genus Roseburia (p < .05) but a significant decrease in the abundance of bacteria in the genus Parabacteroides (p < .05) compared to the SDR + 0 hr group (Table 2). Although Roseburia was not significantly different immediately after SDR, the relative abundance of Parabacteroides was significantly increased in the SDR + 0 hr compared to the HCC + 0 hr mice (p < .05) (Table 2).
Table 2.
Top 30 Genera in the Cecal Contents
| HCC + 0 hr | SDR + 0 hr | HCC + 15 hr | SDR + 15 hr | |
|---|---|---|---|---|
| Clostridium spp. | 35.95 3.53 | 45.91 3.22S† | 44.63 2.46 | 35.37 3.91S† |
| Bacteroides spp. | 12.60 ± 2.15 | 6.79 ± 0.93* | 8.77 ± 0.80 | 14.06 ± 3.48 |
| Oscillibacter spp. | 10.16 ± 2.34 | 11.01 ± 1.96 | 7.23 ± 0.54 | 7.53 ± 0.85 |
| Eubacterium spp. | 9.24 ± 3.25 | 8.29 ± 1.25 | 6.73 ± 0.79 | 4.82 ± 1.13 |
| Alistipes spp. | 5.14 ±1.83 | 4.96 ± 0.92 | 7.05 ± 2.11 | 4.96 ± 0.32 |
| Roseburia spp. | 4.04 ± 1.80 | 2.94 ± 0.69 | 4.65 ± 0.77 | 6.08 ± 0.53** |
| Prevotella spp. | 5.01 ± 1.30 | 2.37 ± 0.41 | 2.66 ± 0.68 | 3.43 ± 0.53 |
| Oscillospira spp. | 3.30 ± 0.83 | 3.57 ± 0.85 | 2.61 ± 0.17 | 2.99 ± 0.99 |
| Moryella spp. | 2.16 ± 0.24 | 2.12 ± 0.25 | 2.26 ± 0.39 | 2.35 ± 0.42 |
| Parasporobacterium spp. | 0.58 ± 0.13 | 0.75 ± 0.23 | 1.88 ± 0.60 | 5.28 ± 3.92 |
| Odoribacter spp. | 1.34 ± 0.29 | 1.63 ± 0.27 | 1.47 ± 0.39 | 1.19 ± 0.30 |
| Paludibacter spp. | 1.00 ± 0.38 | 1.53 ± 0.23 | 0.99 ± 0.15 | 1.15 ± 0.40 |
| Anaerotruncus spp. | 0.69 ± 0.15 | 0.92 ± 0.31 | 0.48 ± 0.12 | 0.84 ± 0.44 |
| Butyrivibrio spp. | 0.64 ± 0.18 | 0.84 ± 0.18 | 0.71 ± 0.13 | 0.60 ± 0.12 |
| Tannerella spp. | 0.72 ± 0.27 | 0.29 ± 0.03 | 0.57 ± 0.19 | 0.97 ± 0.17 |
| Coprococcus spp. | 0.64 ± 0.19 | 0.22 ± 0.04† | 0.85 ± 0.21 | 0.59 ± 0.12† |
| Barnesiella spp. | 0.34 ± 0.17 | 0.53 ± 0.11 | 0.66 ± 0.17 | 0.76 ± 0.08 |
| Dorea spp. | 1.13 ± 0.22 | 0.42 ± 0.17 † | 0.39 ± 0.06 | 0.35 ± 0.15 † |
| Rikenella spp. | 0.26 ± 0.02 | 0.29 ± 0.06 | 0.65 ± 0.17 | 0.77 ± 0.25 |
| Ruminococcus spp. | 0.49 ± 0.21 | 0.41 ± 0.10 | 0.30 ± 0.06 | 0.60 ± 0.31 |
| Pseudomonas spp. | 0.10 ± 0.10 | 0.01 ± 0.01 | 0.44 ± 0.29 | 1.02 ± 0.79 |
| Anaerostipes spp. | 0.25 ± 0.17 | 0.18 ± 0.04 | 0.70 ± 0.36 | 0.44 ± 0.25 |
| Johnsonella spp. | 0.42 ± 0.18 | 0.64 ± 0.16 | 0.29 ± 0.07 | 0.13 ± 0.04 |
| Syntrophococcus spp. | 0.35 ± 0.11 | 0.32 ± 0.13 | 0.10 ± 0.04 | 0.65 ± 0.33 |
| Corneybacterium spp. | 0.63 ± 0.42 | 0.28 ± 0.21 | 0.11 ± 0.08 | 0.39 ± 0.37 |
| Lactobacillus spp. | 0.18 ± 0.06 | 0.10 ± 0.05M† | 0.47 ± 0.15 | 0.21 ± 0.08M† |
| Pseudobutyrovibrio spp. | 0.24 ± 0.10 | 0.04 ± 0.01† | 0.35 ± 0.19 | 0.07 ± 0.02† |
| Parabacteroides spp. | 0.11 ± 0.04 | 0.26 ± 0.04* | 0.22 ± 0.08 | 0.04 ± 0.02** |
| Oribacterium spp. | 0.11 ± 0.08 | 0.30 ± 0.13 | 0.08 ± 0.01 | 0.10 ± 0.05 |
| Sporobacterium spp. | 0.03 ± 0.02 | 0.17 ± 0.08 | 0.33 ± .14 | 0.04 ± 0.02 |
Data are the mean relative abundance standard error.
p < .05 vs. HCC + 0 hr;
p < .05 vs. SDR + 0 hr;
p = .07 vs HCC at the same time point;
main effect for group p < .05;
p = .08.
Stressor exposure had a more generalized effect on the relative abundance of 3 genera. The abundance of bacteria in the genus Coprococcus was lower in mice exposed to SDR as evidenced by a main effect for SDR exposure (F(1, 16) = 4.70, p < .05)(Table 2). Stressor exposure had a similar effect on the relative abundance of Dorea spp. (F(1, 16) = 5.48, p < .05) and Pseudobutyrivibrio spp. (F(1, 16) = 4.72, p < .05) (Table 2). There was a trend for SDR exposure to also reduce the relative abundance of bacteria in the genus Lactobacillus (F(1, 16) = 3.28, p = .08) (Table 2).
Impact of SDR on Circulating Cytokines and Chemokines
Exposure to SDR resulted in a significant increase in circulating cytokine levels as assessed using a multiplexed cytometric bead array. The largest increase was found in IL-6 levels, which were significantly elevated in the SDR animals, both in the + 0 hr and + 15 hr groups (as indicated by a main effect for group (F(1, 16) = 83.5, p < .01) (Fig. 4). Interestingly, changes in IL-6 levels could be predicted by 3 different genera of microbiota as indicated by a statistically significant inverse correlation between log(10) transformed IL-6 levels and the relative abundance of bacteria in the genus Coprococcus (r(18) = −.46, p < .05) (Fig. 5A) and Pseudobutyrivibrio (r(18) = −.47, p < .05) (Fig. 5B) and a marginally significant correlation with bacteria in the genus Dorea (r(18) = −.42, p = .06) (Fig. 5C).
Figure 4.
Exposure to SDR significantly increases circulating levels of IL-6 and MCP-1. A. Mice in the SDR + 0 hr and SDR + 15 hr groups had significantly more IL-6 in their plasma compared to the non-stressed HCC mice at both time points. B. Mice in the SDR + 0 hr group had significantly more MCP-1 than did mice in the HCC + 0 hr group. In all cases * indicates p < .05 vs. HCC at the same time point. n = 5 per group.
Figure 5.
Circulating cytokine levels are inversely associated with the relative abundance of cecal microbiota. Circulating IL-6 was inversely associated with the relative abundance of A. Coprococcus spp., B. Pseudobutyrovibrio spp., and C. Dorea spp. D. Circulating MCP-1 was inversely associated with Coprococcus spp. Data are from HCC + 0 hr, HCC + 15 hr, SDR + 0 hr, and SDR + 15 hr animals (n = 5 per group).
Exposure to the SDR stressor also resulted in elevated circulatory levels of MCP-1 (i.e., CCL2) (time x group interaction : F(1, 16) = 10.72, p < .05). But, here the difference was only evident in the SDR + 0 hr group (p < .05 vs. HCC + 0 hr) (Fig. 4B). There was no difference between the HCC + 15 hr and SDR + 15 hr mice (Fig. 4B), but differences in the MCP-1 levels could be predicted by an inverse correlation with the relative abundance of bacteria in the genus Coprococcus (r(18)=−.54, p < .01) (Fig. 5D). Although circulating levels of TNF-α and IFN-γ were higher in SDR mice compared to HCC mice regardless of time point of investigation as indicated by main effects for TNF-α (F(1, 16) = 5.08, p < .05) and IFN-γ (F(1, 16) = 5.54, p < .05) (Table 3), the relative abundances of bacterial genera were not predictive of these cytokine levels (data not shown). Levels of circulating IL-12 and IL-10 did not reliable exceed the assays limit of detection in either stressed or non-stressed control mice (data not shown).
Table 3.
Circulating Cytokines
| TNF-α | IFN-γ | |
|---|---|---|
| HCC + 0 hr | 9.92 1.54 | 2.21±0.24 |
| SDR + 0 hr | 28.76±17.20* | 2.76±0.33* |
| HCC + 15 hr | 9.01±0.93 | 2.34±0.40 |
| SDR + 15 hr | 16.64±4.11* | 4.55±1.23* |
Data are the mean±st. error.
indicates main effect for group, p < .05.
Impact of Antibiotic Administration on Stressor-Induced IL-6 and iNOS mRNA
The antibiotic cocktail significantly reduced the number of bacteria anaerobically cultured from the stool by approximately 2 log units (i.e., 100 fold; see Supplemental Data). To determine whether reducing microbiota density would affect the stressor-induced increases in circulating IL-6, mice were treated with the antibiotic cocktail (or with vehicle) and either exposed to the SDR stressor or left undisturbed as a non-stressed control. There was a statistically significant interaction between the group and the treatment at both the + 0 hr (F(1, 41) = 4.51, p < .05) and the + 15 hr time points (F(1, 41) = 9.96, p < .05). Post-hoc testing indicated that this was due to a statistically significant increase in IL-6 levels in the vehicle treated mice exposed to SDR at both time points (p < .05) (Fig. 6A and 6B). Post hoc testing also showed that antibiotic administration blocked the stressor-induced increase in circulating IL-6 (Fig. 6A and 6B). Circulating levels of IFN-γ, TNF-α, and MCP-1 could not be reliably detected in either vehicle or antibiotic treated, HCC or SDR exposed mice most likely due to the increased handling involved with the repeated gavages.
Figure 6.
Antibiotic treatment prevents the stressor-induced increase in circulating IL-6. A. SDR + 0 hr mice treated with vehicle had significantly higher levels of IL-6 compared to the vehicle treated HCC controls (* p < .05). Giving SDR + 0 hr mice an antibiotic cocktail prevented this stressor-induced increase in IL-6. n = 9 per group. B. SDR + 15 hr mice treated with vehicle had significantly higher levels of IL-6 compared to the vehicle treated HCC controls (* p < .05). Giving SDR + 15 hr mice an antibiotic cocktail prevented this stressor-induced increase in IL-6. n = 9 per group. C. The effects of stressor exposure on circulating IL-6 is not specific to the SDR stressor. The mean level of IL-6 in vehicle-treated mice was increased in mice exposed to a restraint stressor. Antibiotic administration prevented this increase. This increase was not quite statistically significant (p = .14) with this small sample size, n = 5 per group.
Because SDR is associated with the development of bite wounds, and because the wounding has been reported to be necessary for the immunomodulatory effects of SDR to occur (Avitsur et al., 2001; Bailey et al., 2004a), an experiment involving a restraint stressor (which does not involve wounding) was conducted to rule out the possibility that the antibiotics were simply preventing bacterial contamination of wounds. Mice were treated with antibiotics (or vehicle control) during exposure to a 2 hr restraint stressor repeated on 6 consecutive evenings (to match the timing of the SDR stressor). Treatment with antibiotics reduced the mean level of IL-6 in the circulation of restrained mice (Fig. 6C), although this difference was not quite large enough to be considered statistically significant with this smaller sample size (t(8) = 1.65, p = 0.14). The mean level of IL-6 in the circulation of restrained mice treated with antibiotics was similar to the amount found in non-stressed home cage controls (7.05 ± 3.05 pg/ml for HCC mice given antibiotics vs. 4.50 ± 1.13 pg/ml for restrained mice given antibiotics).
Exposure to SDR has been shown to prime the innate immune system in part by enhancing gene expression for inducible nitric oxide synthase (iNOS), which is necessary for the production of microbicidal reactive nitrogen intermediates (Bailey et al., 2007). In the current experiment, SDR caused a marginally significant increase in iNOS gene expression at the + 0 hr time point (F(1, 37) = 3.27, p = .07)(Fig. 7A). This increase was not evident in the antibiotic treated SDR + 0 hr mice (Fig. 7A). In contrast, iNOS mRNA was not affected in the vehicle-treated SDR + 15 hr group, but antibiotic treatment significantly decreased iNOS mRNA across groups at the 15 hr time point, as indicated by a significant main effect for antibiotic treatment (F(1, 24) = 23.55, p < .01)(Fig. 7B).
Figure 7.
Antibiotic administration affects splenic iNOS mRNA. A. The mean level of iNOS mRNA was increased in the spleen immediately after SDR († indicates p = .07). This increased mean level was not evident in the antibiotic treated mice exposed to SDR. B. Antibiotic treatment reduced iNOS mRNA in the spleen regardless of whether mice were in the SDR + 15 hr or HCC + 15 hr group. * indicates main effect for antibiotic administration, p< .05. n = 9 per group.
Discussion
The results of this study indicate that reducing the indigenous microbiota blocks the stressor-induced increase in circulating IL-6 and iNOS mRNA in the spleen. Although the mechanisms through which this occurs have not yet been systematically studied, it is likely that stressor-induced alterations of the microbiota results in translocation of bacteria and/or bacterial products across the intestinal barrier to act as a priming stimulus for the innate immune system. We previously reported that exposure to the SDR stressor enhances the translocation of intestinal, as well as cutaneous, microbiota to secondary lymphoid organs (Bailey et al., 2006). Although the number of translocating bacteria was low, 47% of the stressor-exposed mice were found to have viable bacteria in the liver (compared to only 14% in the non-stressed controls). Approximately 27% of the translocating bacteria were characterized as Gram-positive bacilli (Bailey et al., 2006). In the current study, the relative abundance of bacteria in the genus Clostridium (which are also Gram-positive bacilli) tended to be increased in the SDR + 0 hr mice. Although the increased abundance of clostridia did not result in overt symptomatology (e.g., diarrhea) an overgrowth of microbiota is one factor that can promote bacterial translocation from the gut (Berg, 1999).
The SDR stressor significantly reduced microbial diversity as calculated by OTU, ACE, and Chao1 estimators. In addition, using the relative percentage of bacteria classified at the genus level and dual hierarchical clustering methods based on Manhattan distance calculations, it was evident that the overall community structure of the microbiota is different immediately following SDR, but 15 hrs later on the morning following SDR exposure, community structure begins to reflect the profile found in the non-stressed home cage controls. This finding is consistent with other reports indicating that the microbiota are relatively resistant to prolonged alterations (Antonopoulos et al., 2009). The unique clustering of the SDR + 0 hr samples separately from the other samples is primarily due to an increase in the relative abundance of Clostridium spp., and lower abundance of Bacteroides spp.
Levels of circulating IL-6 and MCP-1 were directly related to decreases in microbiota abundances, including the abundance of Coprococcus spp., Dorea spp., and Pseudobutyrivibrio spp. These genera have only recently been described as being part of the human microbiome, and their importance to host physiology is not yet known. However, it is possible that when suppressed, they allow for a bloom in other species, such as the clostridia, that are known to induce inflammation (Brook, 2008; Libby and Bearman, 2009). In support of this, there was an inverse relationship between the relative abundance of bacteria in the genus Pseudobutyrovibrio and the genus Clostridium (Spearman ρ (8) = − .85, p < .001; data not shown). There was also a trend for Lactobacillus spp. to be decreased by the stressor, which is consistent with studies showing that stressor exposure suppresses the number of lactobacilli shed from humans and nonhuman primates (Bailey et al., 2004b; Bailey and Coe, 1999; Knowles et al., 2008). Many members of the genus Lactobacillus can suppress the host inflammatory response (Jones and Versalovic, 2009; Lin et al., 2008) and prevent bacterial translocation (Zareie et al., 2006). Thus, stressor-induced reductions in Psudobutyrivibrio and/or Lactobacillus spp. could allow other members of the microbiota, like the clostridia, to translocate and induce an inflammatory response.
The SDR stressor (Stark et al., 2002), as well as other laboratory animal stressors like tail shock (Johnson et al., 2005), is well recognized to induce increases in circulating cytokines and to increase microbicidal activity through the production of reactive nitrogen and oxygen intermediates (Bailey et al., 2007; Campisi et al., 2002; Campisi et al., 2003). Inducible nitric oxide synthase (iNOS) is important for the development of reactive nitrogen and oxygen intermediates, and daily administration of broad spectrum antibiotics significantly reduced splenic iNOS gene expression and circulating IL-6 in the stressor-exposed mice. This effect does not appear to be due to an immunosuppressive effect of the antibiotic, since the dose of antibiotic that was used is below those reported to be immunosuppressive (Fararjeh et al., 2008), and because injecting antibiotic treated and vehicle treated mice with lipopolysaccharide resulted in similar levels of circulating cytokines (namely, IL-6, MCP-1, IFN-γ, IL-12, and TNF-α) (data not shown). This suggests that baseline levels of microbiota are necessary for stressor-induced increases in inflammatory mediators, and is consistent with other studies that have shown that the microbiota provide a constitutive priming signal to innate immune cells through activation of pattern recognition receptors, particularly the nucleotide-oligomerization domain-containing protein-1 (Nod1) and Nod2 receptors (Clarke et al., 2010; Kim et al., 2009). In addition, others have reported that stressor-induced increases in circulating microbial antigen-associated heat shock protein 72 (HSP72) potentiates the innate immune response and enhances iNOS expression (Fleshner et al., 2007). Our data further emphasize the influence that the microbiota have on the immune system, and indicate that when altered during the stress response, the microbiota influence stressor-induced immunomodulation.
The mechanisms by which the stress response can suppress immune functioning are well understood, and are known to involve glucocorticoid mediated suppression of transcription factors, such as NF-κB (Padgett and Glaser, 2003). However, the mechanisms by which the stress response enhances immune activity are not yet fully elucidated. Recent studies indicate that the sympathetic nervous system (SNS) is involved in stressor-induced immune enhancement (Bierhaus et al., 2003; Cole et al., 2010), and blocking the activation of the SNS through the use of the α- and β-adrenergic receptor antagonists has been reported to block the stressor-induced increases in innate immunity (Hanke et al., 2008; Johnson et al., 2005). In addition to direct effects on the immune system, the SNS is also involved in the bidirectional communication between the gut and the brain that facilitates the regulation of gut function. Stressor-induced SNS activity significantly impacts gut secretion and motility (Lomax et al., 2010), which can in turn impact the stability of the microbiota. In addition, SNS-derived catecholamines, particularly norepinephrine (NE) have the capacity to stimulate the growth of many enteric bacteria (Freestone et al., 2000; Lyte et al., 1997; Lyte, 2004), including Clostridium spp. (Lyte, 2004) which were found to be increased in the SDR-exposed mice. Thus, it is possible that stressor-induced SNS activity directly and/or indirectly affects the microbiota and results in increased reactivity of the innate immune system.
It is well recognized that stressor exposure is associated with elevated circulating inflammatory cytokines. Studies in humans demonstrate that both prolonged natural stressors, as well as acute laboratory stressors result in increased circulating cytokines such as IL-6 and TNF-α (reviewed in (Steptoe et al., 2007). Whether stressor-induced cytokines in humans are related to stressor-induced effects on the microbiota is not known, but studies have indicated that humans experiencing stressful situations have altered profiles of intestinal microbiota (Holdeman et al., 1976; Knowles et al., 2008). And, bacterial translocation, as assessed by an increased occurrence of circulating antibodies to microbiota, has been linked to mood disorders, such as depression, presumaby through a cytokine-mediated mechanism (Maes, 2008; Maes et al., 2008). This is consistent with findings in patients with functional and inflammatory bowel disease that have frequent psychiatric co-morbidity and alterations in the intestinal microbiota (Collins and Bercik, 2009). Our studies indicate the microbiota are primarily and interactively involved in stressor-induced immunoenhancement and contribute to the growing literature on the impact that the microbiota have on the health of the host.
Research Highlights
Exposure to the social stressor, called social disruption, significantly changed the community structure of the intestinal microbiota.
Stressor-induced increases in circulating IL-6 and MCP-1 were significantly correlated with stressor-induced changes in 3 members of the microbiota, Dorea spp., Coprococcus spp., and Pseudobutyrivibrio spp.
Administration of a broad spectrum antibiotic cocktail to reduce the microbiota prevented the stressor-induced increase in IL-6 and splenic inducible nitric oxide synthase gene expression.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Mark Hanke for assistance with CBA data analysis. This work was supported by NIH RO3AI069097-01A1 and Ohio State University start up funds to MB, and by Texas Tech University Health Sciences internal grants to ML.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Acosta-Martinez V, Dowd S, un Y, llen V. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biology and Biochemistry. 2008;40:2762–2770. [Google Scholar]
- Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 2008;105 Suppl 1:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Brown RF, Berg RD, Dunn AJ. Bacterial translocation can increase plasma corticosterone and brain catecholamine and indoleamine metabolism. Am. J. Physiol Regul. Integr. Comp Physiol. 2000;279:R2164–R2172. doi: 10.1152/ajpregu.2000.279.6.R2164. [DOI] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitsur R, Stark JL, Sheridan JF. Social stress induces glucocorticoid resistance in subordinate animals. Horm. Behav. 2001;39:247–257. doi: 10.1006/hbeh.2001.1653. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Avitsur R, Engler H, Padgett DA, Sheridan JF. Physical defeat reduces the sensitivity of murine splenocytes to the suppressive effects of corticosterone. Brain Behav. Immun. 2004a;18:416–424. doi: 10.1016/j.bbi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev. Psychobiol. 1999;35:146–155. [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Parry NM, Galley JD, Schauer DB, Lyte M. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 2010;78:1509–1519. doi: 10.1128/IAI.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Engler H, Powell ND, Padgett DA, Sheridan JF. Repeated social defeat increases the bactericidal activity of splenic macrophages through a Toll-like receptor-dependent pathway. Am. J. Physiol Regul. Integr. Comp Physiol. 2007;293:R1180–R1190. doi: 10.1152/ajpregu.00307.2007. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Engler H, Sheridan JF. Stress induces the translocation of cutaneous and gastrointestinal microflora to secondary lymphoid organs of C57BL/6 mice. J. Neuroimmunol. 2006;171:29–37. doi: 10.1016/j.jneuroim.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Kinsey SG, Padgett DA, Sheridan JF, Leblebicioglu B. Social stress enhances IL-1beta and TNF-alpha production by Porphyromonas gingivalis lipopolysaccharide-stimulated CD11b+ cells. Physiol Behav. 2009;98:351–358. doi: 10.1016/j.physbeh.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J. Pediatr. Gastroenterol. Nutr. 2004b;38:414–421. doi: 10.1097/00005176-200404000-00009. [DOI] [PubMed] [Google Scholar]
- Berg RD. Bacterial translocation from the gastrointestinal tract. Adv. Exp Med Biol. 1999;473:11–30. doi: 10.1007/978-1-4615-4143-1_2. [DOI] [PubMed] [Google Scholar]
- Bierhaus A, Wolf J, Andrassy M, Rohleder N, Humpert PM, Petrov D, Ferstl R, von EM, Wendt T, Rudofsky G, Joswig M, Morcos M, Schwaninger M, McEwen B, Kirschbaum C, Nawroth PP. A mechanism converting psychosocial stress into mononuclear cell activation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1920–1925. doi: 10.1073/pnas.0438019100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I. Microbiology and management of abdominal infections. Dig. Dis. Sci. 2008;53:2585–2591. doi: 10.1007/s10620-007-0194-6. [DOI] [PubMed] [Google Scholar]
- Brydon L, Edwards S, Jia H, Mohamed-Ali V, Zachary I, Martin JF, Steptoe A. Psychological stress activates interleukin-1beta gene expression in human mononuclear cells. Brain Behav. Immun. 2005;19:540–546. doi: 10.1016/j.bbi.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Brydon L, Edwards S, Mohamed-Ali V, Steptoe A. Socioeconomic status and stress-induced increases in interleukin-6. Brain Behav. Immun. 2004;18:281–290. doi: 10.1016/j.bbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Brydon L, Steptoe A. Stress-induced increases in interleukin-6 and fibrinogen predict ambulatory blood pressure at 3-year follow-up. J. Hypertens. 2005;23:1001–1007. doi: 10.1097/01.hjh.0000166841.57474.d0. [DOI] [PubMed] [Google Scholar]
- Campisi J, Leem TH, Fleshner M. Acute stress decreases inflammation at the site of infection. A role for nitric oxide. Physiol Behav. 2002;77:291–299. doi: 10.1016/s0031-9384(02)00861-2. [DOI] [PubMed] [Google Scholar]
- Campisi J, Leem TH, Fleshner M. Stress-induced extracellular Hsp72 is a functionally significant danger signal to the immune system. Cell Stress. Chaperones. 2003;8:272–286. doi: 10.1379/1466-1268(2003)008<0272:sehiaf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Arevalo JM, Takahashi R, Sloan EK, Lutgendorf SK, Sood AK, Sheridan JF, Seeman TE. Computational identification of gene-social environment interaction at the human IL6 locus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SM, Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology. 2009;136:2003–2014. doi: 10.1053/j.gastro.2009.01.075. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS. Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC. Microbiol. 2008a;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS. ONE. 2008b;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR. Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST) BMC. Bioinformatics. 2005;6:93. doi: 10.1186/1471-2105-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KM, Burns VE, Ring C, Carroll D. Sex differences in the interleukin-6 response to acute psychological stress. Biol. Psychol. 2006;71:236–239. doi: 10.1016/j.biopsycho.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Fararjeh M, Mohammad MK, Bustanji Y, Alkhatib H, Abdalla S. Evaluation of immunosuppression induced by metronidazole in Balb/c mice and human peripheral blood lymphocytes. Int. Immunopharmacol. 2008;8:341–350. doi: 10.1016/j.intimp.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Fleshner M, Johnson JD, Friedman J. Extracellular Hsp72: A double-edged sword for host defense. In: Asea AA, De Maio A, editors. Heat Shock Proteins: Potent Mediators of Inflammation and Immunity. 1 ed. New York: Springer; 2007. pp. 235–263. [Google Scholar]
- Freestone PP, Lyte M, Neal CP, Maggs AF, Haigh RD, Williams PH. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 2000;182:6091–6098. doi: 10.1128/jb.182.21.6091-6098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontcharova V, Youn E, olcott RD, ollister EB, entry TJ, owd SE. Black box chimera check (B2C2):a Windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. The Open Microbiology Journal. 2010;4:47–52. doi: 10.2174/1874285801004010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke ML, Bailey MT, Powell ND, Stiner L, Sheridan JF. Beta-2 adrenergic blockade decreases the immunomodulatory effects of Social Disruption Stress (SDR) Brain, Behavior, and Immunity. 2008;22:39–40. doi: 10.1016/j.bbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway H, Shipley M, Mullen MJ, Kumari M, Brunner E, Taylor M, Donald AE, Deanfield JE, Marmot M. Social and psychosocial influences on inflammatory markers and vascular function in civil servants (the Whitehall II study) Am. J. Cardiol. 2003;92:984–987. doi: 10.1016/s0002-9149(03)00985-8. [DOI] [PubMed] [Google Scholar]
- Holdeman LV, Good IJ, Moore WE. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 1976;31:359–375. doi: 10.1128/aem.31.3.359-375.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB. The microbiota and allergies/asthma. PLoS. Pathog. 2010;6:e1000549. doi: 10.1371/journal.ppat.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JD, Campisi J, Sharkey CM, Kennedy SL, Nickerson M, Greenwood BN, Fleshner M. Catecholamines mediate stress-induced increases in peripheral and central inflammatory cytokines. Neuroscience. 2005;135:1295–1307. doi: 10.1016/j.neuroscience.2005.06.090. [DOI] [PubMed] [Google Scholar]
- Jones SE, Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC. Microbiol. 2009;9:35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Preacher KJ, MacCallum RC, Atkinson C, Malarkey WB, Glaser R. Chronic stress and age-related increases in the proinflammatory cytokine IL-6. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9090–9095. doi: 10.1073/pnas.1531903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OY, Monsel A, Bertrand M, Cavaillon JM, Coriat P, dib-Conquy M. Translocation of bacterial NOD2 agonist and its link with inflammation. Crit Care. 2009;13:R124. doi: 10.1186/cc7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles SR, Nelson EA, Palombo EA. Investigating the role of perceived stress on bacterial flora activity and salivary cortisol secretion: a possible mechanism underlying susceptibility to illness. Biol. Psychol. 2008;77:132–137. doi: 10.1016/j.biopsycho.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby DB, Bearman G. Bacteremia due to Clostridium difficile--review of the literature. Int. J. Infect. Dis. 2009;13:e305–e309. doi: 10.1016/j.ijid.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm. Bowel. Dis. 2008;14:1068–1083. doi: 10.1002/ibd.20448. [DOI] [PubMed] [Google Scholar]
- Lizko NN. Stress and intestinal microflora. Nahrung. 1987;31:443–447. doi: 10.1002/food.19870310530. [DOI] [PubMed] [Google Scholar]
- Lomax AE, Sharkey KA, Furness JB. The participation of the sympathetic innervation of the gastrointestinal tract in disease states. Neurogastroenterol. Motil. 2010;22:7–18. doi: 10.1111/j.1365-2982.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 2004;12:14–20. doi: 10.1016/j.tim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Lyte M, Arulanandam B, Nguyen K, Frank C, Erickson A, Francis D. Norepinephrine induced growth and expression of virulence associated factors in enterotoxigenic and enterohemorrhagic strains of Escherichia coli. Adv. Exp. Med. Biol. 1997;412:331–339. doi: 10.1007/978-1-4899-1828-4_54. [DOI] [PubMed] [Google Scholar]
- Lyte M, Bailey MT. Neuroendocrine-bacterial interactions in a neurotoxin-induced model of trauma. J. Surg. Res. 1997;70:195–201. doi: 10.1006/jsre.1997.5130. [DOI] [PubMed] [Google Scholar]
- Lyte M, Nelson SG, Thompson ML. Innate and adaptive immune responses in a social conflict paradigm. Clin. Immunol. Immunopathol. 1990;57:137–147. doi: 10.1016/0090-1229(90)90029-p. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann. N. Y. Acad. Sci. 2004;1029:36–43. doi: 10.1196/annals.1309.005. [DOI] [PubMed] [Google Scholar]
- Maes M. The cytokine hypothesis of depression: inflammation, oxidative & nitrosative stress (IO&NS) and leaky gut as new targets for adjunctive treatments in depression. Neuro. Endocrinol. Lett. 2008;29:287–291. [PubMed] [Google Scholar]
- Maes M, Kubera M, Leunis JC. The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuro. Endocrinol. Lett. 2008;29:117–124. [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- Padgett DA, Glaser R. How stress influences the immune response. Trends Immunol. 2003;24:444–448. doi: 10.1016/s1471-4906(03)00173-x. [DOI] [PubMed] [Google Scholar]
- Powell ND, Bailey MT, Mays JW, Stiner-Jones LM, Hanke ML, Padgett DA, Sheridan JF. Repeated social defeat activates dendritic cells and enhances Toll-like receptor dependent cytokine secretion. Brain Behav. Immun. 2009;23:225–231. doi: 10.1016/j.bbi.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Stark JL, Avitsur R, Hunzeker J, Padgett DA, Sheridan JF. Interleukin-6 and the development of social disruption-induced glucocorticoid resistance. J. Neuroimmunol. 2002;124:9–15. doi: 10.1016/s0165-5728(02)00004-8. [DOI] [PubMed] [Google Scholar]
- Steptoe A, Hamer M, Chida Y. The effects of acute psychological stress on circulating inflammatory factors in humans: a review and meta-analysis. Brain Behav. Immun. 2007;21:901–912. doi: 10.1016/j.bbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Tannock GW, Savage DC. Influences of dietary and environmental stress on microbial populations in the murine gastrointestinal tract. Infect. Immun. 1974;9:591–598. doi: 10.1128/iai.9.3.591-598.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- von KR, Kudielka BM, Preckel D, Hanebuth D, Fischer JE. Delayed response and lack of habituation in plasma interleukin-6 to acute mental stress in men. Brain Behav. Immun. 2006;20:40–48. doi: 10.1016/j.bbi.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Zareie M, Johnson-Henry K, Jury J, Yang PC, Ngan BY, McKay DM, Soderholm JD, Perdue MH, Sherman PM. Probiotics prevent bacterial translocation and improve intestinal barrier function in rats following chronic psychological stress. Gut. 2006;55:1553–1560. doi: 10.1136/gut.2005.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.