Abstract
Previous work by us and others reported decreased expression of miR-199a-3p in hepatocellular carcinoma (HCC) tissues compared to adjacent benign tissue. We report here a significant reduction of miR-199a-3p expression in 7 HCC cell lines. To determine if miR-199a-3p has a tumor suppressive role, pre-miR-199a-3p oligonucleotides were transfected into the HCC cell lines. Pre-miR-199a-3p oligonucleotide reduced cell proliferation by approximately 60% compared to control oligonucleotide in only two cell lines (SNU449 and SNU423); the proliferation of the other 5 treated cell lines was similar to control oligonucleotide. A pre-miR-199a-3p oligonucleotide formulated with chemical modifications to enhance stability while preserving processing, reduced cell proliferation in SNU449 and SNU423 to the same extent as the commercially available pre-miR-199a-3p oligonucleotide. Furthermore, only the duplex miR-199a-3p oligonucleotide, and not the guide strand alone, was effective at reducing cell viability. Since a CD44 variant was essential for c-Met signaling (Orian-Rousseau, et al., Genes Dev. 2002) and c-Met is a known miR-199a-3p target, we hypothesized that miR-199a-3p may also target CD44. Immunoblotting confirmed that only the two HCC lines that were sensitive to the effects of pre-miR-199a-3p were CD44+. Direct targeting of CD44 by miR-199a-3p was confirmed using luciferase reporter assays and immunoblotting. Transfection of miR-199a-3p into SNU449 cells reduced in vitro invasion and sensitized the cells to doxorubicin; both effects were enhanced when HA was added to the cell cultures. An inverse correlation between the expression of miR-199a-3p and CD44 protein was noted in primary HCC specimens. The ability of miR-199a-3p to selectively kill CD44+ HCC may be a useful targeted therapy for CD44+ HCC.
Keywords: HCC, cancer, miR-199a*, microRNA mimetic, liver
Introduction
microRNAs (miRNA) are differentially expressed in essentially all solid tumors including hepatocellular carcinoma (HCC)1 [1]. miRNAs with altered expression in HCC include those that are upregulated (miR-221/-222 [2], miR-21 [3]) and downregulated (miR-122a [4], miR-199a-3p (miR-199a*) [5]) in the tumor. miR-199a-3p is of interest to us because it was reduced in several HCC studies [6; 7; 8; 9] and it was shown to target c-Met [10]. c-Met is an important oncogene involved in HCC invasion and metastasis [11]. Restoration of cellular miR-199a-3p levels decreased invasion and proliferation in HCC cell lines [12]. miR-199a-3p is also reduced in alcohol induced steatohepatitis, a known precursor to HCC [13]. miR-199a-3p regulates hepatitis C [12] and hepatitis B viral replication [14], both predisposing factors to HCC.
Often miRNAs with reduced expression in the tumor function as tumor suppressors. Reintroduction of the miRNA to the tumor causes a general reduction in the malignant phenotype (i.e. reduced proliferation, colony formation, invasion and tumor formation in nude mice). Examples include miR-122a in HCC [4] and miR-1/-216 in rhabdomyosarcoma [15]. The expression of miR-199a-3p and miR-199a-5p is enriched in liver tissue and is abundantly expressed in epithelial as well as non epithelial tissues [16]. We report here that miR-199a-3p targets the hyaluronic acid (HA) receptor CD44. Introduction of the miR-199a-3p mimetic reduces the proliferation of HCC cell lines, however only if they express CD44. Our data suggest a potential role for miR-199a-3p targeted therapy of CD44+ HCC.
Methods
Cell Lines
The human HCC cell lines, Huh7, HepG2, SNU182, PLC/PRF/5, Hep3B, SNU423, and SNU449 were purchased from the American Type Tissue Collection (Manassas, VA). All cells were cultured using standard conditions with Huh7, HepG2, and Hep3B cultured in Minimum Essential Media (Invitrogen, Carlsbad, CA) and SNU182, SNU423, and SNU449 cultured in RPMI 1640 media (Invitrogen). Primary hepatocytes (HH-2) were purchased from ScienCell, Carlsbad, CA; short term cultures of these cells were performed using standard conditions as recommended by the supplier.
RNA, protein isolation and tissue procurement
Nine pairs of HCC tumors and adjacent benign liver were available for the study (Stable 1). Tumor samples with matched adjacent benign tissue were collected during surgical resections at the Mayo Clinic, frozen in liquid nitrogen, and stored at -80°C. Following pulverization in a cold mortar and pestle, total RNA was isolated from the tissues using Trizol reagent (Invitrogen).
miR-199a-3p Oligonucleotides for Introduction in Cells
pre-miR-199a-3p mimetic and control oligonucleotide (negative control #1) were purchased from Ambion (Austin, TX). Oligonucleotides representing the guide and passenger strands for miR-199a-3p were synthesized from IDT (Coralville, IA) using the following chemical modifications to enhance stability and assure proper processing. 5’-x-ACCAAUGUGCAGACUACUGt (passenger strand) and 5’ pACAGUAGUCUGCACAUUGGUUA 3’ (guide strand) where p represents phosphate, x SpC3; capital letters RNA; small letters DNA and bold underlined 2’ OMe RNA. Oligos were HPLC purified by the manufacturer. To prepare the passenger strand-guide strand duplex, the equivalent amounts of oligo were combined in water, heated to 56° C for 5 mins, 37° C for 10 min, then cooled to room temperature and placed on ice.
Oligonucleotide transfection and cell proliferation assay
Cells were plated on a 96 well plate at 2,000 cells per well one day before transfection. Oligonucleotides were transfected using Lipofectamine 2000 and Opti-MEM medium (Invitrogen) following the manufacturer’s protocol. The cell proliferation assay was performed using the reagent WST-1 (Roche, Indianapolis, IN). Ninety-six h after transfection, 20 μL of WST-1 was added to the cell culture medium and incubated for 2 h. Sample absorbance was analyzed at 450 nm. All experiments were performed at least in triplicate.
Protein extraction and immunoblotting
Protein was harvested using CelLytic™ M (Sigma) and 1x protease and phosphatase inhibitor (Pierce) using standard techniques. Protein concentration was measured using the BCA Protein Assay Kit (Pierce). Thirty micrograms of total protein extract was separated on a 10% SDS-PAGE gel. Blotting was performed for c-Met (Cell Signaling, Boston, MA) and CD44 (Cell Signaling, 3578). β-actin (Abcam, Cambridge, MA) was used as a loading control. Secondary horseradish peroxidase antibody was detected using ECL Western Blotting Analysis System (Amersham Biosciences, Piscataway, NJ).
qPCR
One microgram of total RNA was used to synthesize cDNA using random primers. cDNA was analyzed for gene expression using gene specific primers (IDT) and the SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA). For the miRNA, 100 ng of total RNA was assayed using the TaqMan miRNA Assays (Applied Biosystems). Data were normalized to 18S rRNA and presented using the comparative CT method. Data were multiplied by 106 to simplify presentation.
Matrigel invasion assay
The in vitro invasion assay was performed using 6.5-mm Transwell chambers (Costar, Cambridge, MA) with porous filters (pore size 8 μm). Matrigel™ basement membrane matrix (BD Biosciences, San Jose, CA) was diluted in cold serum free cell culture media (200 μg/ml) and then 100 μl was added to the filters, and allowed to dry overnight. SNU182, SNU423 or SNU449 cells (50,000 cells/well) were then seeded onto the membrane of the upper chamber in serum free RPMI1640. The lower chamber was filled with 600 μl of NIH-3T3 conditioned media as a chemoattractant. To assess the invasive potential of the HCC cells upon HA, matrigel was prepared with or without 2 mg/ml ultra low or high molecular weight hylauronic acid (R&D Systems, Minneapolis, MN). Twenty four h after incubation, cells were fixed, stained with crystal violet and the image density (mean of 4 visual fields, 20) was captured using Image J software (NIH, Bethesda, MD).
Luciferase Analysis
The full length CD44 3’ UTR was cloned into the psiCHECK-2 Vector (Promega, Madison, WI). SNU449 cells were plated at a density of 100,000 cells/well (24 well) and then incubated for 24 h prior to transfection with either the modified pre-miR-199a-3p miRNA precursor or control oligonucleotide (50 and 100 nM). Luciferase expression was analyzed using the Dual-Luciferase Reporter Assay System (Promega) per the manufacturer’s protocol.
Chemosensitivity
SNU449 cells (1,500 cells/well) were plated onto a 96-well plate, attached overnight and then transfected with the pre-miR-199a-3p or the control oligomer (Ambion). Following a 48 h incubation, cells were exposed to 0, 10, 50 μM of doxorubicin for 48 h. Twenty microliters of WST-1 reagent (Roche) was added per well, the plates were incubated for 2 hs; sample absorbance was analyzed at 450 nm.
Results
Effect of miR-199a-3p mimetic on viability of HCC cell lines
As miR-199a-3p was reduced in HCC tissues [6; 7; 8; 9] and introduction of certain miRNAs to cancer cell lines cause a general reversal of the malignant phenotype, we wanted to study the effect that pre-miR-199a-3p has on the viability of HCC cell lines. qPCR was used to measure the expression of miR-199a-3p in 7 HCC cell lines (SNU449, SNU423, SNU182, HepG2, Hep38, Huh7 and PLC/PRF/5) along with primary hepatocytes (HH-2). The expression of miR-199a-3p was significantly reduced in all 7 of the HCC cell lines compared to the primary hepatocytes (SFig. 1). miR-199a-3p mimetic or control oligonucleotides were transfected into the 7 HCC cell lines. After 96 h, only two of the cell lines demonstrated a reduction in proliferation; SNU423 and SNU449. The proliferation of SNU423 was reduced by 62.5% with miR-199a-3p mimetic (p<0.001, Fig. 1A). SNU449 cell proliferation was also significantly reduced by 56.8% with miR-199a-3p mimetic (p<0.001). The other 5 cell lines showed no change in proliferation from the miR-199a-3p compared to control (Fig. 1A).
Figure 1. miR-199a-3p oligonucleotide reduces cell proliferation in CD44 positive HCC cells.
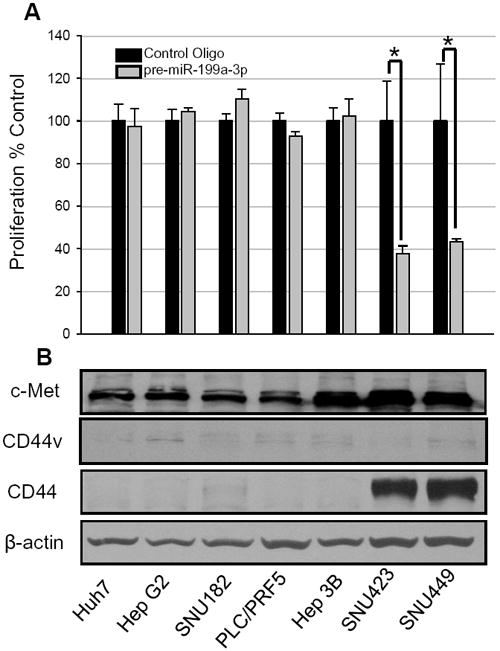
(A) HCC cells were transfected with 100 nM of pre-miR-199a-3p or control oligonucleotides. Following a 96 h exposure, cell proliferation was measured using a WST1 assay. (B) Protein expression in HCC cell line. The expression of c-Met, CD44v, CD44 and β-actin protein was determined by immunoblotting; * p<0.001.
Before proceeding, we wanted to ask an important question that has not been addressed in the literature to our knowledge. It concerns the biological activity of duplex pre-miRNA oligonucleotides compared to the guide strand alone. Ambion Pre-miR™ oligonucleotides are duplex RNA oligonucleotides that use proprietary chemical modifications to enhance stability while preserving passenger strand degradation by miRISC. We had the miR-199a-3p guide and passenger strands synthesized using the chemical modifications described in Materials and Methods. The duplex miR-199a-3p oligonucleotides synthesized with this chemistry were as effective at reducing cell viability as the Ambion pre-miR-199a-3p (SFig. 2A). We then compared the activity (i.e. ability to reduce cell proliferation) of the duplex miR-199a-3p oligonucleotide compared to the guide or passenger strands alone. The duplex miR-199a-3p reduced proliferation by 42% compared to duplex control oligonucleotides (SFig. 2B). Unexpectedly, the cells transfected with either strand alone showed a slight increase in proliferation (SFig. 2B). We conclude that only the duplex miR-199a-3p mimetic is active at reducing cell proliferation.
We were puzzled as to why miR-199a-3p reduced proliferation in only some of the HCC cell lines (Fig. 1A). In our search, we came upon the work of Orian-Rosseau, et al., who demonstrated that a splice variant of CD44 (i.e. CD44v) is a necessary component of c-MET signaling [17]. Since miR-199a-3p had been previously shown to target c-met [18], we wondered if it is possible that miR-199a-3p could target CD44. Examination of the program TargetScan did indeed show that CD44 is a predicted target of miR-199a-3p. Therefore, we measured CD44 protein levels in the cell lines to see if there was a correlation between CD44 protein expression and reduction in cell proliferation due to miR-199a-3p. CD44 was detectable only in SNU423 and SNU449; precisely those cell lines with reduced viability following transfection of pre-miR-199a-3p (Fig. 1B). c-Met protein was present in all 7 cell lines, with slightly higher levels being present in Hep3B, SNU423 and SNU449. CD44v was absent in all cell lines.
CD44 mRNA is a target of miR-199a-3p
Additional experiments were performed to establish that CD44 is a target of miR-199a-3p. A putative miR-199a-3p binding site was located in the 3’UTR of CD44 (Fig. 2A). Luciferase reporter assays were conducted to establish the interaction between miR-199a-3p and the 3’UTR of CD44. Both the low (50 nM) and high (100 nM) concentrations of miR199a-3p mimetic reduced the luciferase expression by greater than 40% (p < 0.05, Fig. 2B). Three cell lines (one that expressed CD44 and two that did not) were transfected with pre-miR-199a-3p or control oligo. In the CD44+ SNU449 cell line, there was a 77% decrease in CD44 protein due to pre-miR-199a-3p compared to control oligonucleotide (Fig. 2C). The luciferase data and the reduction in CD44 protein indicate that CD44 is a target of miR-199a-3p.
Figure 2. CD44 is a target of miR-199a-3p.

(A) Schematic representing the location and conservation of the putative miR-199a-3p binding site within the 3’UTR of CD44 mRNA. (B) Luciferase reporter plasmids containing the wild-type CD44 binding sequences were transiently transfected in SNU449 cells with either pre-miRNA control or miR-199a-3p at 50 nM and 100 nM. Luciferase expression was measure at 48 h after transfection as described in Materials and Methods. The data are the mean ± s. d. of at least 3 independent transfections; * p<0.05 (C) SNU182, SNU449, and HepG2 cells were transfected with pre-miRNA control oligonucleotide (n) or pre-miR-199a-3p at 50 nM for 48 h. CD44 and β-actin protein levels was determined by immunoblotting.
Ability of miR199a-3p mimetic to sensitive the effects of doxorubicin
Pre-miR199a-3p was combined with doxorubicin to determine if the miRNA mimetic can sensitize the effects of a traditional anti-cancer agent. Control and pre-miR-199a-3p oligonucleotides were transfected into SNU449 cells. As demonstrated in our previous proliferation experiments, SNU449 cells transfected with pre-miR-199a-3p alone showed a 42.4% reduction in proliferation (SFig. 3). Doxorubicin alone reduced cell proliferation by 68.6% and 71.2% at 10 and 50 nM, respectively. Combining miR-199a-3p plus doxorubicin reduced the cell proliferation by 80.5% and 82.5% at the 10 and 50 nM concentrations, respectively (SFig. 3). These data demonstrate the ability of miR-199a-3p mimetic to sensitize the HCC cells to doxorubicin.
Reduction of invasion of a CD44-positive HCC cell line by miR-199a-3p
The ability of pre-miR-199a-3p to decrease the invasiveness of HCC cells was studied. HA, a component of the extracellular matrix, is a ligand for the CD44 receptor. We hypothesized that HA would enhance the in vitro invasiveness of CD44+ HCC cells because Lara-Pezzi et al., demonstrated that HA increased cell migration in HCC by binding to CD44 [19]. SNU182 (CD44-) and SNU449 (CD44+) cells were plated with and without high-molecular weight HA. The CD44- SNU182 cells did not invade through the matrigel (Fig. 3A) and invasiveness was not enhanced by HA. The HA increased the invasiveness of the CD44+ SNU449 cells by 32.8%. SNU449 cells plated with HA and were transfected with either the miR-199a-3p mimetic or control oligo. The miR-199a-3p mimetic decreased invasiveness of the HCC cell line by 84.6% (Fig. 3B-C).
Figure 3. miR-199a-3p reduces in vitro invasion of CD44+ HCC cell lines.

(A) Untreated CD44- SNU182 cells did not invade through the matrigel coated chamber. CD44+ SNU449 cells exposed to high-molecular weight HA were transfected with control (B) or miR-199a-3p mimetic (C) 48 h prior to migration through the chamber.
miR199a-3p and CD44 levels in primary HCC and paired benign tissues
To extrapolate our findings to HCC patients, we evaluated the miR-199a-3p and CD44 (mRNA and protein) levels in 9 pairs of HCC and adjacent benign tissues. Seven of the nine pairs had reduced levels of miR-199a-3p in the tumor compared to the unaffected benign tissue (Fig. 4A). The reduction in miR-199a-3p levels in the tumor ranged from 4 to 30-fold. CD44 protein levels from the immunoblot inversely correlated with the miR-199a-3p RNA levels (Fig. 4B,C). Examination of the CD44 mRNA by qPCR showed that the CD44 mRNA positively correlated with the CD44 protein, suggesting that binding of miR-199a-3p to the CD44 3’ UTR suppresses mRNA levels (SFig. 4).
Figure 4. miR-199a-3p and CD44 protein expression in HCC tissues.
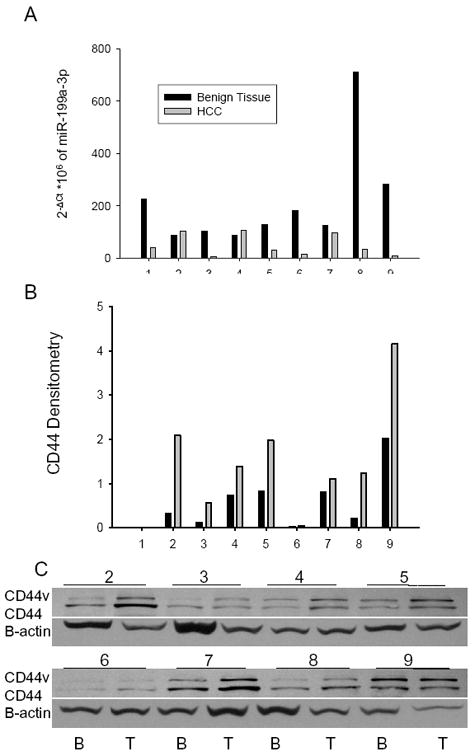
(A) miR-199a-3p levels in the 9 paired HCC and benign tissue specimens were determined by qPCR. Data are presented relative to 18S rRNA. CD44 protein expression in 8 pairs of HCC and benign tissue is presented from the immunoblot (C) and the densitometry values of the immunoblot (B).
Discussion
This study was undertaken to more completely comprehend the relationship between miR-199a-3p and HCC. It was puzzling when we showed a very substantial reduction in cell proliferation following introduction of miR-199a-3p mimetic in some HCC cell lines but not others (Fig. 1A). We showed that miR-199a-3p specifically targets CD44 following binding to conserved sequence within the CD44 3’ UTR (Fig. 2) and only those cell lines that express CD44 protein were sensitive to the antiproliferative effects of miR-199a-3p mimetic (Fig. 1). This effect is more clear cut for CD44 compared to c-met, since the Hep3B, SNU423 and SNU449 cells express similar levels of c-met protein, yet Hep3B was not sensitive to the miR-199a-3p oligonucleotide (Fig. 1).
Of the two cell lines studied, only the CD44+ cells were invasive (Fig. 3). miR-199a-3p mimetic reduced the in vitro invasion of the CD44+ SNU449 cells by 85% (Fig. 3). The inverse relationship between miR-199a-3p and CD44 in patients’ specimens (Fig. 4) suggests that re-expression of miR-199a-3p to the HCC could restore levels to that of the benign condition. While additional in vivo experiments are necessary, these data suggest that treating CD44+ metastatic cells in advanced HCC with miR-199a-3p mimetic may be possible.
The CD44 family of transmembrane glycoproteins act mainly as receptors for HA [20]. Interaction of HA with CD44 is involved with cell-cell interactions, cell adhesion and migration. A number of CD44 isoforms exist due to alternative splicing of the CD44 gene. All isoforms are expressed in cancers and at least one isoform is expressed in 75% of HCC cases [20; 21; 22; 23]. CD44+ HCCs are often poorly differentiated, have a higher rate of reoccurrence and a decreased overall survival [23; 24]. Inhibition of CD44 in CD44+ HCC cell lines using antisense oligonucleotides increased apoptosis, enhanced chemosensitivity, reduced tumorigensis and invasion and [25]. CD44 has also been linked to potential HCC stem cells [26]. Therefore, CD44 appears to play an intricate role in at least a subset of HCC cases. The use of miRNA to block CD44 expression has been recently conducted by Shao, et al. [27]. This group reduced the proliferation of NIH3T3 fibroblasts in a cardiac transplant murine model by CD44 antibodies or by transfecting the cells with a vector expressing an artificial miRNA that regulates CD44 [27]. This study is significant in that it demonstrates the ability of miRNA targeting CD44 as a therapeutic for HCC.
In conclusion, we report that miR-199a-3p mimetic targets CD44 and reduces in vitro proliferation and invasion in CD44+ HCC cells. In general, treating patients with oligonucleotide based drugs (such as antisense, siRNA and miRNA mimetic) is a challenge due to poor cellular uptake and lack of tissue targeting. However, since negatively charged oligonucleotides naturally accumulate in highly perfused organs such as the liver, treating advanced HCC patients with miR-199a-3p mimetic may be feasible.
Supplementary Material
Clinical data of Patient specimens
miR-199a-3p expression in normal hepatocytes and HCC cell lines. The expression of miR-199a* was determined in primary human hepatocytes and in 7 HCC cell lines by qPCR. Data are presented relative to 18S rRNA.
(A) HCC cells were transfected with 100 nM of control oligonucleotide (Ambion), pre-miR-199a-3p (Ambion) or the miR-199a-3p duplex oligonucleotide chemically modified as described in the Materials and Methods. (B) SNU449 cells were transfected with duplex control, miR-199a-3p duplex or the single stranded guide or passenger strands for miR-199a-3p using the chemical modifications described in the text. Following a 96 h exposure, cell viability was assessed using a WST-1 assay; *p<0.005, †p<0.001).
SNU449 cells were transfected with 100 nM control or miR-199a-3p oligonucleotides for 48 h. The cells were exposed to doxorubicin for an additional 48 h and then the proliferation was assessed by the WST-1 assay; *p<0.001).
The expression of CD44 mRNA was measured in 9 pairs of primary HCC and adjacent benign tissues using qPCR. Data are normalized to 18S rRNA.
Acknowledgments
We thank Mark Behlke for his assistance with the design of the custom miR-199a-3p mimetic. This work was funded by a Provost’s Targeted Investment in Excellence award from the Ohio State University.
Footnotes
HCC, hepatocelluar carcinoma; miRNA, microRNA; HA, hyaluronic acid.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–8. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 6.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, Cohen D, Yerushalmi N. hsa-miR-191 Is a Candidate Oncogene Target for Hepatocellular Carcinoma Therapy. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 7.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–93. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 8.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 9.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 11.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–95. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453–60. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–10. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–78. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16:3074–86. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128–36. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 19.Lara-Pezzi E, Serrador JM, Montoya MC, Zamora D, Yanez-Mo M, Carretero M, Furthmayr H, Sanchez-Madrid F, Lopez-Cabrera M. The hepatitis B virus X protein (HBx) induces a migratory phenotype in a CD44-dependent manner: possible role of HBx in invasion and metastasis. Hepatology. 2001;33:1270–81. doi: 10.1053/jhep.2001.1270. [DOI] [PubMed] [Google Scholar]
- 20.Rudzki Z, Jothy S. CD44 and the adhesion of neoplastic cells. Mol Pathol. 1997;50:57–71. doi: 10.1136/mp.50.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci U S A. 1992;89:12160–4. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolg C, Hofmann M, Herrlich P, Ponta H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993;21:1225–9. doi: 10.1093/nar/21.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo K, Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J Hepatol. 2000;32:78–84. doi: 10.1016/s0168-8278(00)80192-0. [DOI] [PubMed] [Google Scholar]
- 24.Yang GH, Fan J, Xu Y, Qiu SJ, Yang XR, Shi GM, Wu B, Dai Z, Liu YK, Tang ZY, Zhou J. Osteopontin combined with CD44, a novel prognostic biomarker for patients with hepatocellular carcinoma undergoing curative resection. Oncologist. 2008;13:1155–65. doi: 10.1634/theoncologist.2008-0081. [DOI] [PubMed] [Google Scholar]
- 25.Xie Z, Choong PF, Poon LF, Zhou J, Khng J, Jasinghe VJ, Palaniyandi S, Chen CS. Inhibition of CD44 expression in hepatocellular carcinoma cells enhances apoptosis, chemosensitivity, and reduces tumorigenesis and invasion. Cancer Chemother Pharmacol. 2008;62:949–57. doi: 10.1007/s00280-008-0684-z. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J, Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067–78. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
- 27.Shao W, Chen JB, Wang F, Xia JJ, Qi ZQ. Combined Application of Blocking Antibodies and MicroRNA Interference in Inhibiting CD44 Expression. Transplant Proc. 2010;42:2777–81. doi: 10.1016/j.transproceed.2010.05.149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical data of Patient specimens
miR-199a-3p expression in normal hepatocytes and HCC cell lines. The expression of miR-199a* was determined in primary human hepatocytes and in 7 HCC cell lines by qPCR. Data are presented relative to 18S rRNA.
(A) HCC cells were transfected with 100 nM of control oligonucleotide (Ambion), pre-miR-199a-3p (Ambion) or the miR-199a-3p duplex oligonucleotide chemically modified as described in the Materials and Methods. (B) SNU449 cells were transfected with duplex control, miR-199a-3p duplex or the single stranded guide or passenger strands for miR-199a-3p using the chemical modifications described in the text. Following a 96 h exposure, cell viability was assessed using a WST-1 assay; *p<0.005, †p<0.001).
SNU449 cells were transfected with 100 nM control or miR-199a-3p oligonucleotides for 48 h. The cells were exposed to doxorubicin for an additional 48 h and then the proliferation was assessed by the WST-1 assay; *p<0.001).
The expression of CD44 mRNA was measured in 9 pairs of primary HCC and adjacent benign tissues using qPCR. Data are normalized to 18S rRNA.


