Abstract
The use of standard nomenclatures for describing the strains, genes, and proteins of species is vital for the interpretation, archiving, analysis, and recovery of experimental data on the laboratory mouse. At a time when sharing of data and meta- analysis of experimental results is becoming a dominant mode of scientific investigation, failure to respect formal nomenclatures can cause confusion, errors, and in some cases contribute to poor science. Here we present the basic nomenclature rules for laboratory mice and explain how these rules should be applied to complex genetic manipulations and crosses.
“These ambiguities, redundancies, and deficiencies recall those attributed by Dr. Franz Kuhn to a certain Chinese encyclopedia called the Heavenly Emporium of Benevolent Knowledge. In its far-away pages it is written that animals are divided into (a) those that belong to the emperor; (b) embalmed ones; (c) those that are trained; (d) suckling pigs; (e) mermaids; (f) fabulous ones; (g) stray dogs; (h) those that are included in this classification; (i) those that tremble as if they were mad; (j) innumerable ones; (k) those drawn with a very fine camel's-hair brush; (l) etcetera; (m) those that have just broken the flower vase; (n) those that at a distance resemble flies.”
El idioma analítico de John Wilkins, Jorge Lius Borges (1942)
Pathologists are trained not only to use defined medical nomenclature but to use words carefully and concisely. This precept extends to the specific animal species we are dealing with; to define accurately the signalment, namely the species (and scientific name if appropriate), strain, age, sex, and any other pertinent information that would help the pathologist make an accurate interpretation (diagnosis/phenotype) of the samples under investigation. Many textbooks attempt to standardize pathology nomenclature as do study groups that focus on organ systems of specific types or groups of diseases e.g. ref.5 and there are now many examples of disease and diagnostic glossaries, controlled vocabularies and thesauri, e.g. refs.3,15 The recent development of disease and phenotype ontologies1,2,8–10 has provided a more structured framework for nomenclature which, as well as being amenable to sophisticated computation, requires strict semantic discipline. Linking such ontologies to data capture systems forces pathologists to use fixed nomenclature which automatically ensures accurate spelling and coding.12,14
The laboratory mouse has, without question, become the premier animal model used in biomedical research today. Genetic engineering technology13 combined with advanced genetic resources such as the Collaborative Cross4 have expanded the value of the mouse far beyond its traditional role in toxicology and other basic and applied research applications. In spite of this, analyses of these mice remain fundamentally the same as for all other species.
A major problem with virtually all biomedical journals including, Veterinary Pathology, is the failure to demand that authors adhere to strictly established nomenclature procedures so that the reader can understand what is actually being reported. More importantly, when the investigator understands the nomenclature system they can recognize what the proper controls are and avoid making serious errors. For example, epicardial and corneal mineralization was reported in severe combined immunodeficiency (SCID) mutant mice.7 Following correct mouse nomenclature, SCID would imply an inbred mouse strain (all letters in capital for the basic strain designation) that at some point during its derivation carried the severe combined immunodeficiency mutation (Prkdcscid). Without the specific mutant allele symbol, one must assume these were all WILDTYPE mice (the correct symbol for a wildtype mouse is +/+) and therefore NOT immunodeficient. The important point here is that although the authors implied that the severe combined immunodeficiency mutation caused the mineralization problem, they should have compared their heart samples to those in true wildtype controls. Had that been done they would have seen identical lesions in both mutant and control mice resulting in the correct interpretation that this was a strain specific lesion unrelated to the severe combined immunodeficiency mutation the mice carried.
Another more common problem is the use of abbreviations as if they are representative of all allelic mutations available for a particular gene. Many image websites illustrate lesions in mice that carry the P53−/− (null) mutation. This is an old and obsolete symbol. The correct gene symbol for the mouse is Trp53 which stands for transformation related protein 53. At the time of this writing there were 222 allelic mutations (each with a distinct and specific symbol that would appear as a superscript showing the type of induced mutation and the initials of the investigator or originating laboratory; i.e. Trp53tm1Att transformation related protein 53; targeted mutation 1, Laura D Attardi vs. Trp53tm1Brd transformation related protein 53; targeted mutation 1, Allan Bradley) that develop lesions ranging from cancers in old age to embryonic lethality) (Fig. 1; Table 1; http://www.informatics.jax.org; 15 March 2010).
Figure 1.
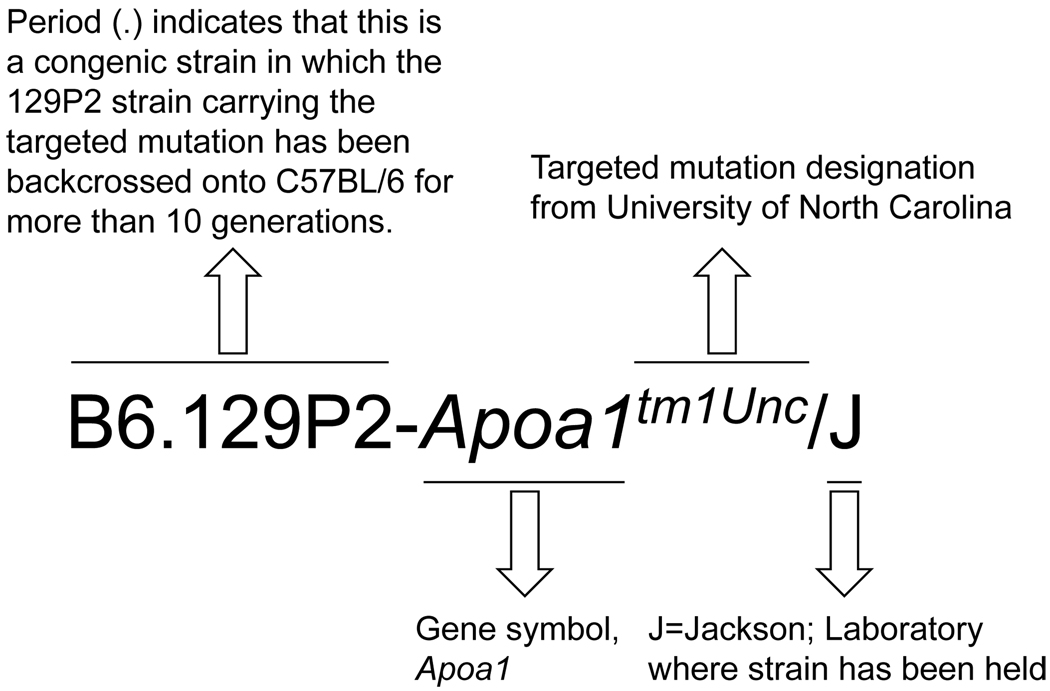
Explanation of the various components of the gene symbol for the targeted mutation (tm, also commonly called knockout or null mutation) for the apolipoprotein A-1 gene.
Table 1.
Examples of different types of mutations in the transformation related protein 53 gene to illustrate various approaches to gene symbols. There are currently 222 allelic mutations reported for this gene in mice (http://www.informatics.jax.org; 15 March 2010). Human gene symbols are all in capital letters. Note in this case the mouse and human symbols are not quite the same. Protein symbols are in capital letters for both species but not in italics. In humans allele designations are limited to an optimum of three characters using only capital letters or Arabic numerals. The allele designation is written on the same line as the gene symbol separated by an asterisk e.g. PGM1*1.
| Allele | Human | Mouse |
|---|---|---|
| Gene Trap | NA | Trp53Gt(CMHD-GT 349E11-3)Cmhd |
| Wildtype Allele | TP53+ | Trp53+ |
| Targeted Allele | NA | Trp53tm1Brd |
| Transgenic Allele | NA | Tg(Trp53A135V)L2Ber |
| Chemically Induced | NA | Trp53Bbl |
| Inversion | NA | Trp53In(11Trp53;11Wnt3)Brd |
NA = not applicable
Committees to Standardize Nomenclature
There are specific committees set up to standardize the nomenclature for mice (International Committee on Standardized Genetic Nomenclature for Mice and Rats (http://www.informatics.jax.org/mgihome/nomen/gene.shtml#genenom). In 2001, the International Committee on Standardized Nomenclature for Mice and the Rat Genome and Nomenclature Committee agreed to establish a joint set of rules for strain nomenclature, applicable to strains of both species. If this discussion is difficult to follow there is an online tutorial that provides more detailed information on this subject (http://jaxmice.jax.org/support/nomenclature/tutorial.html). While many think these are new ideas and rules, they have actually been evolving for a very long time. Earlier versions of the rules, particularly for mice, were published since 194111 and a history can be found on the Mouse Genome Informatics website listed above. One of the major goals initially was to prevent renaming or misnaming of strains and spontaneous mouse mutant locus designations. As mammalian genetics evolved as a discipline, the nomenclature evolved to accommodate the ever increasing complexity of these animals as well as the biotechnology that made these advances possible.
Sources for Assistance for Nomenclature Issues
Nomenclature, both the current gene name and the symbol for the specific allelic mutation under investigation, can be obtained from the vendor’s website if mice were purchased (The Jackson Laboratory, http://jaxmice.jax.org/query; Taconic, http://www.taconic.com; Charles River, http://www.criver.com). The nomenclature can be checked against the most current nomenclature standards on line (go to http://www.informatics.jax.org, search Genes, then Access Data: Genes and Markers Query, and enter the full name or gene symbol for the mutation). Lastly, a symbol proposed by the investigator can be applied for if it is a potentially novel gene, strain, allele, or construct by contacting the nomenclature committee directly. If in doubt, the scientists who maintain the Mouse Genome Informatics website are available to assist with nomenclature questions (click “User Support” at the bottom of the Mouse Genome Informatics Home Page or go to the page directly : http://www.informatics.jax.org/mgihome/support/mgi_inbox.shtml).
Mouse and rat nomenclature web sites are listed above. Human gene nomenclature is governed by the Human Genome Organisation Nomenclature Committee. Rules and names are available online (http://www.genenames.org/). These should be used to ensure that the most accurate and current gene/protein names and symbols are being used.
Inbred Strain Designations
For laboratory mice, inbred strain names are all in capital letters. After the strain name, and equally important, are the laboratory (investigator) and institutional codes that designate substrains. For example, NOD/ShiLtJ is a strain designation that reveals that the nonobese diabetic (NOD) inbred mice originated from inbreeding the Cataract Shionogi (CTS) strain (originally outbred Jcl:ICR mice) selected for on the basis of an elevated fasting blood glucose level in cataract-free mice. The holder of the mice at that time, Shionogi, is designated in the nomenclature of the current strain as Shi. NOD substrains, originally available in Japan, were distributed in the early 1980s to Australia and the United States and eventually breeder pairs were sent to Dr. E. Leiter (investigator’s laboratory symbol: Lt) at The Jackson Laboratory (J; Fig. 1; http://jaxmice.jax.org/strain/001976.html, 9 Feb 2010).
Strain Abbreviations
Abbreviations for commonly used strains are also standardized (see: http://www.informatics.jax.org/mgihome/nomen/strains.shtml#hybrids) (Table 2). B6 specifically refers to the C57BL/6J strain although this is commonly used in the literature to refer to all of the C57BL/6 substrains, many of which carry unique mutations and therefore can be different from each other. By contrast, BALB/cJ mice are abbreviated C and BALB/cByJ mice are CBy. Hybrids or incipient congenics, where a mutated gene is being transferred to another strain, are designated with a semicolon between the strain abbreviations, such at B6;129. This segregating background is in sharp contrast to a congenic strain where the semicolon is replaced by a period to indicate the congenic procedure has been completed (10 backcrosses, N10, onto the new strain). Such mice are designated B6.129. While six backcrosses (N6; an incipient congenic) is commonly accepted by many journals to be adequate, speed congenic technology has clearly shown that this is not adequate.6
Table 2.
Symbols for various types of inbred mouse strains and stocks. See the tutorial for more information (http://jaxmice.jax.org/support/nomenclature/tutorial.html).
| Type of strain/stock |
Symbol | Explanation |
|---|---|---|
| Inbred Strain | C57BL/6J | C57BL/6 strain distributed from The Jackson Laboratory |
| Substrain | C57BL/6N | C57BL/6 strain distributed from the National Institutes of Health |
| Substrain | C57BL/6NCrl | C57BL/6N strain distributed by Charles River Laboratories International, Inc. |
| Substrain | C57BL/6NTac | C57BL/6N strain distributed by Taconic Farms, Inc. |
| Inbred Strain Abbreviation | B6 | Abbreviation for the C57BL/6J strain only. Commonly misused for all C57BL/6 substrains |
| Recombinant Inbred Strain | BXD29/Ty | Cross (X) between C57BL/6J (B) female and DBA/2J (D) male from which an inbred strain (20 brother sister matings) was created by Dr. Benjamin Taylor (Ty). This example was line number 29. |
| Congenic Strain | B6.129P2-Apoa1tm1Unc/J | A strain in which a specific mutation or genetic interval was moved from one strain (129) to another (B6). In this case the 129P2 donor E14TG2a ES cell line was used to create a targeted mutation of the Apoa1tm1Unc/J which was moved onto the B6 background using 10 or more backcrosses. |
| Consomic Strain | C57BL/6J-Chr 13PWD/Ph/ForeJ | This strain strain is one of a panel of inter-species chromosome substitution (consomic) strains (IC-ISS) in which each strain (in this case B6) carries a chromosome (in this case Chromosome 13) from PWD/Ph. |
| Segregating Background | B6;129S | An undetermined mixture of B6 and 129S1/SvlmJ genes is indicated by the semicolon. Often these mice are in the process having a congenic strain created. |
| Hybrid | B6129SF1 | A cross between a C57BL/6J female and a 129S1/SvlmJ male. The F (filial) indicates the generation (first progeny from this type of cross). |
Mutant Gene Symbols
Gene symbols for mouse mutant gene loci were written historically with the first letter capitalized and subsequent letters in lower case for dominant or semidominant mutations in mice, while recessive mutation gene symbols are written in all lower case. This persists for the spontaneous allelic mutations which are now listed as superscripts after the known gene symbol. The gene symbols for mice have the first letter capitalized followed by lower case letter. Human genes have all letters in upper case which makes them easily separated from mouse gene symbols. Gene symbols are written in italics. For example the mouse hairless and rhino mutations are on two different inbred strains and are written HRS/J-Hrhr/Hrhr and RHL/J-Hrrh7J/Hrrh7J, respectively. Human gene symbols are always written in capital letters and in italics so the human hairless gene is written as HR. Protein symbols are similar to gene symbols except that they are all written in capital letters without being in italics for both species (HR for the hairless protein).
Specific nomenclature can be found on the Mouse Genome Informatics website or curators can be contacted directly. Nomenclature standards change over time, as do many of the gene symbols. The nomenclature committees and respective websites maintain the historical synonyms so it is possible to figure out which allelic mutation was studied historically relative to current studies if the original work was annotated correctly. Strict adherence to these nomenclature standards will allow work to be fairly compared and more importantly reviewed accurately.
Mouse genetic nomenclature can get very complicated for some of the highly specialized stocks of genetically engineered mice. Help should be solicited for correctly designating these lines. However, basic knowledge of the difference between strains and mutations needs to be understood.
Nomenclature for Specific Genetic Tools
Historically, inbred strains are brother/sister mated in a carefully controlled manner for 20 generations. The new Collaborative Cross mice are more complicated and 25 or more generations may be needed to inbreed them adequately.4 Congenic mice, created by moving a single mutated gene from one strain to another, also involve 20 crosses but are more complicated. In this case the strain carrying the mutation is crossed with another strain and their F1 offspring are intercrossed to produce F2 mice, 25% of which carry the mutant gene (if recessive). These mutants are then crossed to the strain it is being backcrossed onto to repeat this again for a total of 10 times. At the end of this process, the mutation should be on a background very close to the inbred strain it was being moved onto. The nomenclature for congenic strains reflects the new inbred strain the mutation was moved onto first followed by a period and either the strain it was moved from or simply Cg for congenic, followed by the allelic mutation that was moved (Table 2). For example CByJ.Cg-Foxn1nu/J is a congenic strain in which the nude mutation was moved onto the BALB/cByJ strain. Alternatively, B6.AK-Foxn1nu-str/J is a congenic strain in which the nude-streaker allelic mutation of the Foxn1 gene was moved from the AKR/J inbred strain, on which this mutation originally arose, onto the C57BL/6J inbred strain. In these complex designations common strains are represented in the abbreviated form discussed above. A third type of strain is called a recombinant inbred strain. In this case two unrelated strains are crossed and their offspring are crossed for 20 generations to create a mixture of the two strains. By inbreeding several lines a series of unique strains are created. The nomenclature for these recombinant inbred strains would be BXD-16/TyJ where B represents the C57BL/6J female crossed (X) with the DBA/2J male, line number 16 of 42, which were created by Dr. Benjamin Taylor (Ty) and distributed by The Jackson Laboratory (J). Yet another type is called a consomic strain in which part or an entire chromosome is moved from one inbred strain to another. An example is the C57BL/6J-Chr11.3PWD/Ph/ForeJ created by Dr. Jiri Forejt (ForeJ) by replacing part of the distal end of mouse Chromosome 11 from the PWD/Ph donor strain into the C57BL/6J host strain using a marker-assisted series of backcrosses (Table 2; http://jaxmice.jax.org/, 11 Feb 2010).
Conclusions
At a time when biological data are increasingly archived, searched, recovered, and analysed computationally, adherence to standard forms of nomenclature is vital. Computers are dumb; unlike Journal editors they are unable to deal with the nuances and ambiguities of natural language and failure to make compliance with standards second nature generates more work, limits the utility of publications and database submissions, and, as shown above can produce bad science.
Acknowledgments
This work was supported by the European Commission (Contract number LSHG-CT-2006-037811; CASIMIR, to PNS) and the National Institutes of Health (CA089713, RR17436 to JPS).
References
- 1.Bard J. Ontologies: formalising biological knowledge for bioinformatics. BioEssays. 2003;25:501–506. doi: 10.1002/bies.10260. [DOI] [PubMed] [Google Scholar]
- 2.Bard JB, Rhee SY. Ontologies in biology: design, applications and future challenges. Nat Rev Genet. 2004;5:213–222. doi: 10.1038/nrg1295. [DOI] [PubMed] [Google Scholar]
- 3.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, Carter JS, deCoronado S, Downing JR, Fredrickson TN, Haines DC, Harris AW, Harris NL, Hiai H, Jaffe ES, MacLennan IC, Pandolfi PP, Pattengale PK, Perkins AS, Simpson RM, Tuttle MS, Wong JF, Morse HC. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 6.Markel P, Shu P, Ebeling C, Carlson GA, Nagle DL, Smutko JS, Moore KJ. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat Genet. 1997;17:280–284. doi: 10.1038/ng1197-280. [DOI] [PubMed] [Google Scholar]
- 7.Meador VP, Tyler RD, Plunkett ML. Epicardial and corneal mineralization in clinically normal severe combined immunodeficiency (SCID) mice. Vet Pathol. 1992;29:247–249. doi: 10.1177/030098589202900309. [DOI] [PubMed] [Google Scholar]
- 8.Osborne JD, Flatow J, Holko M, Lin SM, Kibbe WA, Zhu LJ, Danila MI, Feng G, Chisholm RL. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10 Suppl 1:S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snell GD. Biology of the Laboratory Mouse. 1st ed. New York: McGraw-Hill; 1941. [Google Scholar]
- 12.Sundberg BA, Schofield PN, Gruenberger M, Sundberg JP. A data capture tool for mouse pathology phenotyping. Vet Pathol. 2009;46:1230–1240. doi: 10.1354/vp.09-VP-0002-S-FL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sundberg JP, Ichiki T. Genetically engineered mice handbook. Boca Raton: CRC Press; 2005. p. 314. [Google Scholar]
- 14.Sundberg JP, Sundberg BA, Schofield PN. Integrating mouse anatomy and pathology ontologies into a diagnostic/phenotyping database: tools for record keeping and teaching. Mammalian Genome. 2008;19:413–419. doi: 10.1007/s00335-008-9123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vahle J, Bradley A, Harada T, Herbert R, Kaufmann W, Kellner R, Mann P, Pyrah I, Rittinghausen S, Tanaka T. The international nomenclature project: an update. Toxicol Pathol. 2009;37:694–697. doi: 10.1177/0192623309340278. [DOI] [PubMed] [Google Scholar]