Abstract
Over the last decade, the potential of antibodies as therapeutic strategies to treat Alzheimer’s disease (AD) has been growing, based on successful experimental and clinical trials in transgenic mice. Despite, undesirable side effects in humans using an active immunization approach, immunotherapy still remains one of the most promising treatments for AD. In this study, we analyzed the V genes of twelve independently isolated monoclonal antibodies raised against the N-terminal immunodominant epitope of the amyloid β peptide (Aβ or A beta). Surprisingly, we found a high and unusual level of restriction in the VH/VL pairing of these antibodies.
Moreover, these antibodies mostly differ in their heavy chain complementary determining region 3 (HCDR3) and the residues in the antibodies which contact Aβ are already present in the germline V-genes. Based on these observations and or co-crystal structures of antibodies with Aβ, the aim of the current study was to better understand the role of antibody V-domains, HCDR3 regions, key contact residue (H58) and germline encoded residues in Aβ recognition. For that purpose, we designed and produced a range of recombinant Fab constructs. All the Fabs were tested and compared by surface plasmon resonance on Aβ1–16, Aβ1–42 high molecular weight and Aβ1–42 low molecular weight soluble oligomers. Although all the Fabs recognized the Aβ1–16 peptide and the Aβ1–42 high molecular weight soluble oligomers, they did not bind the Aβ1–42 low molecular weight soluble oligomers. Furthermore, we demonstrated that: (1) an aromatic residue at position H58 in the antibody is essential in the recognition of Aβ and (2) Fabs based on germline V-genes bind to Aβ monomers with a low affinity. These findings may have important implications in designing more effective therapeutic antibodies against Aβ.
Keywords: Alzheimer’s disease, Passive immunotherapy, Anti-Aβ antibodies, Restricted antibody repertoire, Antibody engineering, Surface plasmon resonance
1. Introduction
Alzheimer’s disease (AD) is the most common form of dementia in the elderly. It is characterized by neuronal loss leading to cognitive dysfunction and the presence of amyloid plaques and neurofibrillary tangles (Selkoe, 1999). The plaques are mainly composed of a 4-kDa (42 amino acids) amyloid β peptide (Aβ or A beta) derived from the proteolysis of the amyloid precursor protein (APP) (Kang et al., 1987). Aβ has a tendency to self-associate and can adopt several pathological forms including soluble oligomers, protofibrils and insoluble amyloid fibrils (Arimon et al., 2005; Lambert et al., 1998; Walsh et al., 2002).
One potentially powerful strategy for treating AD is immunotherapy, in which antibodies facilitate the clearance of amyloid plaques or neutralize toxic forms of Aβ (conformation-dependant antibodies). The potential of immunotherapy for AD was first realized when mouse monoclonal antibodies (mAbs) raised against the linear N-terminal amino acids of the Aβ peptide not only disaggregated amyloid fibrils and prevented the formation of Aβ fibrils in vitro, but also protected against Aβ-mediated neurotoxicity in vitro (Solomon et al., 1996, 1997; Frenkel et al., 1999). These observations were later confirmed in AD mouse models in which β amyloid plaques were reduced in the brain (Schenk et al., 1999) and performance in memory tasks were improved following immunization with Aβ1–42 fibrils (Janus et al., 2000). Subsequent studies have shown that antibodies which bind amyloid plaques and induce their clearance were those directed against the N-terminal region of Aβ (Bard et al., 2003). These data were rapidly translated into the clinic to evaluate the efficacy of active anti-Aβ vaccination in humans. The active vaccination trial (AN1782) initiated by Elan Pharmaceuticals, consisted of an intramuscular injection of fibrillar human Aβ1–42 formulated in strong Th1 adjuvant, QS-21. The linear immunodominant epitope recognized by antibodies from patient’s sera immunized with AN1792 vaccine is located in the N-terminal region of Aβ (Lee et al., 2005). However, these trials were stopped due to brain inflammation in 6% of the patients (Orgogozo et al., 2003). Nevertheless, other active immunization studies involving the N-terminal linear immunodominant epitope of the Aβ peptide which do not evoke detrimental side effects observed during the first clinical trial are still under development (Frenkel et al., 2000; Agadjanyan et al., 2005).
An alternative strategy, known as passive immunotherapy, involves the systemic infusion of monoclonal antibody directed against Aβ1–42. It has been demonstrated that this approach was equally as effective as active immunization in terms of reducing amyloid plaque (Bard et al., 2000) and improving cognition (Wilcock et al., 2006) in an AD mouse models. Among the antibodies tested using a passive immunotherapeutic approach, only those directed to the linear N-terminal regions of Aβ have demonstrated anti-aggregating properties in vivo (Bard et al., 2000). Therefore, it is not surprising that the most advanced monoclonal antibody, (bapineuzumab in phase III clinical trials) is directed against this region (Agadjanyan and Cribbs, 2009). Bapineuzumab is a humanized version of the antibody 3D6 which recognizes Aβ1–5 and was developed by Pfizer/Johnson & Johnson.
Recently, Basi et al. (2010) solved the structure of three different mAbs (12A11, 10D5 and 12B4) in complex with Aβ1–7 (1DAEFRHD7)(Basi et al., 2010) and demonstrated that they shared high structural and sequence homologies with three other antibody structures (PFA-1, PFA-2 and WO2) described by two other groups (Gardberg et al., 2007; Miles et al., 2008). In this study we compared and analyzed the variable heavy (VH) and light (VL) chain sequences from twelve anti-Aβ mAbs isolated by different research groups (Basi et al., 2010; Miles et al., 2008; Gardberg et al., 2007; Golde et al., 2010; Acton et al., 2006; Shen and Biere-Citron, 2008) and found that they exhibited a highly restricted VH/VL pairing, mostly differing in the VH complementary determining region 3 (CDR3) sequences. Interestingly, these antibodies were obtained following immunization with different forms of Aβ (i.e. monomeric Aβ peptide, soluble Aβ oligomers, protofibrils and fibrils) and some are reported to preferentially bind to the soluble Aβ oligomeric species (Acton et al., 2006). However, despite using different forms of the Aβ as an antigen, only a few mouse monoclonal antibodies have been reported to exhibit a conformational specificity.
In this study we aimed to further understand the effects of particular antibody sequences in their overall affinity for different forms of Aβ. A range of recombinant Fabs (rFab) were designed and produced by variable domain swapping, VH CDR3 loop grafting and amino acid changes. In addition, a rFab with unmutated VH and VL domains (i.e. closest germline genes) was synthesized and expressed (gWO2 rFab) to examine the contribution of germline encoded residues in Aβ recognition. The constructs were assessed by surface plasmon resonance (SPR) for their ability to bind Aβ1–16 monomeric peptide, Aβ1–42 high molecular weight (HMW) and Aβ1–42 low molecular weight (LMW) soluble oligomers.
2. Materials and methods
2.1. Sources of VH and VL sequences
The retrieval of relevant nucleotide and amino acid (AA) sequences from scientific and patent publications has been performed by entering the VH and VL nucleotide and/or amino acid sequences from WO2 in the SciFinder software.
2.2. Analysis of VH and VL sequences and 3D structure modeling of the antibody fragment
The germline usage of different mAbs was determined by comparing the nucleotide sequences to those in IMGT®, the international ImMunoGeneTics information system (Lefranc et al., 2009) (http://www.imgt.org) using the web-based program IMGT/V-QUEST (Brochet et al., 2008) and IMGT/JunctionAnalysis (Yousfi Monod et al., 2004) tools. Amino acid sequences were analyzed using IMGT/DomainGapAlign (Ehrenmann et al., 2010). Contact analysis data are from IMGT/3Dstructure-DB (Ehrenmann et al., 2010). Codons, amino acids and delimitations of the framework regions (FR) as well as complementarity determining regions (CDR) are provided according to the IMGT unique numbering for V domain (Lefranc et al., 2003).
The molecular model of the 20.1 and Ab9 variable domains were obtained by using the web antibody modeling (WAM) algorithm (Whitelegg and Rees, 2000) (http://antibody.bath.ac.uk). Images of the model were generated using PyMOL (PyMOL version 0.82, http://pymol.sourceforge.net) (DeLano Scientific LLC).
2.3. Generation of the recombinant chimeric Fab, production, periplasmic extraction and purification
Cloning of all the constructs into the pGC-Fab vector was performed as described previously (Robert et al., 2009). Transformed E. coli TOP10F’ cells (Invitrogen, Mount Waverley, VIC) were grown at 30 °C in 3 l of 2YT medium containing glucose (0.1%) and ampicillin (100 μg/ml). When the absorbance at 600 nm was 1, isopropyl β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM and the temperature was reduced to 26 °C. After 4 h of protein induction, the cells were pelleted and a periplasmic extract was prepared by sequential extraction with ice-cold TES buffer (0.2 M Tris–HCl, pH 8.0, 0.5 mM EDTA, 0.5 M sucrose) and 0.2× TES buffer. The periplasm extract was clarified by centrifugation (20,000 × g, 30 min) and dialyzed through 0.45 μm membrane (Millipore, Bedford, USA).
The recombinant Fabs were purified by a two-step purification protocol. The cleared periplasmic extract was first loaded onto a 1 ml HisTrap™ FF crude column (GE Healthcare) using the Profinia protein purification system (Bio-Rad) according to the manufacturer’s instructions. The IMAC-purified Fabs were then desalted against phosphate buffered saline (PBS) and loaded onto the size exclusion gel chromatography column (10/30 Superdex 200). The separation was performed with a flow rate of 0.5 ml/min at room temperature with 1× PBS (pH 8) as the elution buffer. Fractions of 500 μl were collected and further analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
2.4. Mutagenesis of the Fab
Site-directed mutagenesis of the WO2 Fab was performed using the Quick Change® Site-directed Mutagenesis Kit II (Stratagene, La Jolla, CA). Mutations were verified by sequencing the Fab variable domains on both strands.
2.5. Enzyme-linked immunosorbent assays (ELISA)
Wells of 96-well Maxisorb Immuno plates (Nunc, Roskilde, Denmark) were coated with 5 μg/ml of a purified Aβ fusion protein (Maltose Binding Protein-Aβ1–42 or MBP-Aβ1–42)(Caine et al., 2007) in carbonate coating buffer. Control wells were coated with 50 μg/ml bovine serum albumin (BSA) or 50 μg/ml MBP overnight at 4 °C. Wells were blocked with 200 μl of 2% milk phosphate buffered saline (MPBS) for 2 h at 37 °C. rFab from crude periplasmic extracts were incubated in wells with MBP-Aβ1–42 for 2 h. Control wells were incubated with the anti-FLAG M2 antibody (Sigma, St Louis, MO), for 1 h at RT. After washing, a 1/2000 dilution of anti-mouse IgG horseradish peroxidase (HRP)-conjugate (Pierce, Rockford, IL) was added to each well and incubated for a further 1 h at 20 °C. The substrate 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS, Roche, Indianapolis, USA) was added and absorbance was read at 405 nm.
2.6. Preparation, purification and characterization of biotinylated Aβ1–42 monomers, biotinylated Aβ1–42 HMW and LMW soluble oligomers
Aβ1–42 synthetic lyophilised peptide (W.M. Keck Laboratory, Yale University, New Haven, CT) or biotinylated Aβ1–42 lyophilised peptide (American Peptide Company, Sunnyvale, CA) was dissolved in 1,1,1,3,3,3-hexafluoro-2-isopropanol (HFIP) (Sigma, St Louis, MO) and aliquoted. HFIP was removed by evaporation under a fume hood. Residual traces of HFIP were removed by drying under vacuum in a SpeedVac (Savant Instrument). The resulting peptide film was stored at −80 °C until required. For the biotinylated Aβ1–42 monomeric preparation, the peptide film was dissolved in dimethyl sulfoxide (DMSO) at 5 mM and then diluted to 100 μM in ice cold 1× PBS. The solution was centrifuged at 16,000 × g for 15 min at 4 °C and the supernatant passed through a 0.2 μm filter to remove any aggregates.
For the biotinylated soluble oligomeric preparations, the Aβ1–42 and biotinylated Aβ1–42 peptide solution were mixed at a 1:4 molar ratio and diluted to 100 μM in ice cold 1 × PBS. The solution was left for 24 h at 4 °C, and then centrifuged at 16,000 × g for 15 min at 4 °C. The biotinylated Aβ1–42 HMW and LMW soluble oligomers in the supernatant were next separated by size exclusion chromatography on a 10 mm × 30 cm Superdex 75 column (Amersham Biosciences, Piscataway, NJ) [Supplemental Fig. 1A] and the corresponding peaks were directly used for immobilization on the surface plasmon resonance (SPR) chip. The biotinylated Aβ1–42 HMW and LMW oligomer fractions were separated by electrophoresis using 12–20% Tris–tricine gels (Invitrogen) and transferred onto 0.2 μm nitrocellulose membranes (Invitrogen). After blocking in 5% MPBS, the membrane was incubated for 2 h with the anti-Aβ4–9 6E10 antibody (Signet labs Inc, Dedham, MA). Following three washes, the membranes were incubated with a goat anti-mouse antibody HRP-conjugate (1 μg/ml) (Pierce) for 1 h. The reaction was developed and visualized with a chemiluminescence detection kit (Amersham Pharmacia Biotech, Uppsala, Sweden) [Supplemental Fig. 1B].
2.7. SPR biosensor binding analysis
2.7.1. General procedures
All SPR experiments were performed at 25 °C using Bio-Rad’s ProteOn XPR36 array biosensor (Bravman et al., 2006). Two different assay formats were designed to analyze the binding of Fab to Aβ: (1) Capture Assay: recombinant Aβ1–16-Im7 fusion protein, used as a “surrogate Aβ antigen” (for details, see Section 3), was injected over Fab captured on the surface of a GLC chip containing previously immobilized Human Fab Binder (GE Healthcare); and (2) Direct Assay: recombinant Fabs were injected over various Aβ forms captured on a NeutrAvidin coated chip (NLC, BioRad). Immobilizations and capture experiments were performed in 1× HBS-P running buffer (10 mM HEPES, 150 mM NaCl, 0.005% (w/v) Tween 20). Binding assays were performed in 1× HBS-EP+ running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% (w/v) Tween 20) containing 0.1% (w/v) BSA.
2.7.2. Capture assay: Aβ1–16-Im7 vs. captured recombinant Fab
All recombinant Fabs analyzed during this study contained human CH1 and CL domains which allowed their capture onto the chip surface via a Human Fab Binder. A standard coupling protocol was employed to immobilize Human Fab Binder onto a GLC chip surface via exposed primary amines. A single lane (ligand direction) on the GLC chip surface was activated by a 5-min injection of freshly prepared 1:1 mixture consisting of 2.5 mM sulfo N-hydroxysuccinimide (sulfo-NHS):10 mM 3-(N,N-dimethylamino)propyl-N-ethylcarbodiimide (EDC). Then 120 μl Human Fab Binder solution (10 μg/ml in 10 mM sodium acetate, pH 5.0) was injected for 5 min at a flow rate of 30 μl/min. To deactivate residual reactive sites, Fab coupling was followed by a 5-min injection of 1 M ethanolamine (pH 8.5). Approximately 6,000 response units (RU; 1 RU=1pg of protein/mm2) of Human Fab Binder were coupled using this method. Purified Fabs were diluted to 20 μg/ml and injected in ligand (vertical) direction over immobilized Human Fab Binder at 100 μl/min for 18 s. This typically resulted in a capture of 300–400 RU of Fab protein. Recombinant Aβ1–16-Im7 fusion protein (O.D. manuscript in preparation) consisting of Aβ1–16 fused to the N-terminus of bacterial Immunity protein 7 (Im7) (Kleanthous et al., 1998) was used as analyte. Thus, five dilutions of Aβ1–16-Im7 protein (diluted 3-fold) and one “zero-buffer” blank were injected simultaneously over captured Fab at a flow rate of 30 μl/min. Analyte-ligand association and dissociation phases were monitored for 120 and 300 s, respectively. The Human Fab Binder surfaces were subsequently regenerated in ligand direction with an 18-s injection of 10 mM glycine pH 2.1 at a flow rate of 100 μl/min.
2.7.3. Direct assay: Fab vs. immobilized Aβ forms
This assay was performed as described previously (Robert et al., 2010). Briefly, various dilutions of biotinylated Aβ1–16 peptide, biotinylated Aβ1–42 LMW oligomers, biotinylated negative control 12-mer peptide and biotinylated Aβ1–42 HMW oligomers were injected in ligand direction in lanes 6, 4, 3 and 2, respectively, resulting in approximate capture levels of 80 RU for Aβ1–16 peptide, 1200 RU for biotinylated Aβ1–42 LMW oligomers, 200 RU for 12-mer peptide and 600 RU for biotinylated Aβ1–42 HMW oligomers. Binding measurements were performed using “classical kinetics” described previously (Robert et al., 2010). Association and dissociation phases were monitored for 120 and 300 s, respectively, using a flow-rate of 30 μl/min. The Aβ1–16 coated chip was regenerated between analyte injections with 10 mM glycine pH 1.5 at 100 μl/min for 18 s.
2.7.4. Data processing and analysis
All binding data were processed using Scrubber-Pro software package (prototype software obtained from D. Myszka, University of Utah). Data were aligned to zero on both x and y axes to establish an origin for all binding steps of interest. Data were internally referenced by subtracting the “interspots” from the reaction spots and then “double-referenced” by further subtracting “zero-buffer” blank injections (Bravman et al., 2006). To determine the kinetic rate constants of the binding interactions, binding data were fit globally to a 1:1 interaction model. The ratio of the rate constants (kd/ka) yielded the value for the equilibrium dissociation constant (KD). Alternatively, for rapidly dissociating interactions, affinity (KD) estimates were derived using steady-state affinity algorithm available within Scrubber-Pro.
3. Result
3.1. Variable genes usage analysis
Recently, a high level of homology in the VH and VL amino acid sequences and structures between six anti-Aβ mAbs was reported (Basi et al., 2010). To investigate if the biased V gene usage and restricted VH/VL pairing occurred in other antibodies, we analyzed the VH and VL nucleotides and AA sequences of twelve anti-Aβ antibodies of which six had been crystallized (PFA-1, PFA-2, WO2, 12A11, 12B4 and 10D5 mAbs) and identified striking similarities in their VH/VL sequences and pairing [Table 1]. Importantly, these antibodies were raised using different forms of the Aβ peptide (Aβ1–28, Aβ1–42 monomers, Aβ1–42-derived diffusible ligands (ADDL), Aβ1–42 protofibrils and Aβ1–42 fibrils) and some were reported to be selective for Aβ oligomers (e.g. 2D6, 4E2 and 20C2).
Table 1.
Analysis of germline gene usage and somatic mutations in VH and VL nucleotide sequences of anti-Aβ antibodies using IMGT/V-QUEST.
| mAb namea | Isotype | Epitope | Immunization | VHb |
R/S FR-IMGTc |
R/S CDR-IMGTc,d |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IGHV gene and allele | IGHD gene and allele | IGHJ gene and allele | FR1 (R/S) | FR2 (R/S) | FR3 (R/S) | CDR1 (R/S) | CDR2 (R/S) | ||||
| WO2 | IgG2a | 2–8 | Aβ1–42 | IGHV8-12*01 (96.6%) | IGHD1-1*01 | IGHJ2*01 (97.7%) | 1/1 | 1/0 | 2/1 | 3/0 | 0/1 |
| 20.1 | IgG2b | 2–8 | Aβ1–42 | IGHV8-12*01 (96.2%) | IGHD2-14*01 | IGHJ3*01 (93.2%) | 5/2 | 0/0 | 1/1 | 0/1 | 0/1 |
| 12B4 | IgG2a | 2–7 | Aβ1–42 | IGHV8-12*01 (96.7%) | IGHD2-13*01 | IGHJ2*01 (100%) | 0/1 | 0/0 | 3/2 | 0/1 | 0/1 |
| 10D5 | IgG1 | 2–7 | Aβ1-28 | IGHV8-12*01 (95.9%) | IGHD1-2*01 | IGHJ2*01 (97.9%) | 1/3 | 0/0 | 4/2 | 2/0 | 0/0 |
| 12A11 | IgG1 | 2–7 | Aβ1–42 | IGHV8-8*01 (98.3%) | IGHD1-1*01 | IGHJ4*01 (90.6%) | 1/1 | 0/0 | 2/0 | 0/1 | 0/0 |
| 20C2 | IgG1 | Conformational | Aβ1–42 ADDL | IGHV8-8*01 (93.1%) | IGHD3-2*01 | IGHJ4*01 (88.9%) | 2/3 | 1/0 | 7/6 | 1/0 | 0/0 |
| 4E2 | IgG1 | Conformational | Aβ1–42 ADDL | IGHV8-8*01 (95.9%) | IGHD1-1*01 | IGHJ2*01 (97.9%) | 1/3 | 0/0 | 5/2 | 1/0 | 0/0 |
| 2D6 | IgG1 | Conformational | Aβ1–42 ADDL | IGHV8-8*01 (95.9%) | IGHD1-1*01 | IGHJ2*01 (97.9%) | 1/3 | 0/0 | 5/2 | 1/0 | 0/0 |
| 2.1 | IgG1 | 1–16 | Aβ1–42 | IGHV8-8*01 (95.2%) | IGHD2-4*01 | IGHJ2*01 (97.9%) | 1/3 | 0/0 | 6/2 | 2/0 | 0/0 |
| Ab9 | IgG2a | 1–16 | Aβ1–42 fibrils | IGHV8-13*1 (99%) | IGHD2-2*01 | IGHJ3*01 (91.7%) | 0/0 | 0/0 | 0/0 | 1/0 | 1/1 |
| mAb namea | VL (V-KAPPA)b |
R/S FR-IMGT |
R/S CDR-IMGTc |
|||||
|---|---|---|---|---|---|---|---|---|
| IGKV gene and allele | IGKJ gene and allele | FR1 (R/S) | FR2 (R/S) | FR3 (R/S) | CDR1 (R/S) | CDR2 (R/S) | CDR3 (R/S) | |
| WO2 | IGKV1-117*01 (94.6%) | IGKJ5*01 (97.4%) | 2/0 | 1/2 | 2/1 | 2/2 | 2/0 | 2/2 |
| 20.1 | IGKV1-117*01 (97.3%) | IGKJ5*01 (100%) | 0/2 | 1/0 | 1/1 | 2/0 | 0/0 | 1/0 |
| 12B4 | IGKV1-117*01 (99.3%) | IGKJ5*01 (100%) | 0/0 | 0/0 | 0/0 | 1/1 | 0/0 | 0/0 |
| 12A11 | IGKV1-117*01 (98.9%) | IGKJ5*01 (100%) | 0/0 | 0/0 | 1/0 | 0/1 | 0/0 | 1/0 |
| 10D5 | IGKV1-117*01 (97.9%) | IGKJ5*01 (97.2%) | 0/0 | 0/0 | 4/0 | 2/0 | 0/0 | 0/0 |
| 20C2 | IGKV1-117*01 (98.3%) | IGKJ5*01 (100%) | 0/1 | 0/1 | 0/1 | 1/0 | 0/0 | 1/0 |
| 4E2 | IGKV1-117*01 (98.9%) | IGKJ5*01 (100%) | 0/0 | 0/0 | 0/2 | 0/1 | 0/0 | 0/0 |
| 2D6 | IGKV1-117*01 (98.9%) | IGKJ5*01 (100%) | 2/0 | 0/0 | 0/0 | 0/1 | 0/0 | 0/0 |
| 2.1 | IGKV1-117*01 (100%) | IGKJ5*01 (100%) | 1/0 | 0/0 | 1/0 | 0/0 | 0/0 | 1/0 |
| Ab9 | IGKV1-110*01 (98.9%) | IGKJ1*01 (100%) | 1/0 | 0/0 | 1/0 | 0/0 | 0/0 | 1/0 |
PFA-1 and PFA-2 were not included in the table because no DNA sequences were available in databases for these clones. However, the analysis of there AA sequences with IMGT/DomainGapAlign revealed that both clones are using the IGHV8-12*01 with IGHJ3*01 and the IGKV1-117*01 with IGKJ5*01 genes.
Closest germline genes (the value between brackets is the percentage of identity)
Somatic hypermutation in complementarity determining region (CDR) and in framework region (FR)–R: replacement mutation; S: silent mutation.
VH CDR3 is not included in the analysis.
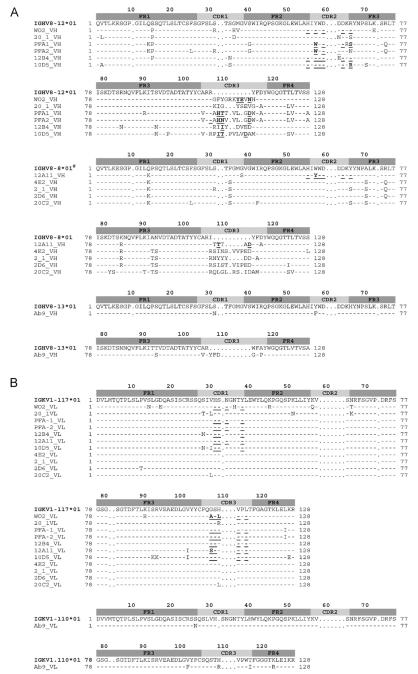
Nucleotide and AA sequences of the VH and VL domains were compared with the closest germline genes using IMGT/V-QUEST and IMGT/DomainGapAlign [Fig. 1A and B]. The IGHV genes of all twelve mAbs analyzed were derived from the IGHV8 subgroup, specifically either IGHV8-12*1, IGHV8-8*1 or IGHV8-13*1 genes and alleles. The nucleotide sequences share a high level of identity, diverging predominantly in the CDR3. Differences between nucleotides in this region appear to be due to a random use of IGHD and IGHJ germline genes during the V-D-J rearrangement (combinatorial diversity), the deletion and stochastic addition of N nucleotides at the V-D and D-J junctions (N-diversity) and somatic hypermutation.
Fig. 1.
Amino acid sequence alignment of anti-Aβ VH (A) and VL (V-KAPPA) (B) domains with the closest germline V, D and J genes identified using IMGT/V-QUEST for the nucleotide sequences and IMGT/DomainGapAlign2 for the amino acid sequences (Yousfi Monod et al., 2004; Lefranc et al., 2009). Numbering and CDR definition are according to the IMGT unique numbering (Lefranc et al., 2003). Dashes indicate identical amino acids, whereas dots indicate no amino acids at the corresponding IMGT numbering positions. Residues contacting the antigen are underlined. (#) The conserved amino acids Q14>K; F35>S; A70>S; K84>R; A92>T; N93>S; I107>R, compared to IGHV8-8*01 suggest the existence of a new IGHV8-8 gene not yet sequenced in BALB/c mice.
The VH CDR3 composition did not reveal any common motif amongst the antibodies. The VH CDR3 length ranged from 9 AA (Ab9) to 17 AA (WO2) and the mean length was 14 AA which correlates with previous results for VH CDR3 derived from mouse antibodies raised against a peptide (Collis et al., 2003).
The distribution pattern of replacement (R) and silent mutations (S) in the CDRs and FRs usually forms the basis of antigenic selection. The R/S ratio was 1.3 for the FR (FR1, FR2 and FR3) while the R/S ratio was 1.85 for the CDR (CDR1 and CDR2). However, the residues which directly contact the antigen as determined by crystal structures are rarely mutated [Fig. 1A; underlined residues].
All clones possessed similar VL domains using the same gene and allele (IGKV1-117*1; IGKJ5*01) with the exception of antibody Ab9 (IGKV1-110*01; IGKJ1*01). The average number of AA changes in the VL domains is low with a R/S ratio of 1.8 and 2.1 for the FR and CDR, respectively. As for the VH, the residues involved in directly binding the Aβ peptide often remained unmutated [Fig. 1B; underlined residues].
3.2. Recombinant Fab small scale expression and activity assessment by ELISA
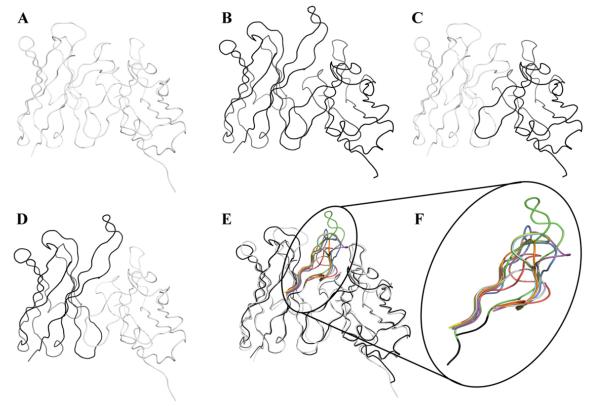
The effect of the VH CDR3 on binding to Aβ was assessed by grafting the CDR3 sequences of well characterized anti-Aβ antibodies (PFA-1, PFA-2, 12A11, 12B4, 10D5 and Ab9) onto the VH frame of our prototype antibodies, WO2 and 20.1. These rFabs are further referred in the text as WO2/PFA-1_HCDR3; WO2/PFA-2 HCDR3; WO2/12A11 HCDR3; WO2/12B4_HCDR3; WO2/10D5_HCDR3; WO2/Ab9_HCDR3; 20.1/PFA-1_HCDR3; 20.1/PFA-2_HCDR3; 20.1/12A11_HCDR3; 20.1/12B4_HCDR3; 20.1/10D5_HCDR3; 20.1/Ab9_HCDR3. In addition, the variable domains (both VH and VL) were shuffled between WO2 and 20.1 [Fig. 2]. The rFab composed of the WO2 VH fused to the 20.1 VL was called WO2_VH/20.1_VL whilst the other consisting of the 20.1 VH fused to the WO2 VL was called 20.1_VH/WO2_VL. Each VH nucleotide sequence was cloned into the pGC Fab vector containing a C-terminal FLAG and 6-HIS tag and expressed in the bacterial periplasm. Production of recombinant antibodies was confirmed by immunoblotting and no significant differences were observed in rFab expression (data not shown). The rFab format allowed us to avoid any avidity effect in the subsequent SPR analysis.
Fig. 2.
Fv ribbon representations of the rFab used in this study: 20.1 (A); WO2 (B); 20.1_VH/WO2_VL (C); WO2_VH/20.1_VL (D); VH CDR3 loops grafted on 20.1 and WO2 (E). Enlargement of the VH CDR3 loops from WO2 (green, PDB ID code 3bkj), 20.1 (red), 10D5 (blue, PDB ID code 3ifo), 12A11 (orange, PDB ID code 3ifl), 12B4 (purple, PDB ID code 3ifp), Ab9 (light blue), PFA-1 (cyan, PDB ID code 2roz) and PFA-2 (yellow, PDB ID code 2row). Codes are from PDB and IMGT/3Dstructure-DB.
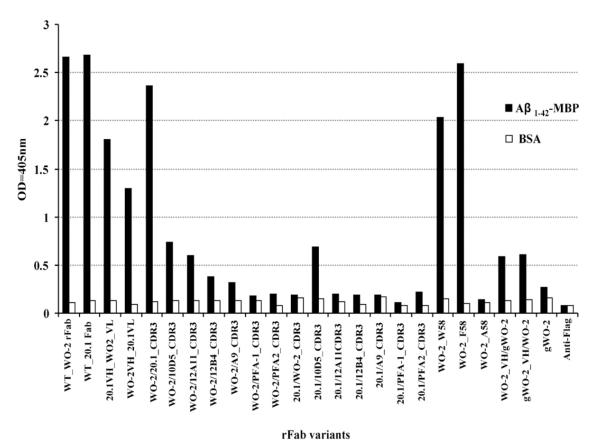
Active clones that bound an Aβ fusion protein (maltose binding protein-Aβ1–42) were identified from crude periplasmic extracts in an ELISA [Fig. 3]. Despite detecting protein bands of the correct size (~50 kDa) for all the rFabs (data not shown), clones WO2/PFA-1_HCDR3; WO2/PFA-2_HCDR3; 20.1/PFA-1_HCDR3, 20.1/PFA-2_HCDR3; 20.1/12A11_HCDR3; 20.1/12B4_HCDR3; 20.1/Ab9_HCDR3; 20.1/WO2_HCDR3 did not exhibit any binding activity in ELISA whereas WO2; 20.1; WO2_VH/20.1_VL; 20.1_VH/WO2_VL; WO2/20.1_HCDR3; WO2/10D5_HCDR3; WO2/12A11_HCDR3; WO2/12B4_HCDR3; WO2/A9_HCDR3; 20.1/10D5_HCDR3 rFabs produced a signal and were selected for a large scale expression and purification and analyzed further by SPR.
Fig. 3.
ELISA of crude periplasmic extract of all recombinant Fabs against Aβ fusion protein (Aβ1–42-MBP) and BSA. The binding activity was measured at 405 nm (each bar represents the average of three data point).
3.3. SPR binding analysis
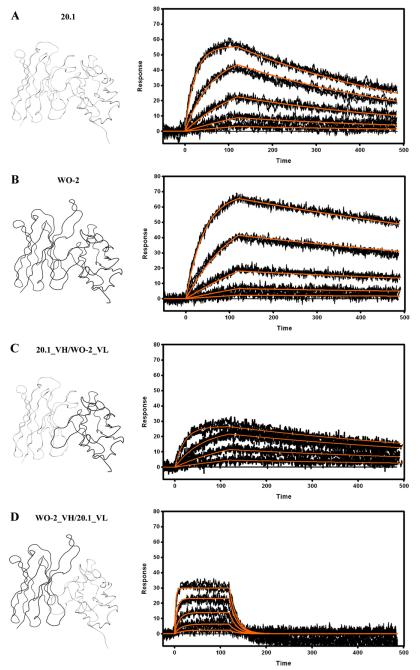
The initial SPR analysis consisted of reversibly capturing the rFab onto the chip surface via previously immobilized human Fab binder and then employing recombinant Aβ1–16–Im7 fusion protein (~14 kDa) as the analyte. We have previously confirmed that Aβ1–16 peptide and Aβ1–16-Im7 fusion protein bound to immobilized WO2 rFab using similar binding parameters (data not shown). The use of the “surrogate Aβ antigen” Aβ1–16-Im7 ensured that a “sufficient” binding (analyte) signal was obtained without having to capture the high levels of rFab required when smaller Aβ1–16 peptide (~2 kDa) was used as the analyte. In fact, we have observed that our ‘human Fab binder-rFab’ capture surfaces drifted significantly when >500 RU of rFab was captured (data not shown), thus making it difficult to estimate binding parameters with Aβ1–16 peptide.
Both WO2 and 20.1 rFab bound strongly to the Aβ1–16-Im7 with KD values in the low nanomolar range [Fig. 4, Table 2]. Interestingly, binding rates of these two rFabs varied significantly. Thus, the association (ka) and dissociation rate constants (kd) were, 5 and 2.5 times faster for 20.1 rFab than for WO2, respectively, which resulted in a 2-fold difference in overall KD. Recombinant Fabs in which the variable domains were shuffled amongst antibodies (VH_WO2/VL_20.1 and VH_20.1/VL_WO2 rFab) were also tested. Unexpectedly, the VL_WO2/VH_20.1 rFab exhibited a similar affinity compared to the parental WO2 and 20.1 Fabs, whereas the VL_20.1/VH_WO2 rFab demonstrated a 15-fold decrease in affinity, mainly due to a much faster dissociation rate constant [Table 2].
Fig. 4.
Surface plasmon resonance measurements of immobilized 20.1 (A) and WO2 (B) Fab, 20.1_VH/WO2_VL (C) and WO2_VH/20.1_VL (D). The rFab were captured via immobilized Human Fab Binder and Aβ1–16-Im7 was injected at concentration of 27–0.333 nM (A), 81–1 nM (B) or 243–3 nM (C and D). Binding responses (black lines) were fit globally to a simple 1:1 interaction model (red lines). Ribbon representations of the Fv, that are part of the Fab and rFab, are shown on the left hand side. Data are representative of three experiments.
Table 2.
Kinetic constants and affinities of various immobilized rFab with Aβ1–16-Im7.
| Analyte | Ligand | ka (×105 M−1 s−1) | kd ( ×10−3 s−1) | KD (nM) |
|---|---|---|---|---|
| Aβ1–16-Im7 | WO2 rFab | 2.84 ± 0.01 | 0.84 ± 0.07 | 3.0 ± 0.2 |
| Aβ1–16-Im7 | 20.1 rFab | 15.7 ± 0.3 | 2.2 ± 0.1 | 1.38 ± 0.04 |
| Aβ1–16-Im7 | 20.1_VH/WO2_VL rFab | 5.4 ± 0.2 | 1.5 ± 0.1 | 2.8 ± 0.2 |
| Aβ1–16-Im7 | WO2_VH/20.1_VL rFab | 15 ± 1 | 61.4 ± 10.9 | 41.0 ± 5.2 |
| Aβ1–16-Im7 | WO2 (Y58>F) mutant rFab | 4.2 ± 0.1 | 1.19 ± 0.03 | 2.8 ± 0.1 |
| Aβ1–16-Im7 | WO2 (Y58>W) mutant rFab | 4.7 ± 0.1 | 16 ± 1 | 34 ± 3 |
| Aβ1–16-Im7 | WO2 (Y58>A) mutant rFab | nd | nd | 4900 ± 0.4 |
| Aβ1–16-Im7 | gWO2_VH/WO2_VL rFab | 3.3 ± 0.1 | 26 ± 1 | 79 ± 2 |
| Aβ1–16-Im7 | WO2_VH/gWO2_VL rFab | 3.9 ± 0.3 | 145 ± 24 | 349 ± 44 |
| Aβ1–16-Im7 | gWO2 rFab | nd | nd | 21300 ± 0.4 |
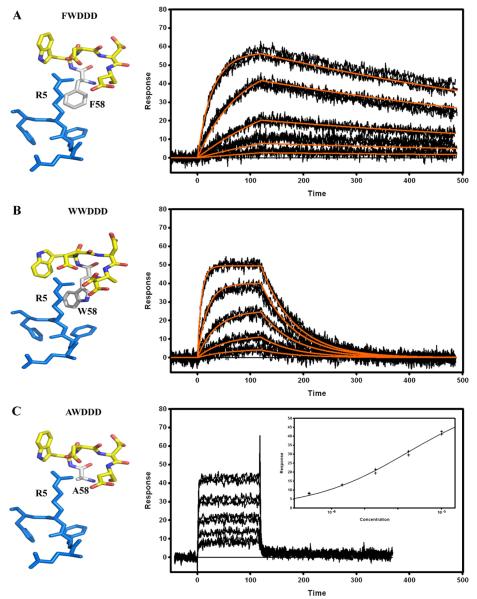
Crystallographic structures of Fab PFA-1, PFA-2, WO2, 12A11, 10D5 and 12B4 in complex with the Aβ1–7 peptide (1DAEFRHD7) containing the Aβ immunodominant epitope indicated that the VH CDR2 region is a key region involved in this interaction and more specifically the 58W(Y)WDD(E)D65 motif [Supplementary information Table 1]. In all the structures reported to date, the residue at position 58 directly interacts with the Aβ peptide residue Arg 5 (R5). The residue 58 is either a tyrosine (Y58) or a tryptophan (W58) but the sequence of the Ab9 revealed that it can also be a phenylalanine (F58). The effect of a single AA change at this position was investigated by replacing the tyrosine (Y58) in the WO2 VH sequence with a tryptophan, a phenylalanine or an alanine (A58) and analyzing binding interactions of these mutants with Aβ1–16 by SPR [Fig. 5]. A ten-fold affinity decrease, mainly driven by a faster dissociation rate constant (kd), was observed for the tryptophan mutant compared to that of the wild type WO2, whereas the affinity of the phenylalanine mutant was not significantly affected [Table 2]. The decrease in affinity observed between the 58YWDDD65 and 58WWDDD65 mutants [Fig. 5A and B] cannot be explained by the extra hydrogen bond formed by the Y58 hydroxyl group with the amine side chain of R5 since no hydroxyl group is present in the 58FWDDD65 motif. Interestingly, the introduction of a very small, non-aromatic and non-charged amino acid residue such as an alanine almost completely abrogated the rFab binding to the Aβ1–16 peptide [Fig. 5C]. By applying the steady state (equilibrium) analysis model to this binding data, we estimated KD to be approximately 5 μM [Table 2]. Hence, the presence of an aromatic amino acid at position 58 in the VH CDR2 appeared to be critical for contacting the Aβ epitope.
Fig. 5.
Surface plasmon resonance measurements of immobilized WO2 (Y58>F) mutant (A), WO2 (Y58>W) mutant (B) and WO2 (Y58>A) mutant (C) rFabs. Recombinant Fabs were captured via immobilized Human Fab Binder and Aβ1–16-Im7 was injected at concentration of 81–1 nM (A), 243–3 nM (B), or 10–0.625 μM (C). Binding responses (black lines) were fit globally to a simple 1:1 interaction model (red lines, A and B). Insert (C): responses at equilibrium were plotted against the analyte concentration and fit to a simple binding isotherm. Stereo views of the 58YWDDD65, 58WWDDD65, 58FWDDD65 and 58AWDDD65 CDR2 motif interacting with the 3EFRH6 part of the Aβ peptide are shown on the left hand side. Data are representative of three experiments.
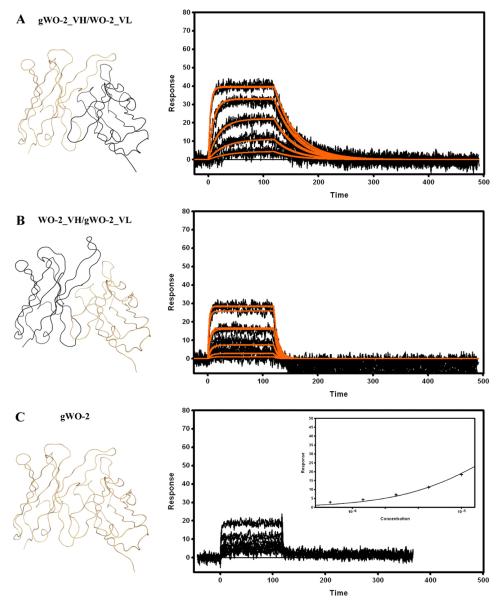
It seemed unlikely that the highly restricted VH/VL pairing observed in the antibodies directed against the N-terminal region of the Aβ peptide occurred randomly. Crystallographic studies have identified the residues involved in the interaction with the 3EFRH6 portion of Aβ. Hence, it is interesting to observe that (1) only the V-KAPPA CDR1 and CDR3, and VH CDR2 and CDR3 are involved [Supplementary information Table 1] and (2) among these regions the V-KAPPA 31HSN34 (CDR1), 108SHVP115 (CDR3) and the VH 58WWDDD65 or 58YWDDD65 (CDR2) motifs are highly conserved and only scarcely mutated from the germline genes. Based on these observations it was hypothesized that an unmutated (i.e. germline) antibody would bind its antigen. To investigate this hypothesis, a rFab (gWO2 rFab) containing germline IGHV, IGHJ, IGHD and IGKV and IGKJ genes (deduced from the analysis of the WO2 DNA sequence) was constructed and expressed. In addition, two other rFab constructs containing the unmutated gWO2_VH paired to the parental WO2_VL (gWO2_VH/WO2_VL) and the parental WO2_VH paired to the unmutated WO2_VL (WO2_VH/gWO2_VL) were also generated. The Aβ1–16 interacting with unmutated rFab was analyzed by flowing Aβ1–16-Im7 over captured rFab on the chip [Fig. 6]. Both the gWO2_VH/WO2_VL [Fig. 6A] and the WO2_VH/gWO2_VL rFab [Fig. 6B] bound Aβ1–16 with affinities that could be estimated by fitting the binding data to a kinetic model [Table 2]. The introduction of either unmutated VH or VL domain clearly destabilized the WO2 rFab/Aβ complex. The weaker affinities of both of these constructs compared to the parental affinities were primarily driven by a faster dissociation rate constant (kd). Finally, the affinity of the gWO2 rFab for Aβ1–16 was low [Fig. 6C] and could only be approximated by applying a steady-state binding model to the data in a process yielding a KD of 21 μM [Table 2].
Fig. 6.
Surface plasmon resonance measurements of immobilized gWO2_VH/WO2_VL (A), WO2_VH/gWO2_VL (B) and gWO2 (C) rFabs. Recombinant Fabs were captured via immobilized Human Fab Binder and Aβ1–16-Im7 was injected at concentration of 729–3 nM (A and B), or 10–0.625 μM (C). Binding responses (black lines) were fit globally to a simple 1:1 interaction model (red lines, A and B). Insert (C): responses at equilibrium were plotted against the analyte concentration and fit to a simple binding isotherm. Ribbon representations of the Fv that are part of the rFab are shown on the left hand side. Data are representative of three experiments.
Further SPR assays involved analysis of rFab (in particular VH CDR3 loop replacements) for binding to various Aβ forms. Due to obvious avidity effects, Aβ multimeric species (Aβ1–42 LMW and HMW oligomers) could not be analyzed in the capture assay described in the previous sections. For this reason, the second SPR analysis strategy involved immobilizing biotinylated Aβ1–16 peptide and biotinylated Aβ1–42 oligomers onto a NeutrAvidin chip and then running various rFabs as analytes. Only those VH CDR3 loop graft rFabs identified as positive binders in ELISA (see previous section) were analyzed in these direct SPR assays [Supplementary information Fig. 2]. For comparative purposes, WO2 and 20.1 Fabs as well as the two chain-shuffled WO2/gWO2 rFabs were utilized in this assay as positive controls. Binding parameters obtained for these positive controls interacting with immobilized Aβ1–16-biotin [Table 3] corroborated well with those obtained for Aβ1–16-Im7 utilized in the capture assay [Table 2], thus validating the fusion protein as a good “surrogate Aβ antigen”.
Table 3.
Kinetic constants and affinities of various rFab with immobilized biotin-Aβ1–16 (bAβ1–16) and biotinylated Aβ1–42 HMW oligomers (bAβ1–42-HMW oligomers).
| Analyte | Ligand | ka (×105 M−1 s−1) | kd (×10−3 s−1) | KD (nM) |
|---|---|---|---|---|
| WO2 rFab | bAβ1–16 | 3.65 | 1.02 | 2.7 |
| 20.1 rFab | bAβ1–16 | 4.6 | 1.4 | 3.0 |
| 20.1_VH/WO2_VL rFab | bAβ1–16 | 0.76 | 0.65 | 8.5 |
| WO2_VH/20.1_VL rFab | bAβ1–16 | 1.44 | 17.8 | 123.8 |
| WO2/20.1_HCDR3 rFab | bAβ1–16 | 3.57 | 23.9 | 66.9 |
| WO2/10D5_HCDR3 rFab | bAβ1–16 | 0.63 | 9.74 | 153.9 |
| WO2/12A11_HCDR3 rFab | bAβ1–16 | 1.71 | 44.9 | 262.5 |
| WO2/A9_HCDR3 rFab | bAβ1–16 | 0.08 | 33.4 | 4200 |
| WO2/12B4_HCDR3 rFab | bAβ1–16 | 0.01 | 69 | 6500 |
| 20.1/10D5_HCDR3 rFab | bAβ1–16 | 0.62 | 5.45 | 87.9 |
| gWO2_VH/WO2_VL rFab | bAβ1–16 | 1.07 | 17.1 | 162 |
| WO2_VH/gWO2_VL rFab | bAβ1–16 | 1.52 | 35.1 | 231 |
| WO2 rFab | bAβ1–42-HMW oligomers | 3.65 | 0.89 | 2.3 |
| 20.1 rFab | bAβ1–42-HMW oligomers | 2.29 | 0.63 | 2.8 |
| 20.1_VH/WO2_VL rFab | bAβ1–42-HMW oligomers | 0.63 | 0.57 | 9.0 |
| WO2_VH/20.1_VL rFab | bAβ1–42-HMW oligomers | 2.19 | 12.7 | 58.1 |
| WO2/20.1_HCDR3 rFab | bAβ1–42-HMW oligomers | 3.18 | 11.2 | 35.28 |
| WO2/10D5_HCDR3 rFab | bAβ1–42-HMW oligomers | 0.61 | 5.46 | 89.24 |
| WO2/12A11_HCDR3 rFab | bAβ1–42-HMW oligomers | 1.33 | 26.1 | 135.3 |
| WO2/A9_HCDR3 rFab | bAβ1–42-HMW oligomers | nd | nd | nd |
| WO2/12B4_HCDR3 rFab | bAβ1–42-HMW oligomers | nd | nd | nd |
| 20.1/10D5_HCDR3 rFab | bAβ1–42-HMW oligomers | 0.74 | 4.67 | 62.7 |
| gWO2_VH/WO2_VL rFab | bAβ1–42-HMW oligomer | 1.30 | 15.7 | 121 |
| WO2_VH/gWO2_VL rFab | bAβ1–42-HMW oligomer | 2.18 | 28.5 | 131 |
Significantly, no major binding differences were observed for all analyzed rFabs binding to the immobilized biotin-Aβ1–16 peptide and biotinylated Aβ1–42 HMW oligomers [Supplementary information Fig. 2; Table 3]. However, none of these rFabs bound to the immobilized biotinylated Aβ1–42 LMW oligomers [Supplementary information Fig. 2]. Binding data further indicated that none of the VH CDR3 loop amino acid changes improved the binding affinity when compared with that of the parental WO2 and 20.1 Fab [Table 3]. This observation was expected since in previous studies, none of the mAbs from which the VH CDR3 were derived, displayed a stronger affinity than the 20.1 and WO2 mAbs (Gardberg et al., 2007; Basi et al., 2010). In fact, in all cases each VH CDR3 replacement generated significantly lower affinities compared with the parental clone. Surprisingly, the VH CDR3 of 20.1 on the WO2 framework (WO2/20.1_HCDR3 rFab) produced the highest affinity (KD of ~66 nM) amongst all the VH CDR3 rFab variants. In contrast when the VH CDR3 of WO2 was grafted onto the 20.1 framework the resulting rFab construct exhibited no binding Aβ1–16. The effect of shuffling 10D5_HCDR3 onto different VH frameworks including that of 20.1 and WO2 did not significantly affect the binding affinity for the Aβ1–16 (KD ~150 and ~88 nM, respectively). These affinities are comparable to those reported for the parental 10D5 antibody (KD ~55 nM) in a similar assay (Basi et al., 2010). Moreover, the 10D5_HCDR3 region is the only one that was functional when grafted into the 20.1 framework. Both WO2/12B4_HCDR3 and WO2/A9_HCDR3 loop graft rFabs bound to immobilized Ab1–16 with very low affinities, which seems to be due to an unusually low association rate constant (ka =8 × 10−3 and 1 × 10−3 M−1 s−1; Table 3). Considering the overall similarity of these two constructs to the parental WO2 Fab, we speculated that active concentrations of these Fab preparations were low, which then resulted in low ka estimates.
4. Discussion
4.1. Genetic analysis revealed a restricted V-genes usage and VH/VL pairing
In this report we described the genetic analysis of mAbs directed against the linear immunodominant B cell epitope of the Aβ peptide 1DAEFRHD7. The study was based on twelve mAb sequences generated in different research groups by immunizing mice with various Aβ forms (monomeric Aβ peptide, soluble Aβ oligomers or ADDLs, protofibrils and fibrils). It was surprising to observe that all these mAbs were characterized by a highly restricted V gene usage and VH/VL pairing. Such restrictions in V gene usage or in VH/VL pairing have been reported in antibodies against certain allergens (Persson et al., 2008), haptens (Diaw et al., 1999), viral (McLean et al., 2005; Zhang et al., 2006), and bacterial antigens (Fernández-Sánchez et al., 2009). It has been postulated that structural constraints could be involved in this phenomenon (Hougs et al., 1999; Andersen et al., 2007; Thomson et al., 2008).
We suspected other mouse antibodies directed against the immunodominant B cell epitope of the Aβ peptide to share the same restrictive variable domain pairing. Hence, we also retrieved the sequence of an anti-Aβ antibody (mAb158) which is reputed to be selective for protofibrils (Gellerfors et al., 2009) and shares the same VL as the antibodies described here. This antibody has been obtained after immunizing mice with Aβ1–42 protofibrils. Likewise, anti-Aβ antibodies isolated by the Intracellular Antibody Capture Technology (IACT) from scFv antibody libraries have been reported to share identity with the PFA-1 and PFA-2 mAbs in their VL genes only (Meli et al., 2009). Meli et al. (2009) have previously suggested creating a database of anti-Aβ antibody sequences in order to link their CDR sequences with their binding proprieties. The work presented here represents another significant step in that direction. It is also known that in rabbit (Miller et al., 2003), dog (Vasilevko et al., 2010), non-human primate Lemere et al., 2004), and human (Lee et al., 2005), antibodies mainly directed against the N-terminal region of Aβ are raised in response to immunization with Aβ fibrils. However, the immunodominant epitope appeared to be different in those four species and is rather located in the 1DAEFR5 region of the Aβ peptide instead of the 3EFRH6 region in mice. An important observation is that antibodies developed in those species did not recognize the full-length APP as opposed to those developed in mice. To our knowledge, no systematic sequencing of the antibodies has been performed to demonstrate if such a restriction occurred in those species.
4.2. rFab variants recognize the Aβ1–16 peptide and the HMW oligomers but not the LMW oligomers
Basi et al. (2010) recently observed that the 12A11, 12B4, 10D5, PFA-1, PFA-2 and WO2 mAbs essentially vary the most in their VH CDR3. More interestingly, they reported that despite their primary sequences (VH and VL) and structural similarities, these mAbs did not recognize oligomeric forms of Aβ and restore cognitive functions in AD mice with the same efficacy. The authors hypothesized that the differences observed in the VH CDR3 loop structure may be responsible for preferential binding of the antibody to different Aβ forms (monomers, soluble oligomers, protofibrils or fibrils).
In order to understand the involvement of this region in Aβ recognition, we designed a number of constructs in which the VH CDR3 loop of antibodies 20.1 and WO2 was replaced by that of previously described antibodies by other groups (12A11, 12B4, 10D5, PFA-1, PFA-2 and Ab9). Interestingly, despite the high homology among the VH sequences of all these antibodies, the affinity for Aβ was either abolished or significantly reduced compared to that of the parental 20.1 and WO2 Fab in most of the VH CDR3 rFab. In the case of the 20.1 frame, only the grafting of the 10D5_HCDR3 loop resulted in a functional rFab, whereas, the grafting of the 20.1_HCDR3, 10D5_HCDR3, 12A11_HCDR3, 12B4_HCDR3, Ab9_HCDR3 loops onto WO2 frame produced functional binders. These data indicate that the WO2 frame can accommodate a variety of HCDR3s, whereas 20.1 is not as tolerant. However, it was not possible to draw any correlation involving the degree of identity of 20.1 or WO2 VH with the VH from which the CDR3 is originally derived and the rate of success of the graft. Hence, it appeared that the correct conformation and stabilization of the VH CDR3 in these mAbs is dependent on specific mutations that occur in the framework regions during the antibody maturation. This finding is consistent with that of others in the field that have demonstrated that the contribution of framework residues can significantly affect antibody affinity, in particular, through mutation of non-contact residues in the periphery of the paratope (Thomson et al., 2008).
Recent evidence suggests that the degree of dementia in AD correlates with the presence of soluble oligomeric forms of the Aβ peptide rather than the presence of plaques that consist of insoluble Aβ fibrils. Synthetic biotinylated Aβ oligomers can be formed in vitro at 4 °C and separated by size exclusion chromatography into HMW-oligomers and LMW-oligomer fractions (Chromy et al., 2003). The affinity of the WO2/20.1_HCDR3, WO2/10D5_HCDR3, WO2/12A11_HCDR3 and 20.1/10D5_HCDR3 rFab variants against different Aβ forms was assessed using SPR. Whilst all variants bound the linear Aβ1–16 peptide and the HMW oligomers immobilized on a chip with a similar affinity, they failed to recognize and bind LMW oligomers. This possibly indicated that the 3EFRH6 linear epitope is not accessible in the LMW oligomers.
4.3. An aromatic residue at position H58 is essential in the recognition
The importance of Y58 in WO2 rFab for binding Aβ was investigated by generating amino acid changes at this position. The results indicated that the interaction with R5 through an aromatic AA is critical for recognition and binding. Given that the AA sequence of the mouse Aβ1–42 differs from the human one at three positions (5, 10 and 13), it is not surprising that (i) the mouse antibodies are directed against the N-terminal region of Aβ, and (ii) the R5 is a highly immunogenic AA and thus is a crucial residue in this antibody/antigen interaction.
4.4. Fab variants based on germline V-genes bind to Aβ with a low affinity
Observations that mouse antibodies directed against the immunodominant portion of Aβ utilized a restricted set of germline IGHV and IGKV genes and that the V-KAPPA CDR1 and VH CDR2 germline-encoded residues make crucial contact with the antigen indicate that the unmutated domains already possess a good starting affinity for the antigen. This is further supported by the low replacement to silent mutation average occurring in the IGHV and IGLV genes. To test this hypothesis, comparative studies were conducted between gWO2 rFab containing unmutated sequences and rFab in which unmutated and affinity matured V domains were shuffled. The affinities of these constructs for the Aβ1–16 peptide clearly indicated that mutations in both WO2 VH and WO2 VL domains significantly contributed to the ~6000-fold increase binding affinity in the mature WO2 Ab as compared to the germline Ab. As it has been well described for a family of antibodies neutralizing the human cytomegalovirus (hCMV), it is probable that the unmutated V genes sculpt the binding site for the 3EFRH6 epitope and that structural constraint allows only a limited set of mutated V domains to bind. However, this hypothesis can only be confirmed by crystallographic analysis by comparing the structure of the parental WO2 and gWO2 antibodies in complex with Aβ.
5. Conclusion
Monoclonal antibodies specifically targeting a conformational epitope are of great interest in order to develop an effective passive immunotherapeutic strategy. Over the past 5 years, many groups (Lee et al., 2006; Lambert et al., 2007) claimed to have isolated such antibodies. However, based on our data, we do believe that most of these antibodies, especially the one obtained after immunizing mice with Aβ1–42 soluble oligomers are still directed against the linear immunodominant B cell epitope and therefore are not truly specific but rather selective for the oligomeric conformation. In order to generate a genuine conformation specific monoclonal mouse antibody and eliminate the humoral response bias towards the immunodominant epitope, new immunizing strategies need to be considered. The most obvious approach is to form Aβ oligomers lacking the 3EFRH6 epitope (N-terminally truncated Aβ oligomers) and to immunize the mice with such Aβ forms. Interestingly, it has been shown that N-terminally truncated Aβ forms are present in the early stage of the AD pathology and represent a large proportion of all Aβ. These forms exhibit enhanced aggregation properties and induce significant neuritic degeneration and cell death in vitro (Pike et al., 1995; Jang et al., 2010). Another approach is to screen human antibody libraries using either phage, ribosome or yeast display technologies. It offers significant advantages including: (i) elimination of the immunization step and the antibody bias resulting from the humoral immune response and (ii) ability to specifically screen and select for antibodies which only recognise a single conformational form of Aβ in soluble oligomeric or fibrils. Recently this technique has been successfully applied to isolate a human scFv that bound specifically to soluble oligomeric Aβ (Zameer et al., 2008; Medecigo et al., 2010).
Supplementary Material
Acknowledgments
The authors would like to acknowledge Ms Meghan Hattarki, Lesley Pearce and Larissa Doughty for their technical assistance, Drs Stewart Nuttall, Rebecca Nisbet and Jo Caine for providing us the Aβ fusion proteins.
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molimm.2010.09.012.
References
- Acton P, An Z, Bett AJ, Breese R, Chang L, Dodson EC, Kinney G, Klein W, Lambert MP, Liang X, Shughrue P, Strohl WR, Viola K. No. US 20060228349 US Patent.
- Agadjanyan MG, Ghochikyan A, Petrushina I, Vasilevko V, Movsesyan N, Mkrtichyan M, Saing T, Cribbs DH. Prototype Alzheimer’s disease vaccine using the immunodominant B cell epitope from beta-amyloid and promiscuous T cell epitope pan HLA DR-binding peptide. J. Immunol. 2005;174:1580–1586. doi: 10.4049/jimmunol.174.3.1580. [DOI] [PubMed] [Google Scholar]
- Agadjanyan MG, Cribbs DH. Active and passive Abeta-immunotherapy: preclinical and clinical studies and future directions: part I. CNS Neurol. Disord. Drug Targets. 2009;8:1–6. doi: 10.2174/187152709787601849. [DOI] [PubMed] [Google Scholar]
- Andersen PS, Haahr-Hansen M, Coljee VW, Hinnerfeldt FR, Varming K, Bregenholt S, Haurum JS. Extensive restrictions in the VH sequence usage of the human antibody response against the Rhesus D antigen. Mol. Immunol. 2007;44:412–422. doi: 10.1016/j.molimm.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Arimon M, Díez-Pérez I, Kogan MJ, Durany N, Giralt E, Sanz F, Fernández-Busquets X. Fine structure study of Abeta 1–42 fibrillogenesis with atomic force microscopy. FASEB J. 2005;10:1344–1346. doi: 10.1096/fj.04-3137fje. [DOI] [PubMed] [Google Scholar]
- Bard F, Cannon C, Barbour R, Burke RL, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Lieberburg I, Motter R, Nguyen M, Soriano F, Vasquez N, Weiss K, Welch B, Seubert P, Schenk D, Yednock T. Peripherally administered antibodies against amyloid β-peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nat. Med. 2000;6:916–919. doi: 10.1038/78682. [DOI] [PubMed] [Google Scholar]
- Bard F, Barbour R, Cannon C, Carretto R, Fox M, Games D, Guido T, Hoenow K, Hu K, Johnson-Wood K, Khan K, Kholodenko D, Lee C, Lee M, Motter R, Nguyen M, Reed A, Schenk D, Tang P, Vasquez N, Seubert P, Yednock T. Epitope and isotype specificities of antibodies to β-amyloid peptide for protection against Alzheimer’s disease-like neuropathology. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2023–2028. doi: 10.1073/pnas.0436286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi GS, Feinberg H, Oshidari F, Anderson J, Barbour R, Baker J, Comery TA, Diep L, Gill D, Johnson-Wood K, Goel A, Grantcharova K, Lee M, Li J, Partridge A, Griswold-Prenner I, Piot N, Walker D, Widom A, Pangalos MN, Seubert P, Jacobsen JS, Schenk D, Weis WI. Structural correlates of antibodies associated with acute reversal of A-beta related behavioral deficits in a mouse model of Alzheimer’s disease. J. Biol. Chem. 2010;285:3417–3427. doi: 10.1074/jbc.M109.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravman T, Bronner V, Lavie K, Notcovich A, Papalia GA, Myszka DG. Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 2006;358:281–288. doi: 10.1016/j.ab.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucl. Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine J, Volitakis I, Cherny R, Varghese J, Macreadie I. Abeta produced as a fusion to maltose binding protein can be readily purified and stably associates with copper and zinc. Protein Pept. Lett. 2007;14:83–86. doi: 10.2174/092986607779117263. [DOI] [PubMed] [Google Scholar]
- Chromy BA, Nowak RJ, Lambert MP, Viola KL, Chang L, Velasco PT, Jones BW, Fernandez SJ, Lacor PN, Horowitz P, Finch CE, Krafft GA, Klein WL. Self-assembly of Abeta(1–42) into globular neurotoxins. Biochemistry. 2003;42:12749–12760. doi: 10.1021/bi030029q. [DOI] [PubMed] [Google Scholar]
- Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- Diaw L, Siwarski D, Coleman A, Kim J, Jones GM, Dighiero G, Huppi K. Restricted immunoglobulin variable region (IgV) gene expression accompanies secondary rearrangements of light chain IgV genes in mouse plasmacytomas. J. Exp. Med. 1999;190:1405–1416. doi: 10.1084/jem.190.10.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenmann F, Kaas Q, Lefranc M-P. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucl. Acids Res. 2010;38:D301–D307. doi: 10.1093/nar/gkp946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Sánchez A, García-Ocaña M, de los Toyos JR. Mouse monoclonal antibodies to pneumococcal C-polysaccharide backbone show restricted usage of VH-DH-JH gene segments and share the same kappa chain. Immunol. Lett. 2009;123:125–131. doi: 10.1016/j.imlet.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Balass M, Katchalski-Katzir E, Solomon B. High affinity binding of monoclonal antibodies to the sequential epitope EFRH of beta-amyloid peptide is essential for modulation of fibrillar aggregation. J. Neuroimmunol. 1999;95:136–142. doi: 10.1016/s0165-5728(99)00003-x. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Katz O, Solomon B. Immunization against Alzheimer’s beta-amyloid plaques via EFRH phage administration. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11455–11459. doi: 10.1073/pnas.97.21.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardberg AS, Dice LT, Ou S, Rich RL, Helmbrecht E, Ko J, Wetzel R, Myszka DG, Patterson PH, Dealwis C. Molecular basis for passive immunotherapy of Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15659–15664. doi: 10.1073/pnas.0705888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellerfors P, Lannfelt L, Sehlin D, Pettersson F. Ekholm, Englund H. No. US 2009/0258009 A1 US Patent.
- Golde TE, Das P, Jansen-west KR, Levites YR. No. US20100104577 US Patent.
- Hougs L, Juul L, Svejgaard A, Barington T. Structural requirements of the major protective antibody to Haemophilus influenzae type b. Infect. Immun. 1999;67:2503–2514. doi: 10.1128/iai.67.5.2503-2514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s disease and Down syndrome. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus C, Pearson J, McLaurin J, Mathews PM, Jiang Y, Schmidt SD, Chishti MA, Horne P, Heslin D, French J, Mount HT, Nixon RA, Mercken M, Bergeron C, Fraser PE, St George-Hyslop P, Westaway D. Aβ peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer’s disease. Nature. 2000;408:979–982. doi: 10.1038/35050110. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, et al. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kleanthous C, Hemmings AM, Moore GR, James R. Immunity proteins and their specificity for endonuclease colicins: telling right from wrong in protein–protein recognition. Mol. Microbiol. 1998;2:227–233. doi: 10.1046/j.1365-2958.1998.00811.x. [DOI] [PubMed] [Google Scholar]
- Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MP, Velasco PT, Chang L, Viola KL, Fernandez S, Lacor PN, Khuon D, Gong Y, Bigio EH, Shaw P, De Felice FG, Krafft GA, Klein WL. Monoclonal antibodies that target pathological assemblies of Abeta. J. Neurochem. 2007;100:23–35. doi: 10.1111/j.1471-4159.2006.04157.x. [DOI] [PubMed] [Google Scholar]
- Lee M, Bard F, Johnson-Wood K, Lee C, Hu K, Griffith SG, Black RS, Schenk D, Seubert P. Abeta42 immunization in Alzheimer’s disease generates Abeta N-terminal antibodies. Ann. Neurol. 2005;58:430–435. doi: 10.1002/ana.20592. [DOI] [PubMed] [Google Scholar]
- Lee EB, Leng LZ, Zhang B, Kwong L, Trojanowski JQ, Abel T, Lee VM. Targeting amyloid-beta peptide (Abeta) oligomers by passive immunization with a conformation-selective monoclonal antibody improves learning and memory in Abeta precursor protein (APP) transgenic mice. J. Biol. Chem. 2006;281:4292–4299. doi: 10.1074/jbc.M511018200. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT®, the international ImMunoGeneTics information system®. Nucl. Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemere CA, Beierschmitt A, Iglesias M, Spooner ET, Bloom JK, Leverone JF, Zheng JB, Seabrook TJ, Louard D, Li D, Selkoe DJ, Palmour RM, Ervin FR. Alzheimer’s disease abeta vaccine reduces central nervous system abeta levels in a non-human primate, the Caribbean vervet. Am. J. Pathol. 2004;165:283–297. doi: 10.1016/s0002-9440(10)63296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean GR, Olsen OA, Watt IN, Rathanaswami P, Leslie KB, Babcook JS, Schrader JW. Recognition of human cytomegalovirus by human primary immunoglobulins identifies an innate foundation to an adaptive immune response. J. Immunol. 2005;174:4768–4778. doi: 10.4049/jimmunol.174.8.4768. [DOI] [PubMed] [Google Scholar]
- Medecigo M, Manoutcharian K, Vasilevko V, Govezensky T, Munguia ME, Becerril B, Luz-Madrigal A, Vaca L, Cribbs DH, Gevorkian G. Novel amyloid-beta specific scFv and VH antibody fragments from human and mouse phage display antibody libraries. J. Neuroimmunol. 2010;223:104–114. doi: 10.1016/j.jneuroim.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli G, Visintin M, Cannistraci I, Cattaneo A. Direct in vivo intracellular selection of conformation-sensitive antibody domains targeting Alzheimer’s amyloid-beta oligomers. J. Mol. Biol. 2009;387:584–606. doi: 10.1016/j.jmb.2009.01.061. [DOI] [PubMed] [Google Scholar]
- Miles LA, Wun KS, Crespi GAN, Fodero-Tavoletti MT, Galatis D, Bagley CJ, Beyreuther K, Masters CL, Cappai R, McKinstry WJ, Barnham KJ, Parker MW. Amyloid-β–anti-amyloid-β complex structure reveals an extended conformation in the immunodominant B-cell epitope. J. Mol. Biol. 2008;377:181–192. doi: 10.1016/j.jmb.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Miller DL, Currie JR, Mehta PD, Potempska A, Hwang YW, Wegiel J. Humoral immune response to fibrillar beta-amyloid peptide. Biochemistry. 2003;42:11682–11692. doi: 10.1021/bi030100s. [DOI] [PubMed] [Google Scholar]
- Orgogozo JM, Gilman S, Dartigues JF, Laurent B, Puel M, Kirby LC, Jouanny P, Dubois B, Eisner L, Flitman S, Michel BF, Boada M, Frank A, Hock C. Subacute meningoencephalitis in a subset of patients with AD after Aβ42 immunization. Neurology. 2003;61:46–54. doi: 10.1212/01.wnl.0000073623.84147.a8. [DOI] [PubMed] [Google Scholar]
- Persson H, Flicker S, Sadegh MK, Greiff L, Valenta R, Ohlin M. A common idiotype in IgE and its relation to recognition of the grass pollen allergen Phl p 2. Mol. Immunol. 2008;45:2715–2720. doi: 10.1016/j.molimm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Pike CJ, Overman MJ, Cotman CW. Amino-terminal deletions enhance aggregation of beta-amyloid peptides in vitro. J. Biol. Chem. 1995;270:23895–23898. doi: 10.1074/jbc.270.41.23895. [DOI] [PubMed] [Google Scholar]
- Robert R, Dolezal O, Waddington L, Hattarki MK, Cappai R, Masters CL, Hudson PJ, Wark KL. Engineered antibody intervention strategies for Alzheimer’s disease and related dementias by targeting amyloid and toxic oligomers. Protein Eng. Des. Sel. 2009;22:199–208. doi: 10.1093/protein/gzn052. [DOI] [PubMed] [Google Scholar]
- Robert R, Streltsov VA, Newman J, Pearce LA, Wark KL, Dolezal O. Germline humanization of a murine Abeta antibody and crystal structure of the humanized recombinant Fab fragment. Protein Sci. 2010;19:299–308. doi: 10.1002/pro.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Liao Z, Lieberburg I, Motter R, Mutter L, Soriano F, Shopp G, Vasquez N, Vandevert C, Walker S, Wogulis M, Yednock T, Games D, Seubert P. Immunization with amyloid-β attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Translating cell biology into therapeutic advances in Alzheimer’s disease. Nature. 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- Shen W, Biere-Citron AL. No. US2008/0292639 A1 US Patent.
- Solomon B, Koppel R, Hanan E, Katzav T. Monoclonal antibodies inhibit in vitro fibrillar aggregation of the Alzheimer β-amyloid peptide. Proc. Natl. Acad. Sci. U. S. A. 1996;93:452–455. doi: 10.1073/pnas.93.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon B, Koppel R, Frankel D, Hanan-Aharon E. Disaggregation of Alzheimer β-amyloid by site-directed mAb. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4109–4112. doi: 10.1073/pnas.94.8.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson CA, Bryson S, McLean GR, Creagh AL, Pai EF, Schrader JW. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27:2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilevko V, Pop V, Kim HJ, Saing T, Glabe CC, Milton S, Barrett EG, Cotman CW, Cribbs DH, Head E. Linear and conformation specific antibodies in aged beagles after prolonged vaccination with aggregated Abeta. Neurobiol Dis. 2010;39:301–310. doi: 10.1016/j.nbd.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- Whitelegg NR, Rees AR. WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng. 2000;13:819–824. doi: 10.1093/protein/13.12.819. [DOI] [PubMed] [Google Scholar]
- Wilcock DM, Alamed J, Gottschall PE, Grimm J, Rosenthal A, Pons J, Ronan V, Symmonds K, Gordon MN, Morgan D. Deglycosylated anti-amyloid-β antibodies eliminate cognitive deficits and reduce parenchymal amyloid with minimal vascular consequences in aged amyloid precursor protein transgenic mice. J. Neurosci. 2006;26:5340–5346. doi: 10.1523/JNEUROSCI.0695-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod M. Yousfi, Giudicelli V, Chaume D, Lefranc MP. IMGT/junction analysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20(Suppl. 1):i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- Zameer A, Kasturirangan S, Emadi S, Nimmagadda SV, Sierks MR. Antioligomeric Abeta single-chain variable domain antibody blocks Abeta-induced toxicity against human neuroblastoma cells. J. Mol. Biol. 2008;384:917–928. doi: 10.1016/j.jmb.2008.09.068. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zharikova D, Mozdzanowska K, Otvos L, Gerhard W. Fine specificity and sequence of antibodies directed against the ectodomain of matrix protein 2 of influenza A virus. Mol. Immunol. 2006;43:2195–2206. doi: 10.1016/j.molimm.2005.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.