Abstract
Multi-tier strategies exist in many biochemical processes to ensure a maximal fidelity of the reactions. In this review, we focus on the two-tier quality control strategy that ensures the quality of the products of the pre-mRNA splicing reactions catalyzed by the spliceosome. The first step in the quality control process relies on kinetic proofreading mechanisms that are internal to the spliceosome and that are performed by ATP-dependent RNA helicases. The second quality control step, spellchecking, involves recognition of unspliced pre-mRNAs or aberrantly spliced mRNAs that have escaped the first proofreading mechanisms, and subsequent degradation of these molecules by degradative enzymes in the nucleus or in the cytoplasm. This two-tier quality control strategy highlights a need for high fidelity and a requirement for degradative activities that eliminate defective molecules. The presence of multiple quality control activities during splicing underscores the importance of this process in the expression of genetic information.
Keywords: exosome, fidelity, nonsense mediated decay, RNA degradation, splicing ATPase
INTRODUCTION
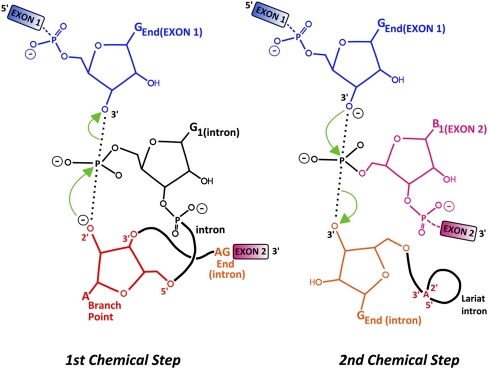
The problem of the fidelity of chemical reactions involving nucleic acids is affected by the catalytic steps of these reactions and by the substrates involved. In the case of pre-mRNA splicing, the splicing machinery has to assemble close to, recognize, and correctly position the substrates of the splicing reactions that will be paired, which are the GU dinucleotide at the 5′ splice site, the branchpoint adenosine, and the AG dinucleotide at the 3′ splice site (Fig. 1; for review, see Wahl et al. 2009). Aside from these three direct substrates of the chemical reaction, the polypyrimidine tract and exonic sequences also contribute to splice sites recognition (Wahl et al. 2009). The substrates recognized by the spliceosome can be located far away from each other on the RNA polymer. In addition, the spliceosome catalyzes two successive chemical steps (Fig. 1), multiplying the risk of generating an incorrect final product if these steps are left unchecked. Thus, the spliceosome faces a daunting task in recognizing and aligning the proper substrates, because of the relatively low sequence complexity of its substrates and because these substrates can be difficult to recognize among many other similar sequences. Indeed, in higher eukaryotes the sequences of the major splice sites and branchpoints used are frequently divergent from those of the consensus sequences, raising the question of how these major sites are selected and recognized as opposed to the incorrect ones. We will not detail here the molecular mechanisms responsible for proper recognition of these signals by the spliceosome. Instead, we will focus on the mechanisms that have evolved to ensure that mistakes made by the spliceosome are corrected. A “mistake” or “error” can be defined as the generation of any noncanonical molecular product, which either results in a nonfunctional protein product, or which induces quality control mechanisms to take action. Naturally, the process of evolution and natural selection owes its progression to numerous “mistakes,” which have proven to create more favorable conditions for their respective organisms, and indeed, some cases of nonproductive splicing can be used to regulate the expression of specific genes (Lareau et al. 2007; Ni et al. 2007; see below). The molecular consequences of a mistake may be considered more limited for an mRNA transcript than for a replicating chromosome, since the mistake is not propagated to the next generation. However, many noncanonical species can be detrimental for the cell, justifying the parallel evolution of quality control mechanisms. Indeed, if an aberrant mRNA is translated highly efficiently, it can generate multiple molecules of incorrect polypeptides, which may interfere with cellular processes. For instance, aberrant or incorrect splicing has been linked to an increasing number of diseases and pathological processes (for review, see Cooper et al. 2009; Ward and Cooper 2010), highlighting the requirement for quality control during splicing. We review below how a two-tier quality control strategy has been selected to ensure the accuracy of splicing reactions and the degradation of molecules that have escaped the fidelity process intrinsic to the spliceosome.
FIGURE 1.
Nuclear pre-mRNA splicing chemical steps.
KINETIC PROOFREADING ACTIVITIES MEDIATED BY SPLICEOSOMAL ATPASES PROVIDE A FIRST QUALITY CONTROL MECHANISM TO LIMIT THE RATE OF MISTAKES MADE DURING SPLICING
The spliceosome has evolved multiple mechanisms to select splicing substrates and to recognize and reject complexes that have assembled onto suboptimal substrates. These built-in proofreading mechanisms have to be complex, and require a multistep process because of the dynamic nature of the spliceosome and of its assembly on the splicing substrates (Wahl et al. 2009; Abelson et al. 2010), and because at least three substrates are involved in two successive chemical steps (Figs. 1, 2). While the initial mechanisms of splice site recognition involve base-pairing recognition with small nuclear RNAs contained in the spliceosome (Staley and Guthrie 1998; Wahl et al. 2009), some proteins also participate directly in the recognition (Chua and Reed 1999; Du and Rosbash 2002; Soares et al. 2006; Wahl et al. 2009). However, despite the presence of multiple mechanisms that promote the selection of the correct substrates, assembly of the spliceosome onto suboptimal or incorrect splicing substrates (aberrant splice site or branchpoint) can occur, especially in the case of metazoan mRNAs for which splice sites and branchpoints often diverge from the consensus.
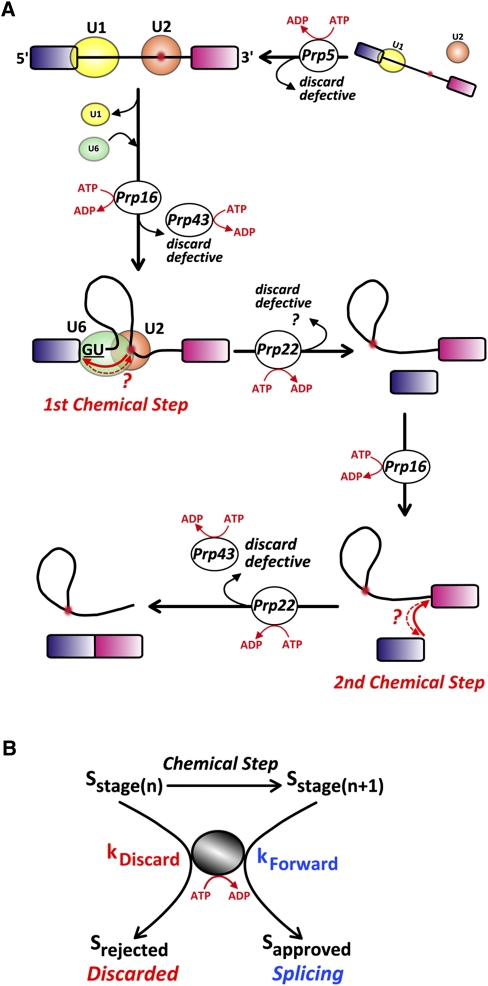
FIGURE 2.
Internal proofreading mechanisms in pre-mRNA splicing. (A) Involvement of splicing ATPases in multiple checkpoints of the splicing pathway. Shown are the spliceosome assembly pathway and the ATPase activities that are involved in checkpoints for incorrectly selected branchpoints (Prp5p, Prp16p) or splice sites (Prp16p, Prp22p, Prp43p), and which trigger a discard pathway. In the case of Prp22, a possible checkpoint of the 5′ splice site is indicated based on genetic evidence that Prp22p mutants can also promote the use of mutated 5′ splice sites. (B) Kinetic proofreading by splicing ATPases. In both cases, the proofreading activity ensures that the rate of the forward reaction is slower than the rate of the rejection reaction in the cases of aberrant substrates.
Several ATP-dependent RNA helicases with conserved DExD/H motifs have been implicated in mechanisms that promote fidelity of the splicing reaction. In the absence of such activities, noncanonical substrates can be used at higher frequencies by the spliceosome. The best documented examples are the DExD/H box ATPases Prp5p, Prp16p, and Prp22p, which are involved in pre-spliceosome assembly, proofreading of the branchpoint sequence, and proofreading of all consensus splice sites, respectively (Fig. 2A; Smith et al. 2008; Koodathingal et al. 2010). Most of the initial evidence showing the involvement of splicing ATPases in proofreading splicing relied on genetic suppression experiments. Prp16p mutants that exhibit decreased ATPase activity allow more efficient splicing of substrates carrying mutated branchpoint sequences (Burgess and Guthrie 1993a). Similarly, Prp5p mutants lacking ATPase activity can suppress the effect of branchpoint mutations, and the extent to which suppression takes place is inversely proportional to the level of ATPase activity (Xu and Query 2007). These observations showed that multiple ATPases are required for a quality control check that ensures the proper conformation of the branched structure (Fig. 2A), highlighting the importance of the integrity of this structure for the splicing process. Work on Prp43p had also revealed the role of this ATPase as a quality-control player in spliceosome assembly, suggesting a role in dissociating aberrant lariat intermediates (Pandit et al. 2006). More recent biochemical analyses have also identified a function for Prp43p (and also Prp16p) in proofreading 5′-splice-site cleavage (Koodathingal et al. 2010), highlighting the role of Prp43p in the discard of pre-mRNAs and lariat intermediates (Mayas et al. 2010). Thus, while initial observations suggested a compartmentalized function for splicing ATPases in specific steps of the splicing pathway, at least a subset of these ATPases can be involved in multiple checkpoints (Fig. 2A). The role of these ATPases is not limited to proofreading of the first chemical step. In the absence of ATP or of functional Prpr22p activity, mutated 3′ splice sites can be used by the spliceosome in vitro, and yeast strains deficient for Prp22p can use mutated 3′ splice sites at higher rates in vivo (Mayas et al. 2006). These observations, and the comparison with GTPases involved in conformation changes during translation, have suggested that ATP hydrolysis by RNA-dependent splicing ATPases provides a kinetic proofreading mechanism that prevents the use of suboptimal splicing substrates (Burgess and Guthrie 1993b; Query and Konarska 2006; Smith et al. 2008). For such substrates, the rate of the rejection reaction is faster than the rate of the forward reaction, promoting higher fidelity (Fig. 2B). In the case of splicing ATPases, the energy of ATP hydrolysis might be used to drive conformational changes that activate discard pathways for aberrant substrates, as shown recently for Prp43p (Koodathingal et al. 2010; Mayas et al. 2010), or drive forward the pathway for normal substrates (Fig. 2B). As of 2008, it was still not clearly known whether the “unwindase” activity of ATPases constituted the mechanistic basis of their activities in the splicing pathway (Smith et al. 2008). Recent research on Prp16p has shown finer details of the competition-like involvement of this ATPase in both forward and discard pathways. Prp16p seems to increase the rate of the rejection pathway prior to the first chemical step for a defective spliceosome, while being much slower-acting on the rejection reaction for intact spliceosomes, allowing the first chemical step to take over (Koodathingal et al. 2010).
Other spliceosomal proteins may possibly assist the proofreading activities of these splicing ATPases. For example, Prp8p has been shown to suppress multiple splice site mutations (Collins and Guthrie 1999; Liu et al. 2007) and may cooperate with ATP-dependent RNA helicases in the general proofreading of the splicing reaction. Prp8p modulates the rate of transition between the two conformations of the spliceosome that are specific to each chemical step (Liu et al. 2007), and may improve the ability of the spliceosome to proofread splicesosomal substrates or products in a conformation specific to each transesterification reaction (Fig. 1).
Finally, a last potential mechanism to ensure accuracy of the splicing reaction relies on the fact that the pre-mRNA splicing reaction is reversible, at least in vitro (Tseng and Cheng 2008). Cheng and colleagues have shown that the forward steps of the splicing reaction are more error-prone that the reverse steps (Tseng and Cheng 2008). In this study, both reverse reactions were shown to be extremely precise, with little or no errors observed in the site of reverse splicing, while more errors were detected during the forward reactions, leading to improper splice site choices even in wild-type situations. Reversibility and the error rates associated with each of the forward and reverse steps could in principle be used in a kinetic manner to ensure that only properly spliced products are generated, if the reverse splicing reactions are faster in the case of incorrectly spliced products. However, in the absence of a direct estimate of the kinetic rates of each of these steps, it is difficult to conclude that reversal participates directly in proofreading of the splicing products, as described above for the splicing ATPases.
EXTERNAL QUALITY CONTROL PATHWAYS PROVIDE ADDITIONAL SPELLCHECKING ACTIVITIES AGAINST ERRORS MADE DURING MRNA SPLICING
Although the mechanisms described above greatly enhance the accuracy of mRNA splicing, they are not sufficient to provide the fidelity required to maintain an error rate compatible with accurate gene expression. In contrast to a process like DNA replication, the rate of mistakes made during splicing is difficult to quantitate because of the diverse nature of the mistakes involved, which include erroneous splice site selection, but also premature escape from the splicing pathway. Regardless of this precise rate, a second line of defense against errors, also known as “spellchecking” in the replication field (Kunkel 1995), which relies on degradative activities external to the spliceosome has evolved to prevent the accumulation of RNA molecules that have escaped the first quality checkpoints of the spliceosome.
In the case of mRNA splicing, “errors” can occur either because the substrate has escaped the splicing machinery, or because it has suffered a mis-splicing event such as the use of an incorrect splice site due to the misassembly of the spliceosome or escape from the proofreading activities mentioned above. Unspliced mRNAs that have prematurely escaped recognition by the spliceosome constitute the first type of frequent “errors” generated during splicing. This escape from the splicing pathway is common for pre-mRNAs containing splicing signal mutations (Legrain and Rosbash 1989; Hilleren and Parker 2003; Mayas et al. 2010), but is also observed in the case of endogenous mRNAs containing suboptimal splicing signals (Sayani et al. 2008). After escaping from the spliceosome or avoiding recognition altogether, the unspliced transcripts can remain in the nucleus or get exported to the cytoplasm. Thus, the second proofreading mechanisms that recognize unspliced pre-mRNAs will depend on the cellular localization of the RNA substrates.
The first mechanism of degradation of unspliced pre-mRNAs is found in the nucleus and is composed of two major nuclease activities: the nuclear exosome, a complex of nine core proteins (Mitchell et al. 1997), with exo- and endonuclease activity, and Rat1p/Xrn2, a 5′–3′ exonuclease (Johnson 1997). These activities have been shown to mediate degradation of unspliced pre-mRNAs (Bousquet-Antonelli et al. 2000; Danin-Kreiselman et al. 2003; Moore et al. 2006). The nuclear exosome can associate cotranscriptionally with some intron-containing genes (Moore et al. 2006; Eberle et al. 2010), suggesting that nuclear RNA degradation of the unspliced precursors may be favored if the substrates are not efficiently recognized in a cotranscriptional manner by the spliceosome, as shown for some transcripts (Moore et al. 2006; Tardiff et al. 2006). In addition to these general nuclear degradation pathways, degradation of specific pre-mRNAs can occur, such as the nuclear turnover of ribosomal protein unspliced mRNAs by the double-stranded endonuclease Rnt1p (Danin-Kreiselman et al. 2003).
The molecular mechanisms by which these RNAs are recognized as unspliced and degraded in the nucleus are unknown, but might be linked to the fact that splicing can deposit proteins onto the spliced mRNA, such as those of the exon-junction complex (Le Hir et al. 2001). Even if proteins from the exon-junction complex have not been directly involved in discriminating spliced mRNAs from unspliced molecules in the nucleus, it is conceptually possible that a similar complex might help identify unspliced pre-mRNAs and target them for degradation. It is also possible that failure to splice may induce defects in capping and/or poly(A) addition, which would induce an export defect and/or render the 5′- or 3′-ends of these RNAs particularly susceptible to degradation by exonucleases. Finally, several proteins such as yeast Mlp1p/Mlp2p have been suggested to function as general retention factors to prevent the premature export of unspliced pre-mRNAs to the cytoplasm (Dziembowski et al. 2004; Galy et al. 2004). These proteins might allow the nuclear retention of these molecules long enough to allow their degradation by nucleases localized in the nucleus (Fig. 3), while mature mRNAs are exported rapidly. However, the activity of these retention factors has been established using an inefficiently spliced reporter system (Dziembowski et al. 2004; Galy et al. 2004), and it has never been demonstrated that these proteins act on endogenous unspliced pre-mRNAs.
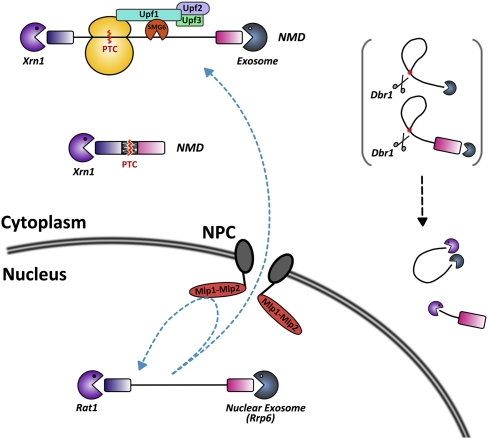
FIGURE 3.
External spellchecking mechanisms for splicing products. RNA degradative pathways for mRNAs that have escaped splicing or contain splicing mistakes. For simplicity purposes, the ribosome stalled at the PTC is not shown for the spliced mRNA that has been incorrectly spliced.
The escape of unspliced pre-mRNAs to the cytoplasm has been demonstrated in the case of endogenous pre-mRNAs and substrates containing splicing signals mutations (Legrain and Rosbash 1989; Hilleren and Parker 2003; Mayas et al. 2010). If unspliced pre-mRNAs escape the nucleus, they are likely to be translated in the cytoplasm, which would result in the production of nonfunctional or truncated proteins. The retention of intronic sequences will likely result in a premature translation termination codon (PTC), either because the intronic sequence itself contains a PTC (Fig. 3), or because the presence of the intron will introduce a translational frameshift generating a PTC in the downstream exon. In addition, splicing signals themselves, such as the 5′ splice site, the branchpoint, and the 3′ splice site can contain translation termination codons (Senapathy 1988; Chanfreau 2010; Mekouar et al. 2010). Intron sequences and lengths are constrained such that there is a significant enrichment for translation termination signals in the introns (Jaillon et al. 2008; Farlow et al. 2010). Therefore, unspliced pre-mRNAs that have escaped to the cytoplasm are prime candidates to be degraded by the nonsense-mediated mRNA decay (NMD), an RNA degradation mechanism that relies on translation and targets PTCs-containing RNA molecules to degradation by the cytoplasmic exonuclease Xrn1 and/or the cytoplasmic exosome (Chang et al. 2007; Isken and Maquat 2008). In higher eukaryotes, degradation by NMD can be triggered by endonucleolytic cleavage by the Smg6 protein (Huntzinger et al. 2008; Eberle et al. 2009). Regardless of its precise mechanism in various eukaryotes, NMD has been shown to induce degradation of inefficiently spliced transcripts in several eukaryotic species (He et al. 1993; Mitrovich and Anderson 2000; Jaillon et al. 2008; Sayani et al. 2008; Mekouar et al. 2010). The widespread accumulation of unspliced pre-mRNAs in NMD mutants or in conditions in which NMD is otherwise inactivated suggests that cytoplasmic turnover provides a large fraction of the RNA surveillance activity devoted to the elimination of unspliced transcripts.
Interestingly, cytoplasmic pathways that are independent from NMD have also been involved in the degradation of aberrant unspliced precursors or splicing intermediates. For example, degradation of a yeast unspliced actin reporter transcript relies on Xrn1p but is independent from NMD, even though this unspliced precursor is translated and contains a premature termination codon (Hilleren and Parker 2003). The reason why some unspliced RNAs are degraded in the cytoplasm independently from NMD is currently poorly understood. Finally, the degradation of lariat intermediates that are released from the spliceosome is also cytoplasmic and requires the debranching enzyme Dbr1p (Fig. 3; Hilleren and Parker 2003; Mayas et al. 2010). The splicing ATPase Prp43p is required for release of these intermediates from the spliceosome and their export out of the nucleus (Mayas et al. 2010).
The second kind of error that can be generated during splicing occurs when an incorrect splice site is used (Fig. 3), which results in transcripts with incorrect genetic information. This type of event results in a change of open-reading frame in two cases out of three; if the splicing results in inclusion of part of the intronic sequence, this sequence may also contain an in-frame PTC (Fig. 2). Because the inserted sequence or the downstream sequences have not been constrained to maintain the translation open-reading frame, the likelihood of encountering a PTC is very high, rendering these aberrantly spliced species also subject to NMD (Fig. 3). Therefore, NMD plays a ubiquitous role in degrading RNA molecules that result from splicing mistakes, either because the substrates have completely escaped splicing or because of the use of incorrect splice sites. While the surveillance of incorrectly spliced RNAs by NMD prevents the accumulation of these aberrant RNAs, it is worth mentioning that this mechanism is also used for regulatory purposes as well, for example to regulate the stability of transcripts containing alternatively spliced exons with in-frame PTCs. Autoregulation of splicing factors relies on alternative splicing of highly conserved exon cassettes containing PTCs, which couples alternative splicing to NMD, and allows fine-tuning by autoregulating of the levels of transcripts encoding these splicing factors (Lareau et al. 2007; Ni et al. 2007). These examples illustrate the fact that so-called splicing errors can indeed be productively used during evolution to contribute to the buffering of biological systems.
SIMILARITIES WITH OTHER SYSTEMS AND REMAINING QUESTIONS
In the previous paragraph we have shown that the spliceosome does not entirely rely on intrinsic mechanisms and quality checks to ensure fidelity of the reactions, and that exogenous machineries are recruited to provide independent proofreading or spellchecking activities. These multiple proofreading steps are necessary to provide the level of fidelity required for accurate gene expression. The presence of multiple fidelity mechanisms relying on similar two-tier strategies is present in other biochemical processes, such as translation or DNA replication. During DNA replication, errors made by DNA polymerases are typically proofread by the 3′–5′ exonuclease activities that are either intrinsic to the polymerase (Kunkel and Bebenek 2000), or associate with them (Shevelev and Hubscher 2002). Following replication, the mismatch repair (MMR) process ensures that errors made by the DNA polymerase that have not been corrected by the proofreading activity of the polymerase are eliminated rapidly during DNA replication (Kunkel and Bebenek 2000; Jiricny 2006; Hsieh and Yamane 2008; Li 2008; Larrea et al. 2010). During protein synthesis, proofreading mechanisms exist not only at the level of tRNAs charging by aminoacyltransferases and during translation (Cochella and Green 2005; Zaher and Green 2009), but also at a second level, by the degradation of nonfunctional polypeptides by various protein machineries, including the proteasome (Ravid and Hochstrasser 2008). The presence of multiple quality control activities in all these biochemical pathways underscores the importance of high levels of quality control in the propagation and expression of genetic information, likely due to the intrinsic inability of cellular enzymes to catalyze biochemical reactions with complete accuracy.
Although the two-tier strategy established to promote high levels of splicing fidelity is well established, multiple questions still remain regarding the precise proofreading mechanisms and the turnover pathways involved in each process. Regarding the first proofreading steps, a major question remaining is to understand the exact mechanism by which multiple splicing ATPases promote checkpoints of the splicing substrates, and their connection to the discard pathways. While it is known that spliceosomes or splicing complexes lacking these ATPase activities can use aberrant substrates at higher rates, the precise molecular fate of the defective substrates or intermediates is unknown, and the molecular identification of the nucleases involved in the discard pathway would be a major advance. Additionally, demonstrating the direct role of the splicing ATPases Prp5, Prp16, Prp22, and Prp43 on endogenous substrates rather than artificial reporters is necessary to understand the full range of their physiological functions. Thus far, most of the studies have focused on the effects of these ATPases on splicing reporters, and it is unclear how these ATPases affect the discard pathways of endogenous mRNAs. The discovery of the reversibility of both steps of the pre-mRNA splicing reaction (Tseng and Cheng 2008) raises the hypothesis that incorrect splicing intermediates or products may be reversed to unspliced precursors at higher rates than correctly processed transcripts, favoring proofreading activities. Such a return of the substrate back to the initial stage is conceptually very attractive, because it would resemble the 3′–5′ exonuclease activities associated with DNA polymerases (Kunkel and Bebenek 2000), which promotes a return of the substrate back to the polymerase active site, once the defective nucleotide(s) have been removed.
Regarding the second tier quality control pathway for splicing, it remains to be established how unspliced molecules are recognized in the nucleus by degradative enzymes such as Rat1p and the nuclear exosome. The absence of proteins deposited by the spliceosome might provide a way to discriminate unspliced molecules from spliced ones, but the identity of such proteins remains to be identified. It is also unclear whether nuclear retention systems such as the one mediated by Mlp1p are active on endogenous RNAs, since their activity has only been demonstrated using an artificial inefficiently spliced reporter. Another major question is to understand the mechanism of transcriptional feedback, as the nuclear exosome seems to induce transcriptional down-regulation of genes encoding pre-mRNAs with splicing mutations (Eberle et al. 2010). With respect to cytoplasmic degradation pathways, it is interesting to note that some unspliced RNAs are degraded in the cytoplasm but are not targeted by NMD (Hilleren and Parker, 2003; Sayani et al. 2008), raising the question why these pre-mRNAs are immune to NMD. It is also unclear what fraction of the surveillance pathways for unspliced RNAs is nuclear, and what fraction is cytoplasmic. Further genomic studies will likely respond to most of these questions.
Finally, most of the studies that have investigated the role of proofreading and spellchecking in the quality control of splicing have done so either in model systems or in standard growth conditions. Recent studies have shown that even in the simple eukaryote S.cerevisiae, splicing can be highly regulated according to environmental conditions (Pleiss et al. 2007). The surveillance activity of the exosome has also been shown to be regulated according to carbon sources (Bousquet-Antonelli et al. 2000). Splicing regulation is even more prevalent and complex in higher eukaryotes (for review, see Licatalosi and Darnell 2010), and future studies are required to understand how RNA degradation is integrated to the regulation of splicing according to changes in environmental conditions and to developmental signal transduction pathways.
ACKNOWLEDGMENTS
We thank Catherine Coffinier, Kevin Roy, Shakir Sayani, and Jonathan Staley for a critical reading of earlier versions of the manuscript, and J. Staley for discussion and communication of results prior to publication. Work in G.C. laboratory is funded by National Institutes of Health Grant GM61518 and American Cancer Society Grant RSG-06-040.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2454711.
REFERENCES
- Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, et al. 2010. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol 17: 504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C 1993a. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1391 [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C 1993b. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci 18: 381–384 [DOI] [PubMed] [Google Scholar]
- Chanfreau GF 2010. A dual role for RNA splicing signals. EMBO Rep 11: 720–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chua K, Reed R 1999. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402: 207–210 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R 2005. Fidelity in protein synthesis. Curr Biol 15: R536–R540 doi: 10.1016/j.cub.2005.07.018 [DOI] [PubMed] [Google Scholar]
- Collins CA, Guthrie C 1999. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev 13: 1970–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G 2003. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell 11: 1279–1289 [DOI] [PubMed] [Google Scholar]
- Du H, Rosbash M 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 23: 4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Hessle V, Helbig R, Dantoft W, Gimber N, Visa N 2010. Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes. PLoS ONE 5: e11540 doi: 10.1371/journal.pone.0011540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow A, Meduri E, Dolezal M, Hua L, Schlotterer C 2010. Nonsense-mediated decay enables intron gain in Drosophila. PLoS Genet 6: e1000819 doi: 10.1371/journal.pgen.1000819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci 90: 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Hsieh P, Yamane K 2008. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev 129: 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, et al. 2008. Translational control of intron splicing in eukaryotes. Nature 451: 359–362 [DOI] [PubMed] [Google Scholar]
- Jiricny J 2006. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 7: 335–346 [DOI] [PubMed] [Google Scholar]
- Johnson AW 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol 17: 6122–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Novak T, Piccirilli JA, Staley JP 2010. The DEAH-box ATPases Prp16 and Prp43 cooperate to proofread 5′splice site cleavage during pre-mRNA splicing. Mol Cell 39: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA 1995. DNA-mismatch repair. The intricacies of eukaryotic spell-checking. Curr Biol 5: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K 2000. DNA replication fidelity. Annu Rev Biochem 69: 497–529 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Larrea AA, Lujan SA, Kunkel TA 2010. SnapShot: DNA mismatch repair. Cell 141:730–e1. doi: 10.1016/j.cell.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Rosbash M 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Li GM 2008. Mechanisms and functions of DNA mismatch repair. Cell Res 18: 85–98 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM 2007. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol 14: 519–526 [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Semlow DR, Staley JP 2010. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci 107: 10020–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekouar M, Blanc-Lenfle I, Ozanne C, Da Silva C, Cruaud C, Wincker P, Gaillardin C, Neuveglise C 2010. Detection and analysis of alternative splicing in Yarrowia lipolytica reveal structural constraints facilitating nonsense-mediated decay of intron-retaining transcripts. Genome Biol 11: R65 doi: 10.1186/gb-2010-11-6-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev 14: 2173–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Schwartzfarb EM, Silver PA, Yu MC 2006. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell 24: 903–915 [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell 27: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2006. Splicing fidelity revisited. Nat Struct Mol Biol 13: 472–474 [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 31: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P 1988. Possible evolution of splice-junction signals in eukaryotic genes from stop codons. Proc Natl Acad Sci 85: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelev IV, Hubscher U 2002. The 3′ 5′ exonucleases. Nat Rev Mol Cell Biol 3: 364–376 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J 2006. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312: 1961–1965 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Cheng SC 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA 2010. The pathobiology of splicing. J Pathol 220: 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Query CC 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]