Abstract
The Cleavage stimulation Factor (CstF) complex is composed of three subunits and is essential for pre-mRNA 3′-end processing. CstF recognizes U and G/U-rich cis-acting RNA sequence elements and helps stabilize the Cleavage and Polyadenylation Specificity Factor (CPSF) at the polyadenylation site as required for productive RNA cleavage. Here, we describe the crystal structure of the N-terminal domain of Drosophila CstF-50 subunit. It forms a compact homodimer that exposes two geometrically opposite, identical, and conserved surfaces that may serve as binding platform. Together with previous data on the structure of CstF-77, homodimerization of CstF-50 N-terminal domain supports the model in which the functional state of CstF is a heterohexamer.
Keywords: mRNA 3′ end maturation, crystallography, dimerization, CstF, D. melanogaster
INTRODUCTION
With the exception of histone mRNAs, all eukaryotic primary mRNA transcripts acquire a poly(A) tail at their 3′ ends. This post-transcriptional modification results from a two-step reaction involving a large cleavage/polyadenylation machinery that recognizes poly(A) signals on the pre-mRNA (Millevoi and Vagner 2010). In humans, ∼75% of the core poly(A) sequences contain a bipartite sequence element defined by a conserved canonical AAUAAA or AUUAAA upstream hexameric motif located upstream of the poly(A) site and a less-defined U- or GU-rich sequence element (DSE) (Zarudnaya et al. 2003; Tian et al. 2005; Nunes et al. 2010). The hexameric sequence is recognized by the Cleavage and Polyadenylation Specificity Factor (CPSF), which contains the endonuclease that catalyzes the first processing step (Keller et al. 1991; Mandel et al. 2006; Kolev et al. 2008). The second canonical sequence element is bound by the Cleavage stimulation Factor (CstF), and this interaction is thought to stabilize the CPSF–RNA interaction (Gilmartin and Nevins 1991; Weiss et al. 1991; Bienroth et al. 1993). CstF complex is composed of three separate protein subunits. Central in the complex is CstF-77 (Rna14p in the yeast Saccharomyces cerevisiae), a HAT-repeat containing protein that serves as a scaffold by bridging CstF-64 (Rna15p in yeast) and CstF-50 (no sequence homolog in yeast) (Takagaki et al. 1990; Minvielle-Sebastia et al. 1994; Takagaki and Manley 1994, 2000). Through its RRM, the CstF-64 subunit binds to RNA with high affinity within the context of the complex (Takagaki and Manley 1997; Noble et al. 2004), whereas CstF-50, a WD-repeat containing protein, binds to the carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) (Fong and Bentley 2001; Fong et al. 2003) and to the BRCA1-associated RING domain 1 (BARD1) (Kleiman and Manley 1999) and PARN upon DNA damage (Cevher et al. 2010).
CstF was initially described as a heterotrimer of about 190 kDa in total size with one copy of each subunit per complex (Takagaki et al. 1990). However, growing evidence coming from Drosophila genetics (Simonelig et al. 1996), in vitro interaction experiments (Takagaki and Manley 2000), and solution studies have led to a more complicated description of the factor. Recently, the crystal structure of two orthologs of human CstF-77 revealed atomic details of homodimerization, and thereby provided the first unambiguous evidence for the subunit stoichiometry within CstF (Bai et al. 2007; Legrand et al. 2007). These studies support a model where each subunit is present in two copies per complex. So far, only far-Western blot analysis supported the self-association of CstF-50 (Takagaki and Manley 2000). In this study, the CstF-50 WD-repeat domain is shown to be dispensable for self-association. In order to provide the structural basis of CstF-50 dimerization and assess the hexameric model of CstF, we have determined the crystal structure of the N-terminal domain of Drosophila CstF-50 (dmCstF-50N) (residues 1–65) at a resolution of 1.4 Å. Our study shows that the N-terminal domain is able to form stable homodimers in solution through phylogenetically conserved residues. We have also identified two conserved surface pockets that may serve as binding sites for yet unidentified factors.
RESULTS
Structure determination of CstF-50
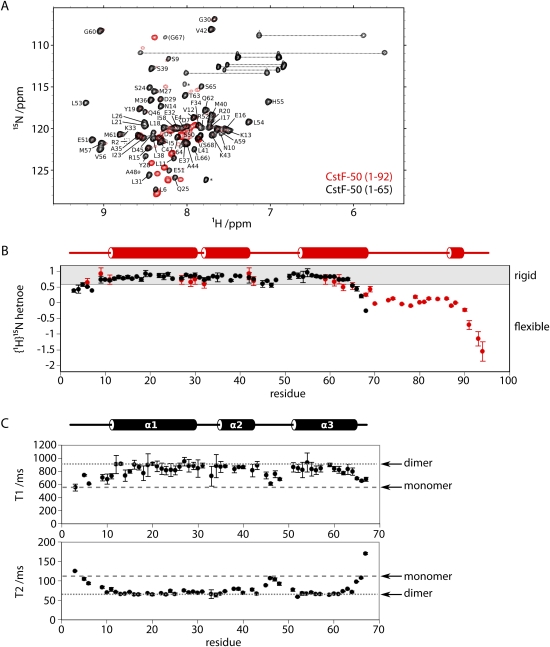
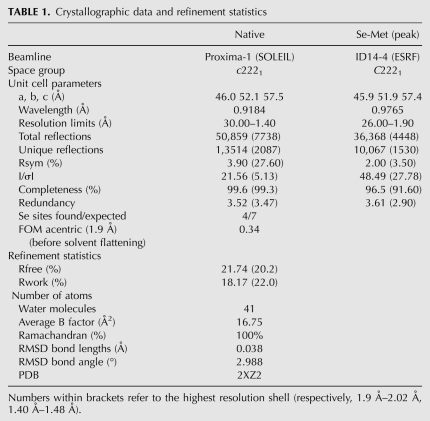
A previous mapping of pairwise interactions by far-Western blot analysis resulted in a domain organization map of the three subunits of human CstF (Takagaki and Manley 2000). In this study, CstF-50 was shown to interact with CstF-77 through its WD-repeat domain, whereas the conserved N-terminal domain was sufficient for self-interaction. To investigate the structural details of this self-association, an initial construct ranging from residue 1 to residue 92 of the Drosophila homolog of CstF-50 (dmCstF-50) was cloned, expressed, and purified (see Materials and Methods). Investigation of this construct by NMR spectroscopy revealed an N-terminal helical domain, followed by a C-terminal region of conformational flexibility beginning around residue 65 (Fig. 1A,B). A shorter construct comprising residues 1–65 of dmCstF-50, thus removing the unfolded C-terminal region, was prepared and crystallized. Phases were obtained from a SAD experiment at the Se-edge with Se-Met-substituted protein crystals. The model has been refined to 1.4 Å resolution to a R-free factor and R-factor of 18.14% and 21.74%, respectively (Table 1).
FIGURE 1.
Characterization of N-terminal peptides of dmCstF-50. (A) Residues 1–65 in dmCstF-50 from a folded domain. Overlay of 1H,15N-HSQC spectra for dmCstF-50 constructs containing either the first 92 (red) or 65 (black) residues. The peaks corresponding to dmCstF-50N have been annotated with the residue type and number. Residues common to both constructs have similar peak positions, whereas the C-terminal residues specific to the larger construct are clustered in the region indicative of disordered peptides (1H values between 8 and 8.5 ppm). (B) C-terminal residues of dmCstF-50 (1–92) are conformationally flexible. 1H,15N-amide heteronuclear relaxation analysis reveals significant peptide backbone flexibility in the C-terminal residues of the larger construct (values <0.6). Secondary structure predicted from the backbone 1H, 13C, and 15N chemical shifts using DANGLE (Cheung et al. 2010) is displayed above. (C) Oligomerization state of dmCstF-50 (1–65) determined by NMR spectroscopy. 15N-amide relaxation experiments conducted at 298 K and 700 MHz reveal T1 and T2 values consistent with those predicted (Daragan and Mayo 1997) for the homodimer (dotted line; 547 msec and 113 msec, respectively) and not of a monomer (dashed line; 929 msec and 64 msec).
TABLE 1.
Crystallographic data and refinement statistics
The N-terminal domain of CstF-50 is sufficient for homodimerization
The protein domain folds around three α-helices arranged in an almost collinear orientation (Fig. 2A). Despite an arrangement of one molecule per asymmetric unit, inspection of the protein packing reveals the presence of a homodimer sitting on a crystallographic axis of symmetry. The oligomeric state of the domain as a dimer was also confirmed in solution by using NMR spectroscopy. Specifically, 15N-amide T1 and T2 relaxation measurements are highly consistent with a globular particle of ∼16 kDa and do not support the existence of a 8-kDa monomer (Fig. 1C).
FIGURE 2.
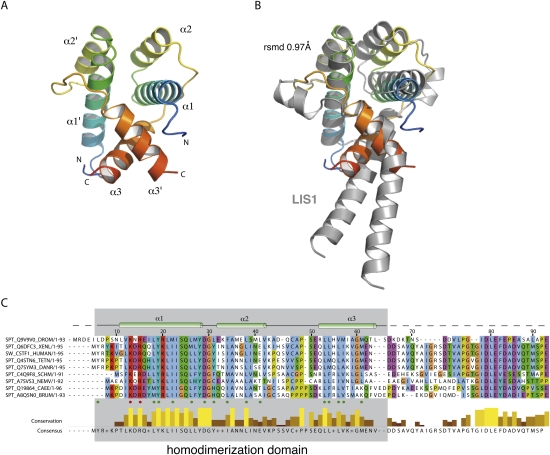
Overall structure of dmCstF-50N. (A) Ribbon representation of dmCstF-50N homodimer (residues 1–65). Each monomer of dmCstF-50N (residues 1–65) is shown according to a rainbow gradient of color (blue to red) from the N terminus to the C terminus. Secondary structure elements are labeled. All drawings were produced with PyMol software (DeLano 2002). (B) dmCstF-50N is structurally homologous to LIS1. The three-dimensional structure of LIS1 (gray) (PDB code 1UUJ) has been superimposed on dmCstF-50N dimer. The overall RMSD on the Cα backbone is 0.97 Å. dmCstF-50N is shown under the same orientation as in A. (C) Sequence conservation among CstF-50 orthologs. Sequence alignment of CstF-50 orthologs was performed with PipeAlign (Plewniak et al. 2003) and rendered using Jalview (Waterhouse et al. 2009). Secondary structure elements dmCstF-50N are shown on top of the alignment. Green and red dots below the alignment correspond to residues involved in the dimer interface, and to solvent-exposed residues, respectively. Sequence conservation and consensus sequence are shown at the bottom of the alignment. (DROM) Drosophila melanogaster, (XENL) Xenopus laevis, (HUMAN) Homo sapiens, (TETN) Tetraodon nigroviridis, (SCHM) Schistosoma mansoni, (NEMV) Nematostella vectensis, (CAEE) Caenorhabditis elegans, (BRUM) Brugia malayi, (DANR) Danio rerio.
The two monomers create an overall antiparallel helix bundle reminiscent of the three-dimensional arrangement found in the N-terminal domain of lissencephaly protein 1 (LIS1), a known dimerization domain (Fig. 2B, RMSD of 0.97 Å) (Kim et al. 2004). No other significant structural similarity was detected by using the SSM server (Krissinel and Henrick 2004). Within the core of the homodimer, α-helix 1 and 1′ as well as 2 and 2′ face each other and form an hydrophobic interface involving residues Leu 6, Leu 18, Tyr 19, Met 22, Leu 26 and Leu 31, Phe 34, Leu 38 and Leu 41 on α-helices 1/1′ and 2/2′, respectively (green dots, Fig. 2A,C). Additional contacts are provided by α-helix 3 and 3′ (residues Glu 51, Leu 53, Leu 54, Val 56, Met 57, Ile 58, and Met 61) that are sandwiched between α-helices 1/1′ and α-helices 3/3′ of the other monomer. The homodimerization of CstF-50 N-terminal domain is supported by the three α-helices altogether, which collectively bury >1500 Å2 per monomer.
Conserved areas at the surface of dmCstF-50N
Sequence conservation is often a hallmark of functional sites within a protein or RNA structure. In order to identify such sites within dmCstF-50N, the sequence alignment displayed on Figure 2C was used with the ESPript server (Gouet et al. 2003) to analyze sequence conservation among species at the surface of the protein complex (Fig. 3).
FIGURE 3.
Sequence conservation of dmCstF-50N. (A,B) dmCstF-50N is a phylogenetically conserved homodimer. dmCstF-50N is depicted under a similar orientation as in Figure 1 with one monomer as a cartoon, whereas the other monomer is shown as a surface representation. Residues involved in homodimer formation in cartooned monomer are labeled. On the right, the cartoon representation of one monomer has been omitted to expose its conserved fingerprint on the other monomer. Sequence conservation is shown from white to red ranging from nonconserved to conserved residues. This figure has been produced using ESPript program (Gouet et al. 2003). Conserved residues involved in protein–protein interaction are labeled. Some secondary structure elements are shown underneath. (C,D) A PEG molecule sits in a conserved surface area of the complex. A PEG molecule coming from the crystallization condition can be modeled into the conserved cavity at the surface of dmCstF-50N. Residue conservation has been calculated as described in Figure 2. The Fobs-Fcalc electron density map contoured at 1.5 σ around the PEG molecule in shown as a blue mesh. Conserved residues are labeled.
The dimerization interface is mostly composed of conserved residues, and those within the hydrophobic core are strictly invariant (Fig. 3A,B). These residues include Leu 18, Tyr 19, Met 22, Leu 26, Asp 29, Phe 34, Leu 38, Leu 53, and Val 56, and this observation supports the hypothesis of CstF-50 homodimerization throughout evolution in higher eukaryotes.
As shown in Figure 3, C and D, a site of invariant residues is present at the surface of the complex. This area is formed by conserved residues of each monomer and extends over both monomers. Due to symmetry, an identical surface is present at the opposite side of the molecule. Side chains of residues Val 12, Lys 13, Arg 15, Tyr 19, Arg 20, Tyr 28, Asp 29, Gly 30, and Val 42 form an open cavity in which a PEG molecule from the crystallization condition is sitting.
DISCUSSION
Within the 3′ end mRNA maturation complex, CstF-50 has been shown to interact with CstF-77 and with the CTD domain of the largest subunit of RNA polymerase II (Takagaki and Manley 2000; Fong and Bentley 2001). Recently, it has also been shown that CstF-50 couples mRNA maturation and DNA damage repair through its interaction with BARD1 and PARN (Kleiman and Manley 1999; Edwards et al. 2008; Cevher et al. 2010). Mapping experiments assigned the WD-repeat region of CstF-50 (residues 92–431) to direct interaction with BARD1 and CstF-77, whereas the N-terminal portion (residues 1–92) is required to make contacts with the pol II CTD (Fong and Bentley 2001). Based on this observation we tried to either cocrystallize peptides harboring one or two heptad repeats YSPTSPS of the CTD, phosphorylated or not, in either position Ser5 or Ser2. We also attempted to characterize the interaction of CstF-50 N-terminal domain with pol II CTD by surface plasmon resonance and NMR titration experiments using biotinylated peptides. None of these assays ended up with a clear answer on the ability of human CstF-50 fragment (1–63) to bind a human CTD heptad repeat. Cocrystallization assays and structure solution did not reveal any extra density that could be accounted for by a bound peptide, although this finding could be due to the position of a well-ordered PEG molecule in a putative CTD-binding site that prevents a weak CTD–protein interaction to occur (Fig. 3). NMR titrations did not reveal significant modification in the chemical shifts of the protein, even at high peptide/protein concentrations. However, pull-down assays confirmed a specific binding of full-length CstF-50 to GST–CTD (data not shown) (Fong and Bentley 2001). The N-terminal portion of CstF-50 might only be weakly involved in CTD binding, since larger constructs of CstF-50 affecting the N-terminal domain folding (residues 36–176) or lacking the N-terminal part (residues 77–176) were still able to interact with the CTD (Fong and Bentley 2001). These data are in favor of the contribution of linker residues 66–92 to CTD interaction rather than in the homodimerization itself. Alternatively, more than two heptad repeats of the CTD might be needed for CstF-50-CTD interaction as observed in the case of Pcf11p and other CTD-binding proteins (Noble et al. 2005).
Concluding remarks
Together with previous data (Takagaki and Manley 2000), this study demonstrates that the N-terminal domain of CstF-50 is a homodimerization module for CstF-50. In contrast to CstF-64 and similarly to CstF-77, CstF-50 is a dimer (Bai et al. 2007; Legrand et al. 2007; this work). CstF-77 is described as the scaffolding subunit of CstF bridging CstF-50 and CstF-64. As a consequence, the number of copies of CstF-77 dictates the number of copies of the remaining subunits of the complex. Through the interaction of CstF-64 with the C terminus of CstF-77, there would exist two copies of CstF-64 and its combination of two RRM domains would provide a higher affinity for target RNA sequence elements (Noble et al. 2004). So far, there is no biological rationale for the dimerization of CstF-50, especially given the homodimerization potential already contributed by CstF-77. One possible explanation is that phylogenetically conserved residues are brought into proximity to form a unique surface that could well be used for protein–protein interaction. This question will need further in vitro and in vivo experiments to fully explore the biological importance of these conserved surfaces.
MATERIAL AND METHODS
Protein expression and purification
The DNA sequence coding for the Drosophila CstF-50 residues 1–92, 1–65 were isolated by PCR amplification and inserted into the NdeI and BamHI sites of a pET-derived (Novagen) plasmid (Romier et al. 2006) to produce N-terminal His-tag fused protein. Proteins were expressed in E. coli BL21(DE3) cells and were purified to homogeneity by an initial affinity column followed by incubation with TEV protease to remove the His-tag and a final size-exclusion chromatography. The protein was concentrated up to 19–24 mg/mL in a buffer containing 50 mM Tris-HCl, 150 mM NaCl, and 1mM DTT. Seleno-substituted protein was produced in B834 E. coli strain and purified in the same way. Protein samples for NMR spectroscopy used BL21(DE3) cells and were expressed in M9 medium supplemented with [13C]-glucose (2 g/L) and/or [15N]-ammonium sulphate (1 g/L) and purified as above.
NMR spectroscopy
Samples contained from 0.4 to 2 mM protein in either 10 mM 2H-Tris, 150 mM NaCl (pH 7.5), or phosphate-buffered saline (PBS, 10 mM sodium phosphate, and 2 mM potassium phosphate, 137 mM NaCl, 2.7 mM KCl at pH 7.4) with 10% 2H2O added for the lock. Spectra were collected on a Bruker Avance 700 MHz spectrometer. Chemical-shift assignment of the peptide backbone 1H, 13C, 15N nuclei used standard 3D HNCO, HNCACO, HNCA, HNCOCA, HNCACB, and CBCACONH spectra. Assignment of the larger construct (residues 1–92) used a sample of 13C,15N-labeled protein in PBS at temperatures of both 295K and 310K, whereas that of the smaller construct (residues 1–65) were only assigned at 295K in 10 mM 2H-Tris, 150 mM NaCl (pH 7.5). Amide 15N-relaxation experiments were conducted at 298K in 10 mM 2H-Tris, 150 mM NaCl (pH 7.5) at 700 MHz using pulse programs as described (Farrow et al. 1994). Titration of 15N CstF-50 (1–65) with the CTD peptide used 0.4 mM protein and increasing addition of CTD in increments of 0.1, 0.2, 0.3, 0.5, 0.75, 1, 1.25, 1.5, 2, 2.5 molar equivalents peptide:protein.
Structure determination
Crystals were initially obtained in 100 mM HEPES sodium salt (pH 7.5), 2% PEG400, 2 M ammonium sulphate, and further refined to 100 mM HEPES sodium salt (pH 7.5), 3% PEG400, 1.4 M ammonium sulphate, and 15%–20% glycerol. Cubic crystals were obtained after a day by the sitting drop method using streak seeding. Crystals belong to the space group c2221, with one molecule per asymmetric unit. The structure was solved by a SAD experiment using crystals of a selenomethionine-substituted protein. The SAD data set was collected on ID14-4 at the ESRF. All data set were reduced using XDS (Kabsch 1993) and four of the seven Se sites were located and refined using SHARP (de La Fortelle and Bricogne 1997). Phasing was performed with SHARP, and automatic building was carried out with ARP/wARP (Perrakis et al. 2001). Manual building and further refinement were performed using Coot and REFMAC5 (Winn et al. 2001; Emsley and Cowtan 2004). The model was refined to an R-free of 18.1% and comprises residues 1–65 (Table 1).
Coordinates
PDB and structure factors have been deposited under the accession number 2XZ2. NMR chemical shifts have been deposited in the BMRB under accession numbers 17308 and 17313 for, respectively, dmCstF-50 (1–65) and dmCstF-50 (1–92).
ACKNOWLEDGMENTS
M.M.M. is supported by a grant from Caixa fundacion and the Association pour la Recherche contre le Cancer. This work is supported by a joint Avenir Inserm/ARC funding, the ICSN, the Aquitaine regional government, and the Fondation pour la Recherche Médicale and the Fondation BNP/Paribas. We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities for assistance in using beamline ID14-4. We acknowledge the Synchrotron SOLEIL and P. Legrand for assistance in using Proxima-1 beamline. We thank M. Simonelig (Institute of Human Genetics, Montpellier, France) for the kind gift of Drosophila CstF-50 cDNA and Dr. Kevin Ryan (City College of New York) for human GST-CTD clone.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2481011.
REFERENCES
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L 2007. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell 25: 863–875 [DOI] [PubMed] [Google Scholar]
- Bienroth S, Keller W, Wahle E 1993. Assembly of a processive messenger RNA polyadenylation complex. EMBO J 12: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE 2010. Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. EMBO J 29: 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MS, Maguire ML, Stevens TJ, Broadhurst RW 2010. DANGLE: A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J Magn Reson 202: 223–233 [DOI] [PubMed] [Google Scholar]
- Daragan VA, Mayo KH 1997. Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog NMR Spectrosc 31: 63–105 [Google Scholar]
- de La Fortelle E, Bricogne G 1997. SHARP: A maximum-likelihood heavy-atom parameter refinement program for the MIR and MAD methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- DeLano WL 2002. The PyMOL User's Manual System. DeLano Scientific, San Carlos, CA [Google Scholar]
- Edwards RA, Lee MS, Tsutakawa SE, Williams RS, Nazeer I, Kleiman FE, Tainer JA, Glover JN 2008. The BARD1 C-terminal domain structure and interactions with polyadenylation factor CstF-50. Biochemistry 47: 11446–11456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003 [DOI] [PubMed] [Google Scholar]
- Fong N, Bentley DL 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes Dev 15: 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bird G, Vigneron M, Bentley DL 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J 22: 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM, Nevins JR 1991. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol 11: 2432–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E 2003. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31: 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J 10: 4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Cooper DR, Oleksy A, Devedjiev Y, Derewenda U, Reiner O, Otlewski J, Derewenda ZS 2004. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure 12: 987–998 [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285: 1576–1579 [DOI] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep 9: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Legrand P, Pinaud N, Minvielle-Sebastia L, Fribourg S 2007. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res 35: 4515–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Keller W 1994. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science 266: 1702–1705 [DOI] [PubMed] [Google Scholar]
- Noble CG, Walker PA, Calder LJ, Taylor IA 2004. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res 32: 3364–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol 12: 144–151 [DOI] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A 2010. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J 29: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS 2001. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr 57: 1445–1450 [DOI] [PubMed] [Google Scholar]
- Plewniak F, Bianchetti L, Brelivet Y, Carles A, Chalmel F, Lecompte O, Mochel T, Moulinier L, Muller A, Muller J, et al. 2003. PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res 31: 3829–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C, Ben Jelloul M, Albeck S, Buchwald G, Busso D, Celie PH, Christodoulou E, De Marco V, van Gerwen S, Knipscheer P, et al. 2006. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: Experimental procedures, database tracking and case studies. Acta Crystallogr D Biol Crystallogr 62: 1232–1242 [DOI] [PubMed] [Google Scholar]
- Simonelig M, Elliott K, Mitchelson A, O'Hare K 1996. Interallelic complementation at the suppressor of forked locus of Drosophila reveals complementation between suppressor of forked proteins mutated in different regions. Genetics 142: 1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 1994. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature 372: 471–474 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 1997. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol 17: 3907–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4: 2112–2120 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EA, Gilmartin GM, Nevins JR 1991. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J 10: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: Primary, secondary and higher-order structures. Nucleic Acids Res 31: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]