Abstract
mRNA decapping is a crucial step in the regulation of mRNA stability and gene expression. Dcp2 is an mRNA decapping enzyme that has been widely studied. We recently reported the presence of a second mammalian cytoplasmic decapping enzyme, Nudt16. Here we address the differential utilization of the two decapping enzymes in specified mRNA decay processes. Using mouse embryonic fibroblast (MEF) cell lines derived from a hypomorphic knockout of the Dcp2 gene with undetectable levels of Dcp2 or MEF cell lines harboring a Nudt16-directed shRNA to generate reduced levels of Nudt16, we demonstrate the distinct roles for Dcp2 and Nudt16 in nonsense-mediated mRNA decay (NMD), decay of ARE-containing mRNA and miRNA-mediated silencing. Our results indicated that NMD preferentially utilizes Dcp2 rather than Nudt16; Dcp2 and Nudt16 are redundant in miRNA-mediated silencing; and Dcp2 and Nudt16 are differentially utilized for ARE-mRNA decay. These data demonstrate that the two distinct decapping enzymes can uniquely function in specific mRNA decay processes in mammalian cells.
Keywords: Dcp2, Nudt16, decapping, mRNA decay
INTRODUCTION
Regulation of mRNA decay is an important step in the control of gene expression. In eukaryotic cells, bulk mRNA decay typically initiates with deadenylation of the 3′ poly(A) tail, followed by degradation of the mRNA body in either the 5′ to 3′ or the 3′ to 5′ direction (Coller and Parker 2004). Degradation from the 3′ end is carried out by the cytoplasmic RNA exosome, which is a multisubunit 3′ to 5′ exoribonuclease complex (Liu et al. 2006), and the residual cap structure is hydrolyzed by the scavenger decapping enzyme DcpS (Liu et al. 2002). In the 5′ to 3′ decay pathway, the monomethyl guanosine (m7G) mRNA cap is cleaved by the Dcp2 decapping enzyme (Dunckley and Parker 1999; Lykke-Andersen 2002; van Dijk et al. 2002; Z Wang et al. 2002) and the monophosphate RNA is degraded progressively by the 5′ to 3′ exoribonuclease, Xrn1 (Decker and Parker 1993; Hsu and Stevens 1993).
The Dcp2 decapping enzyme catalyzes the removal of m7GDP from the mRNA 5′ cap. It has been implicated in various mRNA decay processes, including nonsense-mediated mRNA decay (NMD), ARE-mediated decay, and microRNA directed gene silencing (Franks and Lykke-Andersen 2008). The NMD pathway, which targets mRNAs with premature translational termination for rapid decay, was reported to trigger decapping in both yeast and human cells (Muhlrad and Parker 1994; Lejeune et al. 2003; Couttet and Grange 2004), and a reduction of Dcp2 levels in Hela cells impaired both nuclear-associated and cytoplasmic NMD (Lejeune et al. 2003). All three Upf proteins, key factors in NMD, coimmunopurified with Dcp2 (He and Jacobson 1995; Lykke-Andersen 2002; Lejeune et al. 2003), indicating that these factors recruit decapping protein to degrade nonsense-containing mRNAs.
AU-rich element (ARE)-mediated decay rapidly degrades mRNAs that contain AREs in their 3′ untranslated region (UTRs). Different AREs seem to differentially activate both the 5′ to 3′ and 3′ to 5′ decay pathways (Chen et al. 2001; Gao et al. 2001; Mukherjee et al. 2002; Lykke-Andersen and Wagner 2005; Stoecklin et al. 2006; Murray and Schoenberg 2007). TTP, a key ARE binding factor, interacts with the Dcp2 protein and enhances decapping of a target ARE-containing RNA in vitro (Fenger-Gron et al. 2005). Depletion of the decapping activator Lsm1 by siRNA impairs ARE-mediated decay in human cells (Stoecklin et al. 2006). Finally, decapping has also been implicated in miRNA-mediated gene silencing. Knockdown of Dcp1:Dcp2 decapping complex effectively relieved miRNA-mediated silencing of firefly luciferase expression in Drosophila cells (Rehwinkel et al. 2005). Depletion of several decapping activators including Hedls (also referred to as Ge-1 and Edc4), Dcp1a, Edc3, and Lsm1 in Drosophila S2 cells suppressed gene silencing mediated by several miRNAs (Eulalio et al. 2007b). Moreover, the decapping activator RCK/p54 has also been shown to be important in miRNA-mediated silencing in human cells (Chu and Rana 2006). Furthermore, Dcp2 can preferentially bind to a subset of mRNA substrates (Li et al. 2008). Highly transcript-specific decapping of Dcp2 has been reported in vitro and in mammalian cells (Li et al. 2008, 2009). Collectively, it appears that in addition to a potential general mRNA decay function, Dcp2 can also target various specific mRNA decay processes.
One striking feature of the decapping machinery is that the decapping enzyme Dcp2 and other decapping activators, including Dcp1a, Edc3, Hedls, and the Lsm1-7 complex, and the 5′ to 3′ exoribonuclease, Xrn1 colocalize to distinct cytoplasmic foci called P-bodies (van Dijk et al. 2002; Sheth and Parker 2003; Fenger-Gron et al. 2005; Yu et al. 2005; Franks and Lykke-Andersen 2008). In addition, proteins involved in NMD, ARE-mediated decay, and miRNA-mediated silencing machineries localize to P-bodies (Eystathioy et al. 2003; Ding et al. 2005; Sen and Blau 2005; Sheth and Parker 2006; Franks and Lykke-Andersen 2007). Moreover, mRNA decay intermediates and translational repressed mRNAs can also be found in P-bodies (Sheth and Parker 2003, 2006; Liu et al. 2005; Franks and Lykke-Andersen 2007), suggestive of a role for P-bodies in mRNA decay. However, loss of visible P-bodies do not lead to detectable alterations in mRNA decay (Decker et al. 2007; Eulalio et al. 2007a; Stalder and Muhlemann 2009) and decapping can occur within polysomes that are devoid of visible P-bodies (Hu et al. 2009); therefore the assembly of P-bodies does not seem to be necessary for mRNA degradation. The importance for the decapping machinery to form cytoplasmic foci still remains to be elucidated.
The X29 protein was first identified in Xenopus as a nuclear decapping protein that specifically binds U8 snoRNA (Tomasevic and Peculis 1999; Ghosh et al. 2004). However, we recently demonstrated that the mammalian homolog, Nudt16, is a cytoplasmic decapping enzyme and can regulate the stability of specific mRNA substrates (Song et al. 2010). In this study, we used immortalized mouse embryonic fibroblast (MEF) cell lines with reduced levels of Dcp2 or Nutd16 to study the differential utilization of the two decapping enzymes in NMD, ARE-mediated decay and miRNA-mediated silencing. Our results showed that Dcp2 was preferentially used in NMD; Dcp2 and Nudt16 are redundant in miRNA-mediated silencing; Dcp2 and Nudt16 are differentially utilized for the decay of ARE-containing mRNAs; and loss of Dcp2 or Nudt16 did not appear to impact P-body assembly. These data suggest decapping enzymes could be differentially utilized for different cellular mRNA decay processed in mammalian cells.
RESULTS
Establishment of Dcp2 and Nudt16 knockdown MEF cell lines
We recently generated mice containing a homozygous insertion of the β-galactosidase-neomycin resistance fusion gene (β-geo) within intron 1 of the mouse Dcp2 loci (Dcp2β/β) (Song et al. 2010). However, the mice are hypomorphic for Dcp2 production likely due to background alternative splicing that bypasses the β-geo gene to generate low levels of Dcp2 protein despite the homozygous insertion of the β-geo gene. To characterize the role of Dcp2 in distinct mRNA decay processes, we utilized SV40 large T antigen immortalized mouse embryonic fibroblast (MEF) cells obtained from wild-type or Dcp2β/β embryos, which contain detectable and undetectable Dcp2 protein levels, respectively (Song et al. 2010). The extensively reduced levels of Dcp2 protein in these cells provided us with a Dcp2 knockdown cell line to assess the significance of Dcp2 in mRNA decay. No morphological changes were obvious between the wild-type and Dcp2β/β MEF cell lines except for a slow growth phenotype observed in the Dcp2β/β cells (Supplemental Fig. S1).
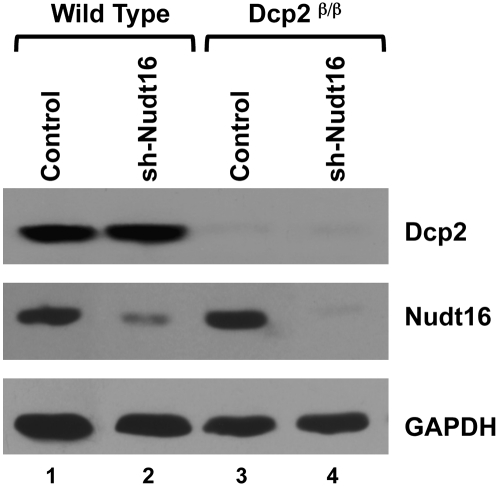
In addition to Dcp2, we also recently identified Nudt16 as a cytoplasmic decapping enzyme in mammalian cells (Song et al. 2010). To investigate the role of both the Nudt16 and Dcp2 decapping proteins in cellular mRNA decay, we utilized stably transformed MEF cell lines with reduced Nudt16 protein expression in wild-type and Dcp2β/β MEF cells. Consistent with Song et al. (2010), >95% reduction of Dcp2 protein level was observed in the Dcp2β/β MEF cells and ∼70% and 90% reduction of Nudt16 was observed in the Nudt16 knockdown cells within the wild-type or Dcp2β/β MEF cells, respectively (Fig. 1). These Dcp2 and Nudt16 individual and double knockdown MEF cell lines were used in the subsequent analyses to assess the significance of Dcp2 and Nudt16 in mRNA decay.
FIGURE 1.
Reduction of Nudt16 protein levels in wild-type and Dcp2β/β MEFs. Extract from immortalized clonal MEF cell lines derived from wild-type embryos or embryos with a homozygous insertion of the β-galactosidase-neomycin resistance fusion gene (β-geo) within intron 1 of the mouse Dcp2 loci (Dcp2β/β) expressing Lentiviral directed empty vector control (lanes 1,3) or Nudt16-specific shRNA (lanes 2,4) (Song et al. 2010), were used in Western blot analysis. Bands corresponding to Dcp2, Nudt16, or the GAPDH loading control protein are indicated on the right.
Loss of Dcp2 and Nudt16 had no apparent impact on P-bodies in MEF cells
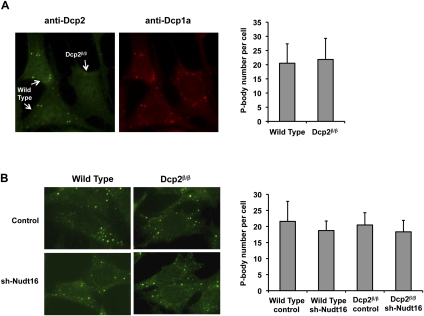
In addition to a dispersed cytoplasmic localization, Dcp2 protein also localizes in cytoplasmic foci termed P-bodies (van Dijk et al. 2002; Sheth and Parker 2003). The function of P-bodies is still unclear, although they have been postulated to be sites of mRNA decay, mRNP storage, and miRNA-directed mRNA decay (Sheth and Parker 2003, 2006; Liu et al. 2005; Franks and Lykke-Andersen 2007, 2008). Disruption of the Dcp2 gene in yeast results in an increase in the size and number of P-bodies (Teixeira and Parker 2007), believed to be a consequence of aberrantly uncapped mRNA-containing mRNPs that aggregate. However, surprisingly, siRNA-directed knockdown of Dcp2 in mammalian cells did not lead to altered P-body states (Yu et al. 2005; Stoecklin et al. 2006). Consistent with these previous observations, the virtually complete knockdown of Dcp2 in mammalian cells also did not reveal appreciable differences in P-bodies within the Dcp2β/β MEF cells. Wild-type and Dcp2β/β MEF cells were mixed and cultured on the same coverslip to ensure similar cell confluency and growth stage. P-bodies were stained with antibodies directed to either Dcp2 or Dcp1a. As expected, both proteins colocalized within P-body foci in the wild-type MEF cells and foci were not detected with the Dcp2-specific antisera in the Dcp2β/β cells (Fig. 2A). The number of P-bodies, as determined by Dcp1a localization, remained comparable regardless of the presence or absence of Dcp2, with 20.2 foci and 21.1 foci per cell respectively. Similar results were also obtained when P-bodies were detected with other P-body markers including antibodies to Hedls (Edc4) or Lsm1 (Supplemental Fig. S2). Analysis of P-bodies in cells expressing reduced levels of Nudt16 protein was also carried out. The number of P-bodies, as determined by anti-Dcp1a immunofluorescence analysis, remained the same in MEF cells with reduced Nudt16 protein or a combination of both Dcp2 and Nudt16 knockdown (Fig. 2B). These data indicate that formation of P-bodies appears independent of both Dcp2 and Nudt16 decapping enzymes in mammalian cells.
FIGURE 2.
P-body numbers are unaltered in MEF cells with undetectable levels of Dcp2 and/or reduced levels of Nudt16 protein. (A) Wild-type and Dcp2β/β MEF cells were simultaneously cultured on the same cover slide and distinguished by immunofluorescence, using affinity purified Dcp2-specific antibody detected with FITC conjugated secondary antibody. Dcp1a was used as a P-body marker and visualized by an anti-Dcp1a antibody detected with Texas Red conjugated secondary antibody. The number of P-bodies per cell, as determined by Dcp1a positive foci, was counted and the average numbers obtained from 30 cells are graphed on the right with standard deviations indicated. (B) P-bodies were visualized by immunofluorescence with anti-Dcp1a antibody detected with FITC conjugated secondary antibody and quantitated as described in A, except wild-type or Dcp2β/β MEF cells expressing a control vector or Nudt16-specific shRNA were used as indicated.
Preferential utilization of Dcp2 in NMD
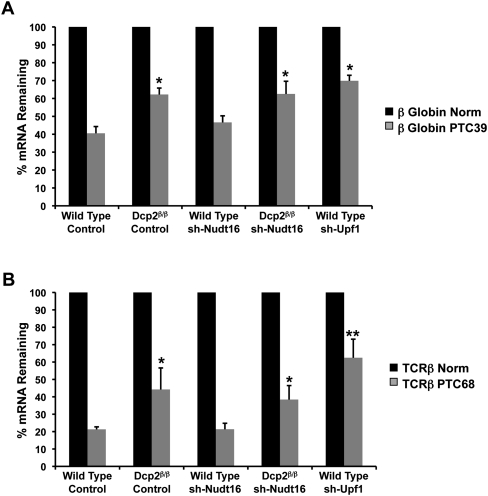
The recent demonstrations that both Dcp2 and Nudt16 possess transcript-specific decapping properties (Ghosh et al. 2004; Li et al. 2008, 2009) prompted us to test whether various mRNA decay processes differentially utilized the two decapping enzymes and assessed their roles in nonsense-mediated decay (NMD), ARE-mediated decay, and noncoding RNA-mediated gene silencing. The effect of Dcp2 on the reduction of nonsense mRNA levels was assessed by transiently transfecting MEF cells with plasmids encoding either a normal β-globin transcript lacking a nonsense mutation (Norm) or its nonsense mutation-containing version (PTC39). A Renilla luciferase plasmid was cotransfected for normalization of transfection efficiencies. Total RNA was isolated 24 h post-transfection, reverse transcribed, and analyzed by quantitative real-time PCR. A comparison of nonsense-containing transcript levels to that of the nonsense-lacking transcript levels are presented in Figure 3A. Consistent with Lejeune et al. (2003), PTC39 mRNA levels were significantly reduced in the wild-type MEF cells with 43% of the mRNA remaining relative to Norm mRNA. In contrast, a statistically significant increase in PTC39 mRNA levels was detected in Dcp2β/β MEF cells where 64% of mRNA remained relative to the Norm mRNA levels.
FIGURE 3.
Contribution of decapping enzymes to NMD. (A) mRNA levels derived from transgenes expressing a β-globin transcript (Norm), or nonsense codon-containing β-globin transcript (PTC39) in wild-type and Dcp2β/β MEF cells, transduced with or without Nudt16-specific shRNA or in wild-type MEF cells transduced with Upf1 shRNA-expressing virus as indicated, were determined by quantitative real-time PCR. mRNA levels were determined 24 h post-transfection and normalized to Renilla luciferase mRNA levels expressed from a cotransfected plasmid. The normalized mRNA value for the sense β-globin transcript was arbitrarily set to 100%. Nomenclature of the cell lines is as described in the legend to Figure 1. The average of three independent experiments is shown with standard deviation indicated. Asterisks denote P < 0.05 (ANOVA followed by Tukey-Kramer Multiple Comparisons Test). (B) The level of TCRβ mRNA (Norm) or nonsense codon-containing TCRβ transcript (PTC68) was determined as described in A. The normalized mRNA value for the sense TCRβ transcript was arbitrarily set to 100%. The average of three independent experiments is shown with standard deviation indicated. Asterisks denote P < 0.05 and double asterisks denote P < 0.01 (ANOVA followed by Tukey-Kramer Multiple Comparisons Test).
Analysis of the same reporter constructs was also carried out in MEF cells containing reduced levels of Nudt16. Interestingly, a reduction of Nudt16 levels did not result in a detectable difference in PTC39 mRNA levels compared to that observed in control knockdown cells (Fig. 3A). Similarly, knockdown of Nudt16 in the Dcp2β/β cell background also did not result in altered PTC39 mRNA levels and were indistinguishable from levels obtained with reduced Dcp2 in the Dcp2β/β cells. Similar results were observed using a PTC-containing TCRβ reporter. MEF cells were transfected with plasmids encoding a normal TCRβ transcript (Norm) or its nonsense mutation-containing version (PTC68). A twofold increase in PTC68 mRNA levels was detected in Dcp2β/β MEF cells relative to Norm mRNA, while a reduction of Nudt16, either in the wild-type or in the Dcp2β/β MEF background, did not significantly alter PTC68 mRNA levels (Fig. 3B). Knockdown of the Upf1, an NMD core factor, was used as a positive control for NMD directed decay of each PTC transcript. However, these are likely underestimations since we were only able to obtain 65% knockdown of Upf1 (data not shown). Collectively, these data demonstrate that Dcp2 is preferentially utilized over Nudt16, in the demise of these two PTC-containing transcripts.
Redundancy of Dcp2 and Nudt16 in miRNA-mediated gene silencing
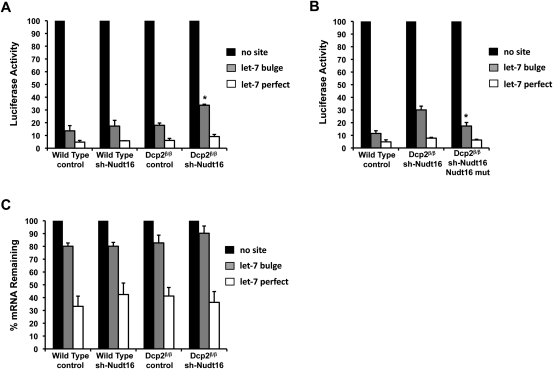
Dcp2 has been implicated in miRNA-mediated gene silencing, where a knockdown of the Dcp1:Dcp2 decapping complex impaired miRNA-mediated silencing of firefly luciferase expression in Drosophila cells (Rehwinkel et al. 2005). To determine the significance of Dcp2 in miRNA- and siRNA-mediated gene silencing in mammalian cells, we assessed the level of Renilla luciferase activity from transcripts containing or lacking miRNA target sites. Wild-type or Dcp2β/β MEF cells were transiently transfected with plasmids encoding Renilla luciferase lacking an miRNA target sequence in the 3′ UTR or containing either a let-7 miRNA target sequence (3X let-7 bulge) or a let-7 perfectly complementary sequence. The three reporter constructs were each co-transfected with a plasmid expressing the pri-let7 miRNA and firefly luciferase plasmid for normalization of transfection efficiencies. Cells were harvested and assayed for luciferase activity 24 h post-transfection. Renilla luciferase activities are presented in Figure 4A with the activity obtained from the reporter lacking a let-7 target sequence arbitrarily set to 100%. Interestingly, significant differences were not detected in Renilla luciferase activities with either the let-7 bulged or perfect match sites when comparing the results of wild-type and Dcp2β/β MEF cells. An 89% reduction in luciferase activity was observed in wild-type MEF cells, while an 87% reduction was detected in the Dcp2β/β cells when assaying luciferase activities from transcripts harboring a bulged let-7 target site. Similarly, a 95% and 94% reduction in luciferase activity were detected in the wild-type and Dcp2β/β cells, respectively, when assaying luciferase activities from transcripts harboring a perfect match to the let-7 site. Therefore, in contrast to the situation in Drosophila, where Dcp2 was found to contribute to miRNA directed silencing, the almost complete absence of Dcp2 does not appear to have a significant impact on miRNA- or siRNA-mediated gene silencing with this reporter system in mammalian cells.
FIGURE 4.
Dcp2 and Nudt16 are redundant in miRNA-mediated gene silencing. (A) Plasmids expressing mRNAs encoding the Renilla luciferase with three let-7 miRNA bulge sites, a let-7 perfectly complementary site, or no let-7 site in the 3′ UTR were transfected into the indicated MEF cell lines and luciferase activity was analyzed 24 h post-transfection. Renilla luciferase activity was normalized to firefly luciferase activity derived from a cotransfected plasmid. The normalized value of Renilla luciferase with no let-7 site was arbitrarily set to 100%. The average of three independent experiments is shown with standard deviation indicated by the error bars. Cell-line nomenclature is as described in the legend to Figure 1. An increase in Renilla luciferase activity was detected only in cells with a reduction of both Dcp2 and Nudt16 protein. Asterisks denote P < 0.01 (ANOVA followed by Tukey-Kramer Multiple Comparisons Test). (B) A similar analysis as that presented in A is shown except the Dcp2 and Nudt16 double knockdown was complemented with an shRNA-resistant Nudt16 expression construct. Luciferase activity was normalized as in A and the average of three independent experiments is shown with standard deviation. Asterisks denote P < 0.02 (ANOVA followed by Tukey-Kramer Multiple Comparisons Test). (C) Renilla luciferase mRNA levels from an analysis as presented in A above were determined by real-time quantitative PCR and normalized to mRNA levels derived from a cotransfected firefly luciferase expression construct. The average of three independent experiments is shown with standard deviation indicated by the error bars.
To assess the role of Nudt16 or the combined effect of Dcp2 and Nudt16, the same reporter constructs were tested in Nudt16 knockdown MEF cells. As shown in Figure 4A, the reduction of Nudt16 alone had no detectable consequence on Renilla luciferase activity with either the let-7 bulged or perfectly matched sites. Importantly, although a reduction of either decapping enzyme individually had no consequence, a double knockdown of both Nudt16 and Dcp2 significantly increased Renilla luciferase activity of the reporter construct containing the let-7 bulged sites. In contrast to the 11% of Renilla activity detected in the control virus-infected cells containing wild-type levels of both Dcp2 and Nudt16, the activity was increased approximately threefold to 31% in the double knockdown conditions (Fig. 4A). Importantly, this increase can be reversed by overexpression of Nudt16 (Fig. 4B) indicating the presence of at least one of the decapping enzymes is sufficient to promote maximal silencing. Interestingly, Renilla luciferase activity of the reporter construct containing a let-7 perfect match target site showed no significant change despite the low level of both Dcp2 and Nudt16. These data indicate that although Dcp2 and Nudt16 do not contribute to siRNA-directed gene silencing in our assays, they demonstrate a redundant contribution to miRNA directed gene silencing where an effect was only detected when the levels of both proteins were simultaneously reduced. This suggests that miRNA-mediated silencing utilizes either the Dcp2 or Nudt16 decapping proteins.
To determine whether the increased luciferase activity in the Dcp2β/β/sh-Nudt16 MEF cell line was due to an increase in luciferase mRNA levels or an indirect consequence of reduced Dcp2 and Nudt16, Renilla luciferase mRNA levels were determined relative to the mRNA from the cotransfected control firefly luciferase reporter. As shown in Figure 4C, despite a considerable decrease in luciferase activity from the let-7 bulge sites containing mRNA, only a modest decrease of the mRNA was observed. Furthermore, a statistically significant difference in steady-state Renilla luciferase mRNA containing the let-7 bulge sites was not detected in the Dcp2 and Nudt16 double knockdown MEF cells relative to control cells. These data indicated that the detected decrease in luciferase activity in Figure 4A is not a direct consequence of a corresponding decrease in mRNA levels. Furthermore, the observed increase in luciferase activity from the let-7 bulge site-containing mRNA in the double knockdown cells are not a result of increased steady-state mRNA levels and therefore appear to be an indirect consequence of Dcp2 and Nudt16 decapping on translation.
Differential utilization of Dcp2 and Nudt16 in the decay of ARE-containing mRNAs
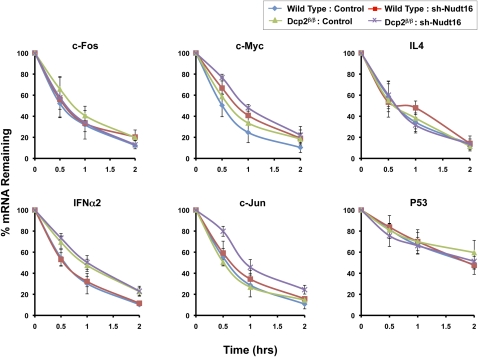
Dcp2 has been reported to function in ARE-mediated decay where the destabilizing ARE-binding factors, TTP and BRF-1, can recruit the Dcp2 decapping complex in vitro to promote ARE-dependent decapping (Fenger-Gron et al. 2005; Lykke-Andersen and Wagner 2005). Consistent with previous reports, we can show that a reporter construct containing a TNFα ARE was responsive to Dcp2 and was stabilized in the Dcp2β/β cell (Supplemental Fig. S3). To further examine the consequence of the lack of detectable Dcp2 on the decay of ARE-containing mRNAs, we investigated the stability of endogenous ARE-containing mRNAs in the Dcp2β/β cells. Wild-type and Dcp2β/β cells were treated with actinomycin D and the decay of specific ARE-containing mRNAs was analyzed by quantitative real-time PCR for up to 2 h following transcriptional arrest. Six different ARE-containing mRNAs with detectable expression in MEF cells were tested. As shown in Figure 5, stability of the IFNα2 mRNA, was significantly increased in Dcp2β/β cells, with a twofold increase in half-life upon reduction of Dcp2. No significant difference in mRNA half-lives was detected in the stability of c-Fos, IL4, c-Jun, and P53 mRNAs when Dcp2 was individually reduced. However, a contribution of Dcp2 was detected on the stability of c-Myc and c-Jun mRNAs when its levels were reduced in conjunction with Nudt16 (see below). These data indicate that Dcp2 is differentially utilized in the decay of ARE-containing mRNAs where a subset, but not all ARE-containing mRNAs appear to be responsive to Dcp2 activity.
FIGURE 5.
Differential utilization of Dcp2 and Nudt16 in ARE-mediated decay. mRNA levels of the indicated mRNAs were followed up to 2 h post-actinomycin D treatment. Levels of the denoted endogenous ARE-containing mRNAs remaining at the indicated time points were determined by quantitative reverse transcription and real-time PCR. The values of three independent RNA preparations normalized to the β-actin mRNA are shown, with ± standard deviation. Cell-line nomenclature is as described in the legend to Figure 1.
The same mRNAs were also analyzed in the absence of Nudt16. Interestingly, a reduction of Nudt16 protein levels did not alter the half-lives of the c-Fos, IL4, or P53 mRNAs (Fig. 5). However, the c-Myc mRNA was stabilized by 50% from a 30 to 45 min half-life upon reduction of Nudt16 and importantly, it was further stabilized to a 1-h half-life in the double knockdown conditions, indicating an additive role for Nudt16 and Dcp2 in decay of the c-Myc mRNA. These results were not restricted to actinomycin D directed transcriptional inhibition and similar results were obtained using DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) as the transcription blocker (Supplemental Fig. S4). Redundant roles of Dcp2 and Nudt16 were also observed with the c-Jun mRNA where an individual reduction of either Dcp2 or Nudt16 protein levels had no effect on the mRNA half-life, while a simultaneous reduction of both proteins lead to an almost doubling of the c-Jun mRNA half-life from 30 to 50 min. These results indicate that Dcp2 and Nudt16 fulfill redundant roles in regulating c-Jun mRNA decay. Collectively, our analysis of ARE-containing mRNAs reveals a complex and transcript-specific utilization of the decapping enzymes by ARE-containing mRNAs ranging from no consequence of reduced Dcp2 or Nudt16 levels to synergistic roles of both decapping enzymes.
DISCUSSION
In this study, we investigated the utilization of Dcp2 and Nudt16 decapping enzymes in distinct cellular mRNA decay events in mouse embryonic fibroblast cell lines individually or simultaneously reduced for Dcp2 and/or Nudt16 proteins. Our results suggest that Dcp2 and Nudt16 are selectively involved in specific mRNA decay processes and do not uniformly function on all mRNAs. The NMD substrate tested preferably used Dcp2; while an miRNA-mediated silencing reporter appears to use both decapping enzymes redundantly and ARE-containing mRNAs differentially utilized the two decapping enzymes.
NMD has been shown to evoke decapping in both yeast and mammalian cells (Muhlrad and Parker 1994; Lejeune et al. 2003; Couttet and Grange 2004). The key NMD factor Upf1 can form a complex with the Dcp2 protein in both organisms (He and Jacobson 1995; Lykke-Andersen 2002; Lejeune et al. 2003). Knockdown of Dcp2 by siRNA impaired the removal of nonsense codon containing transcript in Hela cells, indicating that Dcp2 decapping is utilized in NMD (Lejeune et al. 2003). Our data with MEF cells containing virtually undetectable levels of Dcp2 were consistent with previous reports where clearing of nonsense containing β-globin or TCRβ reporter mRNA was compromised in the absence of Dcp2 (Fig. 3). Interestingly, we did not detect a role for Nudt16 on the same mRNA (Fig. 3), suggesting Dcp2, but not Nudt16, is specifically utilized with these NMD substrates. As expected, a knockdown of Upf1 resulted in a more pronounced increase in the PTC containing mRNA levels, however, this is likely an underestimation since only a 65% reduction in Upf1 mRNA levels were obtained (unpublished observations). Considering that PTC containing transcripts are subjected to NMD-directed clearing by multiple pathways, including deadenylation (Chen and Shyu 2003; Zheng et al. 2008) and Smg6 endonucleolytic decay (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009), only a modest increase of the PTC transcript would be expected in the Dcp2β/β cells. Thus, Dcp2 decapping contributes to, but is not essential for NMD, while Nudt16 decapping may not be a major contributor to NMD.
Decapping has been implicated in miRNA-mediated silencing, since depletion of Dcp1:Dcp2 in Drosophila cells partially relieved miRNA-mediated repression of luciferase activity (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006). However in our analyses, depletion of Dcp2 protein alone in MEF cells did not significantly affect miRNA-mediated silencing of luciferase activity (Fig. 4A). Only when Dcp2 and Nudt16 were simultaneously reduced did we observe a significant restoration of luciferase activity (Fig. 4A). This indicates that Dcp2 and Nudt16 are redundant in miRNA-mediated repression and that the miRNA silencing machinery could use either Dcp2 or Nudt16 in mammals. The lack of an obvious ortholog for Nudt16 in Drosophila (Taylor and Peculis 2008) might be one explanation. One surprising finding in our studies was that although we detect a simultaneous dependence of both decapping enzymes, miRNA target reporter mRNA levels were not significantly altered under reduced decapping enzyme conditions. This could either be due to residual levels of decapping enzyme still present in the knockdown conditions or an indication of an indirect role of the decapping enzymes in mammalian miRNA directed silencing in this system. The latter possibility suggests a potential role of Dcp2 and Nudt16 in the regulation of mRNA(s) that encode facilitators of translational silencing. Future efforts will address this possibility. Furthermore, since decapping activators including Dcp1a, Edc4, Edc3, and Lsm1-7 are differentially required for distinct miRNA targets in Drosophila (Eulalio et al. 2007b), analysis of additional miRNA substrates will be necessary to determine whether Dcp2 and/or Nudt16 may also fulfill a direct function in regulating mRNA levels for a subset of miRNA targets in mammal cells. In contrast to the miRNA-directed silencing, siRNA-directed silencing was not impacted by the loss of either Dcp2 or Nudt16 (Fig. 4A,C), consistent with siRNA predominantly evoking an endonucleolytic cleavage mechanism for mRNA decay (Zamore et al. 2000; Liu et al. 2004).
An important finding revealed from our data is that ARE-containing transcripts can differentially utilize the two different decapping enzymes to promote their decay. Several lines of evidence have previously implicated decapping in ARE-mediated decay with the focus exclusively on Dcp2. For example, the ARE-binding factors TTP and BRF-1 can interact with the decapping complex and TTP could specifically activate Dcp2 decapping in vitro (Fenger-Gron et al. 2005; Lykke-Andersen and Wagner 2005), suggesting ARE-mediated decay involves Dcp2 decapping. In addition Dcp2 was shown to contribute to decay of the c-Fos mRNA (Murray and Schoenberg 2007). However, the first line of evidence suggesting that Dcp2 may not be the only decapping enzyme involved in ARE-mediated decay was provided by Stoecklin et al. (2006), where intriguingly, depletion of Xrn1 significantly stabilized a GM-CSF reporter mRNA, while depletion of Dcp2 had no detectable effect on the decay of the same mRNA. These finding suggested the GM-CSF mRNA is degraded from the 5′ end but in a Dcp2 independent manner.
Our results demonstrate that decapping enzymes could be differentially utilized for the decay of ARE-containing mRNAs, where both Dcp2 and Nudt16 can individually or synergistically be involved (Fig. 5). The IFNα2 mRNA selectively utilizes the Dcp2 protein for decay, while degradation of the c-Myc mRNA involves both Dcp2 and Nudt16. The c-Jun mRNA was only stabilized when both Dcp2 and Nudt16 were reduced, suggesting Dcp2 and Nudt16 are redundant in the decay of this mRNA. The decay of c-Fos, IL4 and P53 mRNAs were insignificantly affected by a depletion of either Dcp2 or Nudt16. This is not surprising since an ARE may also promote exosome-mediated 3′ to 5′ decay (Chen et al. 2001; Mukherjee et al. 2002). The differential requirement of decapping enzymes in ARE-mediated decay is likely a consequence of distinct ARE-binding proteins that interact with each ARE and recruit one or the other decapping enzyme. Future efforts to identify Nudt16 binding partners will begin addressing this possibility.
Curiously, Nudt16 was recently reported to be an inosine diphosphatase linked to the prevention of single-stranded DNA breaks (Iyama et al. 2010). Whether the diphosphatase activity and mRNA decapping activity of Nudt16 constitute distinct activities and whether mRNA stability functions in single-stranded DNA breaks remain to be determined. The demonstration that at least two different decapping enzymes can initiate mRNA 5′ end decay in mammalian cells and that distinct mRNA decay processes differentially utilize these enzymes, provides new avenues to pursue mRNA turnover. The utilization of distinct decapping enzymes in specific decay may be a broadly utilized process as evident by a 5′-end quality control mechanism with decapping endonuclease activity on aberrantly capped mRNAs (Jiao et al. 2010). Future efforts addressing how each decay process and each transcript recruits and modulates the decapping proteins will greatly advance our understanding of regulated mRNA decay.
MATERIALS AND METHODS
Plasmids
The pmCMV-G1 Norm and pmCMV-G1 39Ter NMD reporter plasmids that encode the normal or nonsense mutation-containing versions of the β-globin transcripts, respectively, were kindly provided by Dr. L. E. Maquat (University of Rochester). The β-495 and β-496 NMD reporter plasmids that encode the normal or nonsense mutation-containing versions of the TCRβ transcripts, respectively, were provided by Dr. Miles Wilkinson (UCSD) (J Wang et al. 2002). The microRNA reporter plasmids encoding the Renilla luciferase (pCMV-RL), Renilla luciferase with three let-7 imperfectly base-paired target sites in the 3′ UTR (pCMV-RL-3Xbulge), the Renilla luciferase with a complementary let-7 target site in the 3′ UTR (pCMV-RL-perfect), and the pri-let7 expression plasmid (pcDNA3-priLet7) were kindly provided by W. Filipowicz (Friedrich Miescher Institute for Biomedical Research) (Schmitter et al. 2006). The pRL-SV40 plasmid encoding the Renilla luciferase reporter protein was obtained from Promega Corp. and the pSV2ALΔ5′-Luc plasmid expressing the firefly luciferase gene has previously been described (de Wet et al. 1987). The plasmid pLKO.1-puro-mUpf1 expressing shRNA directed against Upf1 was obtained from Sigma-Aldrich. To construct the retroviral plasmid pBMN-Nudt16mut, which overexpresses a shRNA-resistant Nudt16 mutant, pGEM-Nudt16 was used as a template with the following PCR primers: Nudt16Mu587F-2 and Nudt16Mu567R-2 (Supplemental Table 1) to generate pGEM-Nudt16mut. pGEM-Nudt16mut was subsequently used as a template with primers Nudt16F2 and Nudt16R2. The PCR product was purified and digested by Xho I and Not I and inserted into pBMN-I-GFP (Addgene).
Endogenous mRNA decay assay by real-time PCR
mRNA half-lives were determined with the use of actinomycin D (5 μg/mL) or 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB; 30ug/mL) to inhibit transcription in mouse embryonic fibroblast (MEF) cells and harvested at the indicated times. Total RNA was isolated by TRIzol reagent (Invitrogen) following the manufacturer's instructions. RNAs were reverse transcribed by M-MLV reverse transcriptase (Invitrogen) with random primers according to the manufacturer's instructions. The mRNA levels were quantified by real-time PCR using SYBR green PCR core reagent (Applied Biosystems) and an ABI Prism 7900HT sequence detection system. The specificity of the amplified PCR products was assessed by a melting curve analysis after the last cycle by the manufacturer's suggested program. Each mRNA was amplified using the appropriate specific primers and values were normalized to the stable β-actin mRNA. Primers used in this study are listed in Supplemental Table 1. Data were analyzed by the comparative Ct method as previously described (Livak and Schmittgen 2001). The mRNA levels at time 0 h were arbitrarily set to 100%.
Reporter assays
Transfections were performed in six-well plates using Lipofectamine 2000 reagent (Invitrogen Corp.) according to the manufacturer's recommendations. For the NMD reporter assays, transfections were carried out with 1 μg of pmCMV-G1-Norm or pmCMV-G1-39Ter plasmid or 0.5 μg of β-495 or β-496 plasmid, along with 1 μg pRL-SV40 plasmid. Total RNA was isolated 24 h post-transfection, β-globin or TCRβ mRNA levels analyzed by real-time reverse transcription-PCR as described above, and normalized to the Renilla luciferase mRNA levels. Analysis of microRNA silencing was carried out with 0.3 μg of pCMV-RL or pCMV-RL-3Xbulge or pCMV-RL-perfect plasmids transfected into MEF cells in six-well plates together with 1.5 μg pcDNA3-priLet7 and 1μg pSV2ALΔ5′-Luc plasmids. Nudt16 complementation studies were carried with the transduction pBMN-Nudt16mut-derived retrovirus according to the manufacturer's instructions to express an shRNA-resistant Nudt16 24 h prior to transfection of the dual luciferase reporter construct. Renilla and firefly luciferase activities were detected using a Dual Luciferase Assay System (Promega) 24 h post-transfection according to the manufacturer's guidelines using GloMax-Multi Luminescence Module (Promega). Analysis of endogenous mRNA levels following a block in transcription with Actinomycin D or DRB with gene-specific quantitative RT-PCR was carried out as above.
Western blot
Cells were sonicated in PBS and protein extract was resolved by 12.5% SDS-PAGE. Nudt16 polyclonal antibody (1:100 dilution [Song et al. 2010]), monoclonal anti-eIF4E antibody (1:1500 dilution [Transduction Laboratories]), affinity purified Dcp2 polyclonal antibody (1:500 dilution [Z Wang et al. 2002]), and monoclonal anti-GAPDH antibody (1:2000 dilution [Abcam Inc.]), were used for Western blot analysis and visualized using secondary antibodies coupled to horse radish peroxidase (Jackson ImmunoResearch) and chemiluminescence (ECL; GE Healthcare Life Science).
Immunofluorescence assay
MEF cells were grown on glass coverslips in six-well plates to 50% confluency. The coverslips containing the cells were treated as previously described (Liu et al. 2004). Affinity-purified Dcp2 rabbit polyclonal antibody was used as the primary antibody to detect endogenous Dcp2 and monoclonal Dcp1a antibody (1:200 dilution [Abnova]) was used to detect Dcp1a containing P-bodies. Goat anti-rabbit or goat anti-mouse secondary antibodies conjugated with Texas Red or FITC (Jackson ImmunoResearch) were used at a 1:200 dilution. The nuclei were stained with 1 μg/mL DAPI for 10 min. The images were obtained with a Zeiss Axiovert 100 M microscope. P-body numbers per cell were counted and the average number of 30 cells was graphed.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank L. Maquat for the β-globin NMD reporter constructs, M. Wilkinson for the TCRβ NMD reporter plasmids, W. Filipowicz for the Renilla luciferase reporter with let7 target sites, and members of the Kiledjian lab for assistance with experimental techniques and helpful discussions. This work was supported by NIH grant GM67005 to M.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2439811.
REFERENCES
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 23: 4805–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210 doi: 10.1371/journal.pbio.0040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukarryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Couttet P, Grange T 2004. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol 7: 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell 19: 437–447 [DOI] [PubMed] [Google Scholar]
- Dunckley T, Parker R 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007a. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E 2007b. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21: 2558–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE-binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wilusz CJ, Peltz SW, Wilusz J 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J 20: 1134–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Ghosh T, Peterson B, Tomasevic N, Peculis BA 2004. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol Cell 13: 817–828 [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9: 437–454 [DOI] [PubMed] [Google Scholar]
- Hsu CL, Stevens A 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Abolhassani N, Tsuchimoto D, Nonaka M, Nakabeppu Y 2010. NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res 38: 4834–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Li Y, Song MG, Kiledjian M 2008. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol 28: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ho ES, Gunderson SI, Kiledjian M 2009. Mutational analysis of a Dcp2-binding element reveals general enhancement of decapping by 5′-end stem-loop structures. Nucleic Acids Res 37: 2227–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rodgers ND, Jiao X, Kiledjian M 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 21: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Jiao X, Liu H, Gu M, Lima CD, Kiledjian M 2004. Functional analysis of mRNA scavenger decapping enzymes. RNA 10: 1412–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581 [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, and Wilusz J 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. Embo J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EL, Schoenberg DR 2007. A+U-rich instability elements differentially activate 5′-3′ and 3′-5′ mRNA decay. Mol Cell Biol 27: 2791–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W 2006. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 34: 4801–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM 2005. Argonaute2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol 7: 633–636 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Li Y, Kiledjian M 2010. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell 40: 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2009. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep 7: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Peculis BA 2008. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res 36: 6021–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18: 2274–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasevic N, Peculis B 1999. Identification of a U8 snoRNA-specific binding protein. J Biol Chem 274: 35914–35920 [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF 2002. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J Biol Chem 277: 18489–18493 [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11: 1795–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]