Abstract
Translation initiation in eukaryotes involves more than a dozen protein factors. Alterations in six factors have been found to reduce the fidelity of start codon recognition by the ribosomal preinitiation complex in yeast, a phenotype referred to as Sui−. No small molecules are known that affect the fidelity of start codon recognition. Such compounds would be useful tools for probing the molecular mechanics of translation initiation and its regulation. To find compounds with this effect, we set up a high-throughput screen using a dual luciferase assay in S. cerevisiae. Screening of over 55,000 compounds revealed two structurally related molecules that decrease the fidelity of start codon selection by approximately twofold in the dual luciferase assay. This effect was confirmed using additional in vivo assays that monitor translation from non-AUG start codons. Both compounds increase translation of a natural upstream open reading frame previously shown to initiate translation at a UUG. The compounds were also found to exacerbate increased use of UUG as a start codon (Sui− phenotype) conferred by haploinsufficiency of wild-type eukaryotic initiation factor (eIF) 1, or by mutation in eIF1. Furthermore, the effects of the compounds are suppressed by overexpressing eIF1, which is known to restore the fidelity of start codon selection in strains harboring Sui− mutations in various other initiation factors. Together, these data strongly suggest that the compounds affect the translational machinery itself to reduce the accuracy of selecting AUG as the start codon.
Keywords: translation initiation, start codon, compound screen, ribosome, yeast
INTRODUCTION
Translation of mRNA into functional protein is energetically expensive and must be highly accurate. The initiation phase of protein synthesis establishes the reading frame of translation and commits the machinery to begin the elongation phase. For most mRNAs, the start codon is an AUG. We and others have previously demonstrated that codons that vary from AUG in one position (near-cognates) can be used up to ∼8% as efficiently as AUG codons as start sites in S. cerevisiae (Zitomer et al. 1984; Clements et al. 1988; Donahue and Cigan 1988; Kolitz et al. 2009). Translation initiation at non-AUG codons has been shown to occur naturally in both mammals and yeast. For example, CAPC, a protein overexpressed in some cancers, initiates translation using a non-AUG start codon (Anaganti et al. 2009). In yeast, two tRNA synthetase genes, GRS1 and ALA1, use non-AUG start codons for normal expression (UUG and ACG, respectively) (Chang and Wang 2004; Tang et al. 2004). Additionally, using ribosomal profiling Ingolia et al. (2009) identified 143 actively translated upstream open reading frames (uORFs) that appear to have non-AUG start codons in yeast. Interestingly, translation from these small uORFs is increased upon amino acid starvation, although neither the reason for this effect nor its mechanism is yet understood (Ingolia et al. 2009).
Recent studies have elucidated core events involved in start codon selection. Briefly, the 40S subunit with eukaryotic initiation factor (eIF) 1, eIF1A, the ternary complex (TC: eIF2, initiator methionyl-tRNA, and GTP), and eIF5 (the GTPase activating protein for eIF2) is loaded onto the 5′ end of the mRNA and scans to locate the start codon. In vivo, eIF4F, eIF4B, and eIF3 are involved in loading of this 43S ribosomal preinitiation complex (PIC) onto the 5′ end of the mRNA and subsequent scanning of the message. After the initiator tRNA anti-codon base pairs with the mRNA start codon, eIF1 is released from the complex. Loss of eIF1 in turn allows inorganic phosphate to be released from eIF2, converting the factor into its GDP-bound form. The release of eIF1 also produces a conformational change in the complex that is thought to prevent further scanning. At this stage, the large ribosomal subunit joins the small ribosomal complex with the help of eIF1A and eIF5B, producing an 80S initiation complex that can enter the elongation phase of the cycle (for reviews of the mechanism of eukaryotic translation initiation, see Sonenberg and Hinnebusch 2009; Jackson et al. 2010; Lorsch and Dever 2010).
Although many steps involved in locating the start codon have been elucidated, the mechanistic details of this process are still a mystery. Many components of the translation machinery are known to impact the fidelity of start codon selection. Mutations in eIF1 (Yoon and Donahue 1992), eIF1A (Fekete et al. 2005; Saini et al. 2010), eIF2 (Donahue et al. 1988; Castilho-Valavicius et al. 1990), eIF5 (Huang et al. 1997), eIF3 (Valasek et al. 2004), and eIF4G (He et al. 2003) are known to decrease the fidelity of start codon selection in vivo (Sui− phenotype) and have been important tools to study the steps involved in translation initiation. While mechanistically very different, the selection of the start codon in the P-site during translation initiation can be related to selection of tRNA in the A-site during elongation; both processes are dependent on matching codon:anti-codon base pairing, which triggers downstream events (Cigan et al. 1988a; Ogle et al. 2001; Kolitz et al. 2009). Small molecules such as the aminoglycoside family of antibiotics have been crucial tools to probe the mechanism of tRNA selection in the ribosomal A site during the elongation phase of translation (Rodnina and Wintermeyer 2001; Ogle et al. 2002; Ogle and Ramakrishnan 2005). Although mutations in eukaryotic initiation factors have been studied, no chemical modulators of start codon selection exist to help elucidate the mechanism of this complicated process. Compounds that increased or decreased misreading during initiation could provide unique insights into the mechanism of start codon recognition.
In this study we set up a high-throughput screen in S. cerevisiae using a dual luciferase reporter to find compounds that alter the fidelity of start codon selection in vivo. We screened ∼55,000 compounds and identified two structurally related molecules that increase the use of non-AUG codons as initiation sites, thus chemically inducing Sui− phenotypes. Our data indicate that these compounds act within the cell and that they increase initiation at a natural uORF with a near-cognate start codon, as well as on the luciferase reporter mRNA. The compounds can also increase growth on medium lacking histidine of a Sui− strain of yeast (sui1-1), in which the AUG start codon of the HIS4 gene has been changed to a near-cognate codon, such that initiation must occur at a non-AUG (his4-303). This effect demonstrates the feasibility of chemically ameliorating a genetic defect caused by mutation of an initiation codon.
RESULTS
Rationale and screen design
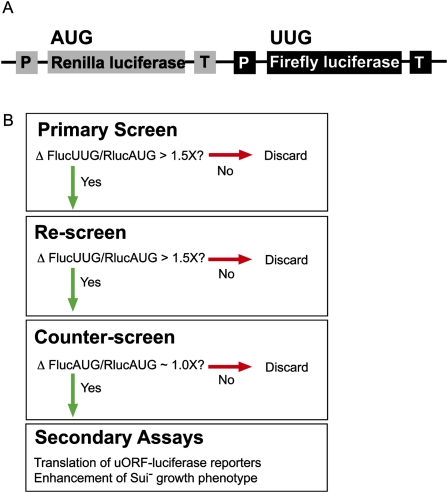
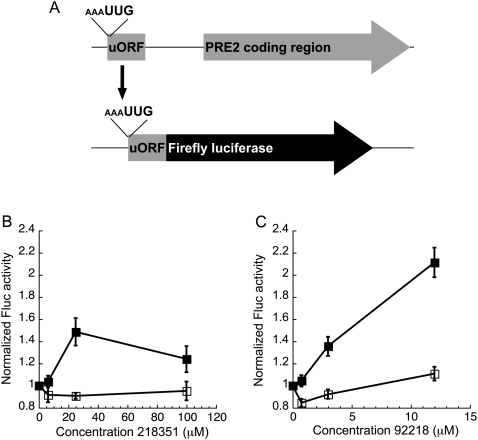
We previously developed a dual luciferase assay to measure the efficiency of translation in yeast and have used the assay to measure translation from near-cognate start codons in wild-type and Sui− mutant yeast strains that exhibit defects in fidelity of start site selection (Cheung et al. 2007; Kolitz et al. 2009). In the assay, Renilla luciferase (Rluc) and firefly luciferase (Fluc) genes are expressed from a single plasmid, each under the control of a separate constitutive promoter and terminator, allowing transcription of the two genes as separate mRNAs. The Fluc mRNA has a non-AUG start codon (Fig. 1A). The Rluc mRNA has an AUG start codon and acts as an internal control for effects on global gene expression and cell growth (as well as pipetting errors or differences in lysis efficiency), enabling us to detect effects specific to initiation on the Fluc mRNA. In both WT and Sui− strains, Fluc activity is detectable when the AUG start codon is changed to near-cognate codons that differ from AUG by only one base (UUG, GUG, CUG, AUA, AUC, AUU, ACG), except AAG and AGG. UUG is used ∼5% as well as AUG in wild-type yeast (Kolitz et al. 2009), and almost as well as AUG in some Sui− strains (Cheung et al. 2007). We chose to use UUG as the near cognate start site in the screen, allowing us to easily identify compounds that either increase (chemically reproducing a Sui− phenotype) or decrease expression (enhancing fidelity) of UUG relative to AUG in the dual luciferase assay.
FIGURE 1.
Overview of the dual luciferase assay and screen. (A) Schematic of the coding region of the plasmid used for the dual luciferase assay. (P) Promoter; (T) terminator. The ADH promoter and HIS terminator were used to produce Renilla luciferase mRNA and the GPD promoter and CYC terminator were used for firefly luciferase mRNA. (B) Flow-chart for identifying compounds that alter the fidelity of start codon recognition.
The scheme for identifying compounds that altered the fidelity of translation initiation is shown in Figure 1B. If a compound appeared to be toxic in the primary screen (both luciferase values near background levels), it was screened again at lower concentrations. Compounds that altered the UUG/AUG ratio by greater than 1.5-fold were rescreened. If the UUG/AUG ratio change was reproducible, the compounds were counter-screened using the dual luciferase assay in which both reporters had AUG start codons. The counter-screen is very powerful, eliminating any compounds that have effects unrelated to the fidelity of start codon recognition. For example, a compound that alters the activity of one of the luciferase enzymes or that generally affects translation of the Fluc mRNA will show an effect in the counter-screen assay as well as the primary screen, whereas compounds that specifically affect initiation from non-AUG codons will not. Compounds that produced similar effects in both the initial screen and the counter-screen (for example, increased both UUG/AUG and AUG/AUG) were not pursued further in this work. For the few compounds that passed the counter-screen, the luciferase ratios, both UUG/AUG and AUG/AUG, were measured at various concentrations of each drug to demonstrate concentration dependence. The compounds were then tested in secondary assays.
Assay validation
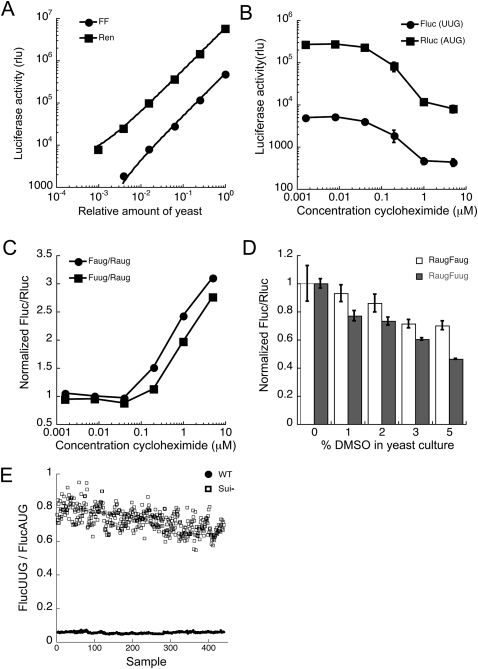
To demonstrate that the dual luciferase assay could be adapted to a high-throughput screen, we characterized the activities of each reporter and the ratio of Fluc to Rluc under screening conditions. For screening, yeast were grown in 96-well plates, then added to Passive Lysis Buffer (Promega) in a luciferase reading plate, and the activity of both reporters measured. Luciferase detection is linear over at least 3-orders of magnitude (Fig. 2A), indicating that an increase or decrease in expression of either reporter should be detectable. When the cells were grown under screening conditions in the presence of cycloheximide, an inhibitor of translation elongation, the raw luciferase values dropped to ∼25% of the control values (Fig. 2B); cycloheximide also caused a change in both ratios, FlucUUG/RlucAUG and FlucAUG/RlucAUG (Fig. 2C), possibly because of differences in the half-lives of the two proteins. The Fluc protein has a half life of 1.5 h in yeast, but the half life of Rluc has not been measured (McNabb et al. 2005). Since cycloheximide altered the FlucUUG/RlucAUG ratio, general translation inhibitors can be identified in the primary screen. However, such compounds will fail the counter screen because the FlucAUG/RlucAUG ratio is also altered (Fig. 2C, circles).
FIGURE 2.
Evaluation of the dual luciferase assay under screening conditions. (A) Increasing volumes of yeast (strain BY4741) culture expressing FlucUUG (●) and RlucAUG (■) from plasmid pFuugRaug were added to lysis buffer, and the resulting luciferase activities measured. Points are fit to a straight line with R > 0.99 for both Renilla and firefly luciferase activities. (B) BY4741 transformed with pFuugRaug was grown with various concentrations of cycloheximide, and the luciferase activities were measured after 4 h (●, FlucUUG; ■, RlucAUG). (C) BY4741 with pFuugRaug, as above, as well as with pFaugRaug, was grown with cycloheximide and the UUG/AUG (■) and AUG/AUG (●) ratios were measured. The Fluc/Rluc ratio of the solvent-only control was used to normalize the treated samples so that both ratios (UUG/AUG and AUG/AUG) equal 1 in the absence of drug. (D) Effect of DMSO on the Fluc/Rluc ratio for AUG/AUG (white bars) and UUG/AUG (filled bars). BY4741 with pFuugRaug or pFaugRaug were grown with 3% DMSO under screening conditions. The effect of DMSO on the ratio was controlled for by normalization with the DMSO-only control during the screening analysis. (E) The FlucUUG/FlucAUG ratios were measured using a wild-type (TD76-8D, closed circles) and a Sui− strain (301-4D, open squares). This data set was used to calculate the Z′ factor (see Materials and Methods).
The compound stocks from the NCI DTP library are dissolved in DMSO. Addition of DMSO alone to growth media resulted in a slight change in the FlucUUG/RlucAUG ratio (Fig. 2D). DMSO controls were included in the screen and were used to correct for the effect of DMSO on the FlucUUG/RlucAUG and FlucAUG/RlucAUG ratios. Unfortunately, no compound that alters the fidelity of start codon recognition is currently known, and thus a positive control was not available to include in the screen. However, the Sui− mutant strains, which decrease the fidelity of start codon recognition by up to 20-fold, can serve as proxy positive controls to evaluate the quality of the assay. The UUG/AUG ratio of the sui1-1 mutant (eIF1 D83G, which increases translation from UUG almost to the level of AUG) (Donahue et al. 1988; Cheung et al. 2007) compared with the UUG/AUG wild-type ratio (Fig. 2E) gives a Z′-factor of 0.68 (see Materials and Methods). The Z′ factor is a statistical characterization to evaluate the quality of a screening assay (Zhang et al. 1999). According to these parameters, the dual luciferase assay is well suited to identify compounds that change the fidelity of translation initiation.
Screen results
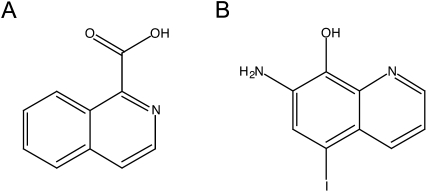
Approximately 55,000 compounds from four libraries were screened. The main source of compounds was the National Cancer Institute (NCI) Developmental Therapeutics Program (DTP) library. Of the >200,000 compounds in this library, we have screened 49,840. We also screened three smaller libraries: (1) ∼2500 compounds that have passed phase I clinical trials (gift of Dr. Jun Liu, Johns Hopkins University School of Medicine); (2) ∼2500 natural products (Dr. Jerry Pelletier); (3) ∼500 microbial growth media extracts (gift of Dr. Scott Strobel, Yale University). With the liberal cut-off of a ≥1.5-fold change in the FlucUUG/RlucAUG ratio, ∼2% of the compounds screened passed the primary screen, but only two compounds, both from the NCI DTP library, passed the counter-screen. The structures of these two compounds, isoquinoline-1-carboxylic acid (NSC218351) and 7-amino-5-iodo-8-quinolinol hydrochloride (NSC92218), are shown in Figure 3. NSC218351 is commercially available through Sigma Aldrich. NSC92218 is not commercially available, and was supplied by the NCI for further studies. High-resolution mass spectrometry confirmed the purity and atomic composition of this compound (see Materials and Methods).
FIGURE 3.
Structures of the compounds from the NCI DTP library that passed the counter-screen. (A) NSC218351, isoquinoline-1-carboxylic acid. (B) NSC92218, 7-amino-5-iodo-8-quinolinol.
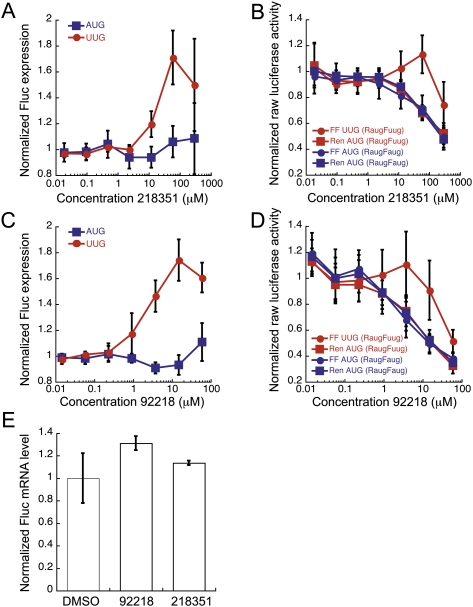
NSC218351 increases the FlucUUG/RlucAUG ratio 1.8-fold (Fig. 4A). The raw FlucUUG value increases slightly as the RlucAUG value decreases (Fig. 4B, red circles and squares, respectively). At high concentrations all values decrease, suggesting a general effect on translation and/or toxicity. The FlucAUG/RlucAUG ratio does not change in the presence of drug (Fig. 4A, blue squares), even when both raw values decrease at high concentrations of drug (Fig. 4B, blue circles and squares). NSC92218 affected the dual luciferase assay in a similar manner to NSC218351. The FlucUUG/RlucAUG ratio increases 1.8-fold, while the FlucAUG/RlucAUG ratio does not change (Fig. 4C, cf. circles with squares). The raw FlucUUG value does not change, or increases slightly, at concentrations that decrease the other luciferase values (Fig. 4D, cf. red circles with red squares and blue circles with blue squares).
FIGURE 4.
The effects of NSC218351 and NSC92218 in the dual luciferase assay. (A,C) BY4741 expressing Fluc with either an AUG or UUG start codon was treated with various concentrations of NSC218351 and NSC92218, respectively, and normalized to the internal Rluc control with an AUG start codon. The Fluc/Rluc ratio from each sample was then normalized to the appropriate DMSO-only control, so that each ratio equals 1 in the absence of compound (normalized FlucUUG, red circles; normalized FlucAUG, blue squares). Points are the averages of at least seven independent experiments ±SE. (B,D) Raw luciferase activity values with increasing concentrations of NSC218351 and NSC92218. Red symbols indicate luciferase values from pFuugRaug plasmid, and blue symbols indicate luciferase values from pFaugRaug counter-screening plasmid (Fluc, squares; Rluc, circles). Luciferase activites are normalized to the DMSO control values in each experiment, and the normalized values of at least six independent experiments are averaged (± average deviation). (E) Relative levels of Fluc/Rluc mRNA measured using RT-q–PCR, from strain BY4741 expressing FlucUUG and RlucAUG treated with 2 μM NSC92218 or 50 μM NSC218351. The Rluc mRNA levels were used to normalize the Fluc mRNA levels, and the Fluc/Rluc ratio of the DMSO sample was used to normalize the samples treated with compounds. Data are the averages of duplicate samples.
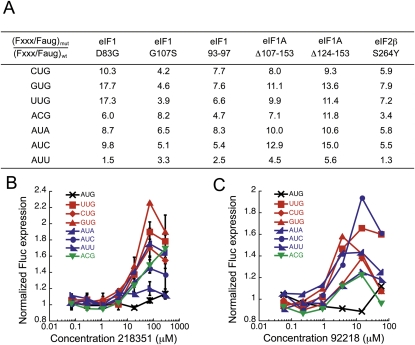
The compounds increase initiation at most near-cognate codons, indicating that their effects are not limited to UUG codons (Fig. 5B,C). AAG and AGG are not used detectably as Fluc initiation codons in yeast (Kolitz et al. 2009) and were thus not tested. The one exception is that NSC218351 does not increase initiation at AUU codons. Interestingly, AUU is generally the near cognate codon whose use as an alternative start site is increased the least by Sui− mutants (Fig. 5A). Efficiency of use of near cognate codons as initiation sites was measured with the dual luciferase assay in several Sui− mutant strains. Translation from each alternative start codon in the Sui− strain was normalized to translation from that codon in a wild-type strain. Sui− mutations increase the use of near cognates up to 18-fold (GUG in eIF1 D83G strain). However, in each mutant strain, translation from AUU is increased the least. Although we do not understand why AUU behaves anomalously, it is interesting that NSC218351 mimics this aspect of the behavior of the Sui− mutations in initiation factors.
FIGURE 5.
The effect of Sui− mutants and NSC218351 and NSC92218 on all near-cognate start codons in the dual luciferase assay. (A) The FlucXXX/FlucAUG ratio was measured for several Sui− mutants, where XXX is the start codon that varies from AUG by 1 bp. This ratio was normalized to the same ratio in the wild-type control [(Fxxx/Faug)Sui-/(Fxxx/Faug)wt] to illustrate the magnitude of the effect of the Sui− mutations on initiation at each codon relative to a wild-type strain. See Table 3 for strain details. (B,C) Fluc expression (as in Fig. 4A,C) from reporters with different near-cognate start codons in cells treated with NSC218351 or NSC92218 (the black X is AUG, changes to position 1 of the start codon are in red, changes to the second position are in green, and changes to the third position are in blue).
At high concentrations, the compounds slow yeast growth; however, the FlucUUG/RlucAUG ratio increases at concentrations of compounds that do not affect growth of wild-type yeast (50 μM NSC218351 and 2 μM NSC92218) (Fig. 4A,C). To demonstrate that the compounds were not affecting mRNA levels at these concentrations, RT-q–PCR (reverse-transcription, quantitative PCR) was used to measure relative levels of the luciferase mRNAs. The levels of the reporter messages did not change significantly (Fig. 4E), indicating that the compounds do not affect synthesis or degradation of these mRNAs. As an additional test for effects on mRNA stability, we used the compounds in the dual luciferase assay with a strain deficient for nonsense-mediated decay (NMD), upf1Δ (He et al. 1993). Premature termination can target a message for NMD. If a message lacks the appropriate start codon, use of an upstream or out-of-frame codon for initiation can lead to premature termination, and potentially NMD (Amrani et al. 2006). Expression from the UUG codon in Fluc mRNA was not changed in the upf1Δ strain relative to the WT strain (in the absence of compound), indicating that NMD does not influence expression of this reporter when it lacks an AUG start codon (Kolitz et al. 2009). If the compounds were inhibiting the NMD pathway, we would not expect to see an effect of the compounds in the assay in a strain where NMD was already blocked. Using the dual luciferase assay, the effects of the compounds on the UUG/AUG ratio were still observed in the upf1Δ strain (data not shown). These results, in combination with the RT-q–PCR experiments and the fact that no effect is seen on the FlucAUG/RlucAUG expression ratio, indicate that the compounds increase expression of the reporter from non-AUG start codons at the translational level.
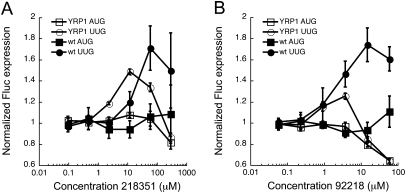
Based on the measured activities of Sui− mutants (Fig. 5A; Cheung et al. 2007), yeast cells are viable even when translation from near cognate start codons is increased almost 20-fold relative to WT cells. To investigate whether approximately twofold is the maximal effect of the compounds on expression from the Fluc reporter, we determined whether time of incubation with drug altered the UUG/AUG expression ratio. Under screening conditions, yeast was grown with compounds for 4 h. We monitored the UUG/AUG and AUG/AUG ratios from 1 to 8 h of growth with compound. At 1 h, no effect of either compound is observed. Approximately 20% of the maximal effect is achieved at 2 h. The maximal effect is reached at 4 h, and further incubation does not result in an additional decrease in the fidelity of start-site selection (data not shown). Another factor possibly contributing to the magnitude of the UUG/AUG ratio change is bioavailability of the molecules. A wild-type strain was used in the screen, which may restrict some compounds from entering the cells effectively. We therefore also tested a strain deficient in efflux pumps (YRP1: snq2Δ, pdr5Δ, erg6Δ) to see whether the magnitudes of the effects of the compounds or their effective concentrations would change if efflux from the cells decreased (Kung et al. 2005). The minimal effective concentration for both compounds is lower in the YRP1 strain than the WT (Fig. 6A,B), suggesting that the compounds act inside of the cell rather than affecting translation by binding to its surface or by altering some other external property. Interestingly, the magnitudes of the UUG/AUG ratio change decreased, rather than increased, in the YRP1 strain. A potential explanation for this phenomenon is that the toxic concentrations of the compounds are lowered in the YRP1 strain because of the loss of the efflux pumps, and the leftward shift of the toxicity curve is greater than the shift of the efficacy curve. If this were true it would suggest that the targets of the compounds that produce toxicity are different from those that alter the UUG/AUG initiation ratio. Alternatively, the differences in the effects of the compounds on the UUG/AUG expression ratio could be due to differences in genetic backgrounds of the wild-type strain used in the screen (BY4741) and the YRP1 strain.
FIGURE 6.
Normalized Fluc(AUG) (squares) and Fluc(UUG) (circles) expression in WT yeast (BY4741; closed symbols) and a strain deficient in drug efflux pumps (YRP1: snq2Δ, pdr5Δ, erg6Δ; open symbols) treated with compounds NSC218351 (A) and NSC92218 (B). Points for YRP1 are averages of data from three separate transformants ± average deviation. Data were analyzed as in Figure 4A, and are plotted with BY4741 data from Figure 4, A and C, for comparison.
Structure-activity analysis
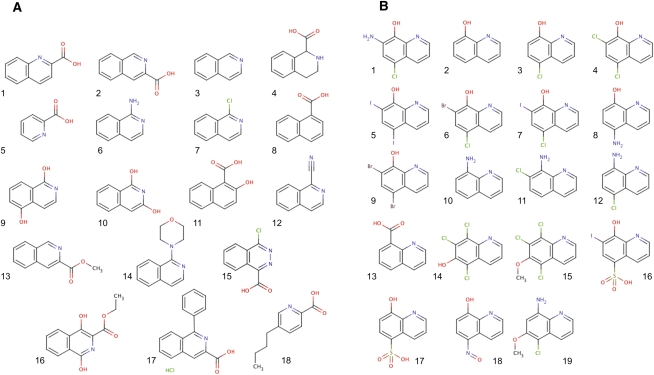
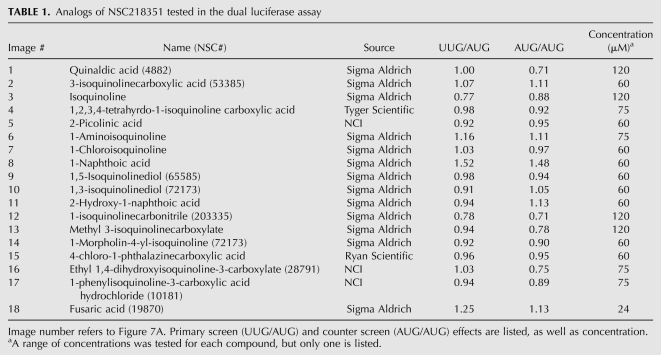
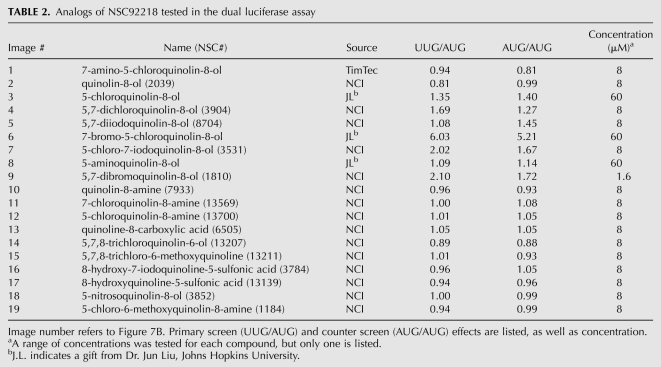
The two active compounds were identified in different sections of the NCI DTP library, but bear striking resemblance in structure and activity. Both compounds increase the UUG/AUG ratio by 1.8-fold, but the minimal concentration required to achieve this effect is fourfold higher for NSC218351 than for NSC92218 (60 μM and 15 μM, respectively). Little information is available about the biological activity of either of these compounds. Studies indicate that compounds related to NSC92218 have antifungal activity through an unknown mechanism (Gershon et al. 1991). NSC218351 and derivatives have been implicated in inhibition of protein kinases (Lu et al. 1996). To learn about the important chemical features of these compounds, we obtained ∼20 analogs of each, and tested them in the dual luciferase assay. We found that changing key functional groups of NSC218351 (moving or removing the N or acid group, eliminating aromaticity or removing one of the rings) results in a loss of the effect on start codon recognition (Fig. 7A; Table 1). Some analogs do change the UUG/AUG expression ratio, but these compounds have similar effects on the AUG/AUG expression ratio, indicating that they are not specifically altering the fidelity of translation initiation (compare columns UUG/AUG and AUG/AUG). Fewer close analogs of NSC92218 were available, but changing -I to -Cl, or removing the amine and -I resulted in a loss of the effect (Fig. 7B; Table 2). These data indicate that the activities of NSC92218 and NSC218351 in altering the fidelity of start codon recognition are very specific and are not possessed by many structurally similar molecules. It should be noted that few analogs consisting of the parent structures with additional functional groups on them were available, and thus we cannot yet say whether these scaffolds can be elaborated into more active molecules.
FIGURE 7.
Analogs of NSC218351 (A) and NSC92218 (B) that were tested at various concentrations in the dual luciferase assay to measure their effects on initiation at UUG and AUG codons.
TABLE 1.
Analogs of NSC218351 tested in the dual luciferase assay
TABLE 2.
Analogs of NSC92218 tested in the dual luciferase assay
Secondary assays
Ribosomal profiling identified over 100 small upstream open readings frames (uORFs) that appear to be translated from non-AUG start codons in S. cerevisiae (Ingolia et al. 2009). uORFs are coding regions that are sometimes used to regulate the translation of a downstream ORF encoding a protein (Meijer and Thomas 2002). To demonstrate that the compounds have a general effect on the fidelity of start codon recognition, rather than a specific one on the use of the start codon in Fluc mRNA, an uORFs identified by Ingolia and colleagues (2009) from PRE2 mRNA was fused to the firefly luciferase coding sequence. This construct was used to assess the effect of the compounds on translation mediated by the non-AUG start codons of the uORFs. The PRE2 uORF has a UUG start codon, with the consensus sequence (-3)AAA(-1) directly upstream of UUG (Fig. 8A). In the control reporter, the luciferase coding region is out-of-frame from the start codon of the uORF. In wild-type yeast, NSC218351 and NSC92218 increase expression of the reporter ∼1.5-fold and twofold, respectively (Fig. 8B,C), similar to their effects in the dual luciferase assay. The out-of-frame controls are not well translated, and incubation with either compound does not improve luciferase signal. RT-q–PCR showed that the compounds do not increase mRNA levels of these reporters (data not shown), indicating that the increase in luciferase signal is due to a decrease in the fidelity of start codon selection.
FIGURE 8.
Both compounds increase translation of a luciferase reporter fused to the small, endogenous uORF from PRE2 beginning with a UUG codon. (A) Schematic of the reporter. As a control, the uORF was fused out of frame from the luciferase coding region. BY4741 expressing the reporter was treated with NSC218351 (B) or NSC92218 (C), and Fluc activity was measured. Closed squares are the in-frame reporters, and open squares are the out-of-frame controls. Fluc activity with DMSO alone was used to normalize the activity with compound. Points are averages of two independent experiments ± average deviation.
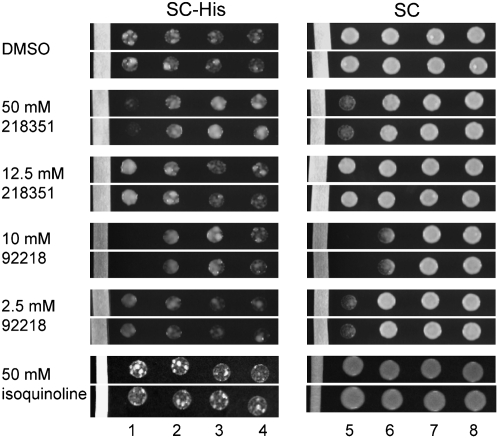
Chemically increasing use of near-cognate codons as start codons mimics the Sui− phenotype. Sui− mutations were originally identified in a screen that requires translation initiation at a non-AUG start codon in a mutant of the HIS4 gene (his4-303 allele) for growth on SC-His media (Donahue et al. 1988). Compounds that increase the use of non-AUG start codons to a small degree should further increase growth of Sui− strains with the his4-303 allele on SC-His media, chemically enhancing the Sui− phenotype. To test this, a paper strip saturated with compound was placed onto a plate of media (SC or SC-His). Yeast with the his4-303 allele was spotted on the plate at various distances from the compound- or solvent-containing strips. DMSO alone does not affect growth of the Sui− strain (sui1-1) on complete media (SC) or selective media (SC-His) (Fig. 9). Close to the compound source, 10 mM NSC92218 prevents growth and 50 mM NSC218351 slows growth (Fig. 9, columns 1,5). In the presence of 50 mM NSC218351 on the paper strip, growth of sui1-1 on selective media is inhibited close to the drug source, but is enhanced relative to the DMSO farther from the source (Fig. 9, cf. columns 1,3,4). With 12.5 mM NSC218351, growth of the sui1-1 strain is enhanced in the column closest to the compound source on SC-His (column 1). In columns 2 and 3, 10 mM NSC92218 enhances growth of the sui1-1 strain on SC-His, especially when normalized for the inhibitory effect on growth on the SC plate (column 2 vs. 6). With 2.5 mM NCS92218, growth enhancement occurs in column 1 on SC-His relative to the inhibitory effect on SC (column 1 vs. 5). Thus, both compounds enhance the phenotype of a Sui− strain of yeast at discrete distances from the compound source on SC-His media. The analog isoquinoline (Fig. 7A, compound 3) does not cause any enhancement of growth in this assay, further indicating specificity of the effects produced by NSC218351 and NSC92218.
FIGURE 9.
Both compounds increase the growth of Sui− strain sui1-1 (eIF1 D83G) on media lacking histidine. Compound or solvent-soaked paper strips were placed on agar plates. Yeast were spotted onto the plate as four rows increasingly distant from the paper strip and grown for 4 d on SC-His and 2 d on SC. Two rows from one representative experiment are shown (the 50 mM isoquinoline data are from a separate experiment). The results were consistent in all rows and in three independent experiments.
Role of start codon context
In addition to identifying compounds that alter the fidelity of start codon recognition, this screen has the potential to identify compounds that affect recognition of the consensus sequence elements flanking start codons in yeast. The yeast consensus sequence is AAAA directly upstream of the initiation codon (Hamilton et al. 1987; Shabalina et al. 2004). In mammals, the consensus sequence is GCC(A/G)CCAUGG, and can have up to a 20-fold effect on use of the codon as a start site (Kozak 1986, 1987). In yeast the consensus sequence generally has a small or no effect on initiation from AUG start codons (Cigan et al. 1988b; Donahue and Cigan 1988), but it does have a strong effect on use of non-AUG start codons (Zitomer et al. 1984; Chen et al. 2008). If the firefly reporter with a UUG start codon lacks the consensus sequence (GCTC instead of AAAA), the mRNA is not detectably translated (data not shown). Since non-AUG codons are more sensitive to this sequence than are AUG codons, the screen might identify compounds that diminish or enhance the influence of the flanking region. Such an effect has not previously been noted in yeast, by genetic mutation or chemical treatment.
In the reporters used for the screen, both the AUG and UUG start codons had AAAA directly upstream (positions −4 to –1). Although the sequence GCTC does not detectably promote translation from a UUG start codon, changing the −3 position to an A in this context (GATC) restores 18% of the signal observed with AAAA (data not shown). Changing any other single upstream position to A does not allow detectable translation from UUG. The Fluc reporter with this minimal stimulatory flanking sequence, GATC, was used to test whether the compounds increase the influence of the sequence upstream of the start codon. The effects of the compounds on Fluc-UUG expression was unchanged when the full (AAAA) upstream sequence was replaced with the minimal version (GATC; data not shown), indicating that the compounds' abilities to increase use of UUG as an initiation codon are not altered by changing the strength of the sequence context around the start codon, and that upstream bases at the −1, −2, and −4 positions are not required for the activity of the compounds.
Investigating interactions of the compounds with the translation initiation machinery
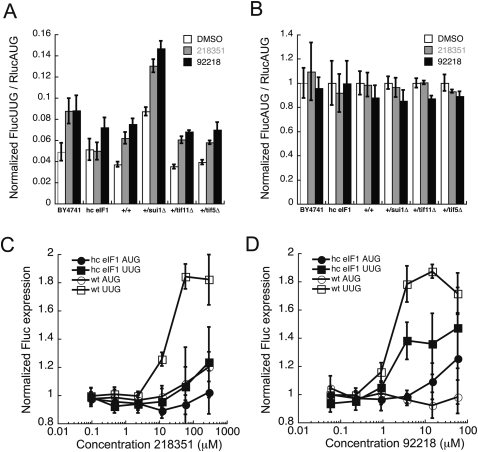
Genetic approaches enabled us to look for synthetic effects, either enhancement or suppression, of the compounds with particular yeast genes. eIF1, eIF1A, and eIF5 play crucial roles in start codon selection (Mitchell and Lorsch 2008). To examine the importance of these factors on the effects of the compounds, we measured translation from the UUG start codon of Fluc using the dual luciferase assay in diploid strains haplo-insufficient for these factors (Open Biosystems) (deletion of any one of these factors in haploids is lethal) (Winzeler et al. 1999). Western analysis demonstrated that the level of each protein in the haplo-insufficient diploid strains decreased approximately twofold relative to the level in the diploid wild-type strain, as expected (data not shown). Haplo-insufficiency of eIF1A (+/tif11Δ) or eIF5 (+/tif5Δ) did not have an effect on translation from UUG (FlucUUG/RlucAUG), but eIF1 haplo-insufficiency (+/sui1Δ) increased use of UUG approximately twofold (Fig. 10A, white bars), consistent with its role as a master switch that controls the response to start codon recognition (Lorsch and Dever 2010). In all of these strains, the compounds still increased the UUG/AUG ratio approximately twofold (Fig. 10A, gray and black bars). This indicates that the effects of eIF1 haplo-insuffciency and the presence of either compound on start codon selection are additive, and that a haplo-insufficiency of eIF1A or eIF5 does not alter the effect of the compounds. The FlucAUG/RlucAUG ratio is not altered in any of these strains or conditions (Fig. 10B), indicating that the effects of the compounds in these strains are specific to translation from a near cognate start codon and that haploinsufficiency of eIF1 reduces the fidelity of start codon recognition rather than generally affecting Fluc expression or activity.
FIGURE 10.
Effect of NSC218351 and NSC92218 on start site selection in strains of yeast with altered levels of eIF 1, eIF 1A, and eIF 5. (A) FlucUUG/RlucAUG expression ratio and (B) FlucAUG/RlucAUG expression ratio, in the presence of DMSO (white bars) or compounds (NSC218351, gray bars; NSC92218, black bars), was measured in strain H3984 (hc eIF1) and compared with wild-type (BY4741) and haplo-insufficient diploids for eIFs 1 (+/sui1Δ), 1A (+/tif11Δ), and 5 (+/tif5Δ), and compared with diploid wild-type BY4743 (+/+). H3984 data are an average of two independent experiments. Error bars represent average deviation. The concentration of NSC218351 was 60 μM and NSC92218 was 3.8 μM. Haplo-insufficiency results are averages of data from two separate transformants. The concentration of NSC218351 was 33 μM, and NSC92218 was 7 μM. FlucAUG/RlucAUG expression ratio with DMSO alone was used to normalize the Fluc/Rluc ratio for both FlucAUG and FlucUUG for each strain (normalized FlucAUG/RlucAUG with DMSO alone is 1 for each strain). Concentration dependence of the effect of NSC218351 and NSC92218 on Fluc expression from UUG (squares), AUG (circles) in hc eIF1 (closed symbols), and wild-type (open symbols) yeast is shown in C and D. Data, from two independent experiments were analyzed as in Figure 4, A and C.
Although a deficiency in eIF1 decreases the fidelity of translation initiation, overexpression of eIF1 does not increase the fidelity of start-site selection in WT yeast (Fig. 10A, cf. white bars of BY4741 and hc eIF1; Cheung et al. 2007). However, overexpression of eIF1 has been shown to suppress the Sui− phenotypes of a number of mutations in eIF1 and other factors (eIF1A, eIF5, eIF3, eIF2β, and eIF4G) (He et al. 2003; Valasek et al. 2004; Cheung et al. 2007; Saini et al. 2010). Interestingly, overexpression of eIF1 also suppresses the effects of both compounds on the fidelity of start codon recognition. The effect of NSC218351 is completely suppressed in a strain overexpressing eIF1 (Fig. 10A, cf. gray bars of BY4741 and hc eIF1; Fig. 10C, cf. open and closed squares), and the effect of NSC92218 is reduced by ∼50% relative to the effect in wild-type cells (Fig. 10A, cf. black bars of BY4741 and hc eIF1; Fig. 10D, cf. open and closed squares). Because overexpression of eIF1 alone does not increase the fidelity of start codon selection, this suppression is specific to the Sui− phenotype, whether mutationally or chemically induced, and provides evidence that the compounds affect the fidelity of translation initiation by altering the function of the 40S ribosomal subunit or one of the initiation factors that participate in start codon selection.
Ribosomal profiling has shown that initiation at uORFs with non-AUG start codons increases under amino acid starvation conditions (Ingolia et al. 2009). In the amino acid starvation response, the kinase Gcn2 phosphorylates eIF2α, which in turn results in reduced ternary complex formation and delivery of initiator tRNA to the ribosome (Hinnebusch 2005). It is currently unknown whether and how phosphorylation of eIF2 affects start codon selection. To investigate whether there is a relationship between the effect of the compounds and the Gcn2 pathway, a strain lacking Gcn2 was used for the dual luciferase assay. In the absence of Gcn2, the compounds were still able to reduce the fidelity of start codon recognition (data not shown), indicating that the Gcn2 pathway is not crucial for their effects. As a further test of the possible involvement of increased phosphorylation of eIF2α in the reduction of the fidelity of start codon recognition mediated by the compounds, we monitored the levels of eIF2α and phosphorylated eIF2α by Western blot. Neither compound increases the level of eIF2 phosphorylation in a wild-type strain (data not shown), indicating that the compounds do not induce the starvation response. Although the relationship, if any, between phosphorylation of eIF2α and start codon selection has not been established, the compounds do not appear to be acting via this pathway.
DISCUSSION
In this study we adapted a dual luciferase assay into a high-throughput screen to identify compounds that alter the fidelity of start codon recognition in yeast. Of the >55,000 compounds screened, 2% passed the primary screen and only two compounds passed the counter-screen. These structurally related compounds increase translation from near-cognate start codons approximately twofold in the dual luciferase assay (at 60 μM NSC218351 and 15 μM NSC92218) and in the uORF-Fluc assay, and enhance the Sui− phenotypes conferred by the sui1-1 D83G eIF1 mutation and by haploinsufficiency of WT eIF1. Thus, the compounds decrease the fidelity of start codon selection in three separate in vivo assays. Additionally, the increased use of UUG as a site of initiation caused by NSC218351 and NSC92218 is suppressed, completely and partially, respectively, by overexpression of eIF1, providing strong evidence that the compounds act in a mechanistically similar manner to Sui− mutations in initiation factors.
eIF1 is a key controller of start codon selection (Mitchell and Lorsch 2008). It binds tightly to the 43S PIC and is important for maintaining a scanning-competent conformation of the ribosome. Upon start codon recognition, eIF1 is released from the PIC, causing a reversion to the closed, scanning arrested state of the ribosome and triggering release of Pi from eIF2 (Lorsch and Dever 2010). Most Sui− mutations in eIF1 act by decreasing the factor's affinity for the PIC and thus increasing the rate of eIF1 release at non-AUG codons. Overexpression of these eIF1 mutants can partially suppress the Sui− phenotype (Cheung et al. 2007). Suppression of the effects of NSC218351 and NSC92218 by hc eIF1 is consistent with the idea that they act by altering eIF1 affinity for the PIC. However, we could not detect an effect of either compound on the affinity of eIF1 for the 40S subunit (±eIF1A) or on the rate of release of the factor upon start codon recognition by the PIC (data not shown). These data suggest that the compounds act on another step in the pathway, and that eIF1 overexpression can suppress the effect on this step. This is consistent with the fact that hc eIF1 can also suppress the Sui− phenotypes of mutations in eIF1A, eIF5, eIF3, eIF4G, and eIF2 (He et al. 2003; Valasek et al. 2004; Cheung et al. 2007; Saini et al. 2010). Determining the mechanism of action of the compounds will require additional in vivo and in vitro approaches.
In addition to providing new insight into the complicated mechanism of start codon selection, the compounds identified in this screen could serve as leads for the development of new drugs targeting translation initiation. For example, modulators of the fidelity of start codon recognition could be used to treat variants of genetic diseases that are caused by mutations of the start codon or region surrounding the start codon. A compound that functions analogously in translation termination has shown promise for clinical use. The compound, PTC124, specifically increases read-through of nonsense codons in vivo, and is currently in clinical trials to treat cystic fibrosis, Duchenne myscular dystrophy, and hemophilia (Welch et al. 2007). In addition to treating genetic diseases, compounds that reduce the fidelity of start codon recognition might be developed into anticancer agents, as rapidly reproducing cells could be less tolerant of mistranslation than are quiescent cells. Alternatively, if the effects of the compounds identified here are specific to yeast, they might be developed into novel antifungal agents.
This study is the first screen to find compounds that alter the fidelity of start codon selection in eukaryotes. The two compounds identified bear striking resemblance to each other, yet the ability to decrease the fidelity of start codon selection appears to be very specific to these two molecules. Future efforts will focus on elucidating the compounds' mechanisms of action and increasing their potency.
MATERIALS AND METHODS
Strains and plasmids
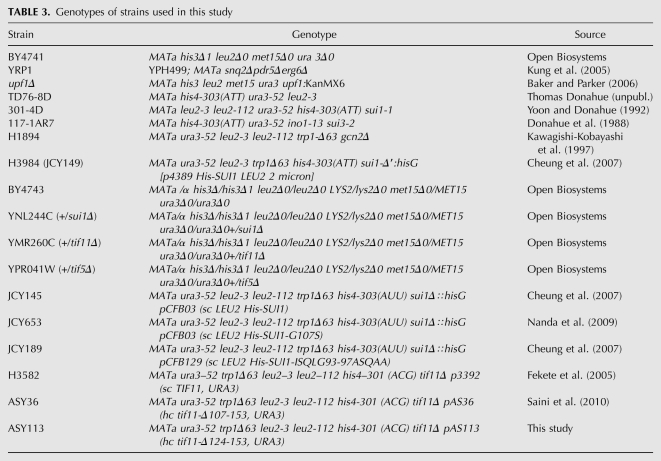
For strain information, see Table 3.
TABLE 3.
Genotypes of strains used in this study
To construct the uORF-luciferase fusion reporters, the PRE2 uORF was fused to the firefly luciferase coding region with a PDR5 3′UTR as Not1/Spe1 fragments in pRS313. The (Spe1)-Fluc-3′UTR fusion was cloned by PCR with oNTI226 (GCAactagtGGAAGACGCCAAAAACATAAAG) and oNTI227 (GCTttaattaaTTACACGGCGATCTTTCCG) on pGL3basic and PCR with oNTI228 (GCAttaatTAATAGAATTTTGAATTTGGTTAAGAAAAG) and oNTI229 (GCTgggcccATCAGAGCTGGTAAATTCAAG) from yeast genomic DNA. A 3-way Spe1/Pac1/Apa1 ligation fused Fluc with the PDR5 3′UTR in the pRS313 background. The in-frame PRE2 uORF plasmid (pNTI33) was made by PCR with primer oNTI248 (GCAactagtTCTATTCAATTTAATAGTAAATTTGTTATT), and the out-of-frame plasmid (pNTI32) was made using the primer oNTI249 (GCAactagtATCTATTCAATTTAATAGTAAATTTGTTAT) in combination with oNTI247 (GCTgcggccgcGTTACTATCAAGATGTATCAAACAATG) and subcloned into the Fluc-PDR5 3′UTR plasmid as a Not1/Spe1 fragment.
TIF11 mutant alleles were constructed by fusion PCR using p3390 containing WT TIF11 as template, as described previously (Olsen et al. 2003). The fusion PCR products were inserted between the EcoRI and SalI sites of YCplac111 (sc) or YCplac181 (hc), and the subcloned fragments of all mutant constructs were confirmed by DNA sequencing. Yeast strains harboring the mutant constructs were constructed from strains H3582 (his4-301) (Fekete et al. 2005) by plasmid shuffling.
Dual luciferase assay
The dual luciferase assay was carried out as in Kolitz et al. (2009), with the following modifications to screening conditions: An overnight culture of wild-type yeast (BY4741 transformed with pJDRaugFuug) was diluted to an OD600 of ∼0.2 in SC-Ura, and 50 μL were aliquoted to each well of a 96-well plate. BY4741 expressing the RaugFaug plasmid was included in the first and last column as an additional control. Compounds to be tested were supplied from the NCI DTP library in 96-well format at 1 mM in DMSO. DMSO was included in all control wells. A total of 1.5 μL of compound (or DMSO) was added to each well of the yeast plate, followed by incubation at 30°C for 4 h while shaking. To measure luciferase activity, 1 μL of culture was added directly to 50 μL of 1X Passive Lysis Buffer (Promega), incubated 40–60 min at room temperature, then luciferase activity measured using a Turner Modulus Microplate Reader (Kolitz et al. 2009). The same protocol was used when testing the other libraries with the appropriate solvent controls. The Fuug/Raug ratio of each sample with drug was compared with the average Fuug/Raug ratio of the solvent only controls on the same plate.
The Z′-factor was calculated using the formula Z′ = 1-(3σc+ + 3σc-)/(|μc+ − μc-|) where c+ is the FlucUUG/AUG ratio from the sui1-1 strain (301-4D) and c- is the same ratio from the wild-type strain (TD76-8D). σ represents standard deviation and μ represents average (Zhang et al. 1999).
uORF luciferase assay
BY4741 was transformed with pNTI33 or pNTI32. Transformants were incubated with compound and luciferase activity measured as described for the dual luciferase assay. The solvent-only firefly luciferase activity was used for normalization.
Growth assays
Yeast with the his4-303 allele from an overnight culture were washed with water and diluted to an OD600 between 0.1 and 1. Strips of sterile Whatman paper were soaked with compound or solvent and placed on an agar plate of the appropriate medium. The yeast culture was spotted onto the plate at various distances from the paper strips. The plates were incubated at 30°C.
RT-q–PCR
RNA was purified from yeast (grown with 50 μM NSC218351 or 2 μM NSC92218) using acid phenol extraction (Fazzio et al. 2001), and DNase treated (DNase I, Roche). The iScript cDNA kit (BioRad) was used to make the cDNA, and the SYBR green protocol was used for the qPCR reactions in a BioRad CFX96 Real-time PCR detection system.
Chemicals
After initial characterization of NSC218351 from the NCI DTP library, additional material was purchased from Sigma Aldrich for further studies. It behaved identically to the compound in the library. NSC92218 could not be obtained commercially, and thus came only from the NCI DTP. The identity and purity of NSC92218 were confirmed by mass spectrometry at the University of Illinois at Urbana Champaign facility. One species with an exact ionized mass of 287.1 was detected by LR ESI, and HR Q-tof gave the possible atomic composition of C9H8N2OI or C7H9N2ONaI. The expected atomic mass, 286.1, and composition C9H8N2OI, exactly matched NSC92218. Cycloheximide was obtained from Sigma Aldrich.
ACKNOWLEDGMENTS
We thank Silke Dorner and Jeff Corden for advice on developing the luciferase assay, Miranda Darby, Kip Bitok, and Kamau Fahie for technical assistance, and the members of the Lorsch and Hinnebusch labs for helpful discussions and comments on the manuscript. We are also grateful to Kristian Baker for the upf1Δ mutant, Karl Kuchler for strain YRP1, Jun O. Liu for the Phase I compound library, Scott Strobel and Marina Santiago for the microbial extract library, and Jonathan S. Weissman for reagents. This work was supported by National Institutes of Health grant DK078633 (J.R.L.) and by the Intramural Research Program of the National Institutes of Health (A.G.H.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2475211.
REFERENCES
- Amrani N, Sachs MS, Jacobson A 2006. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 [DOI] [PubMed] [Google Scholar]
- Anaganti S, Hansen JK, Ha D, Hahn Y, Chertov O, Pastan I, Bera TK 2009. Non-AUG translational initiation of a short CAPC transcript generating protein isoform. Biochem Biophys Res Commun 380: 508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Parker R 2006. Conventional 3′ end formation is not required for NMD substrate recognition in Saccharomyces cerevisiae. RNA 12: 1441–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho-Valavicius B, Yoon H, Donahue TF 1990. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics 124: 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KJ, Wang CC 2004. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J Biol Chem 279: 13778–13785 [DOI] [PubMed] [Google Scholar]
- Chen SJ, Lin G, Chang KJ, Yeh LS, Wang CC 2008. Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J Biol Chem 283: 3173–3180 [DOI] [PubMed] [Google Scholar]
- Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan AM, Feng L, Donahue TF 1988a. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science 242: 93–97 [DOI] [PubMed] [Google Scholar]
- Cigan AM, Pabich EK, Donahue TF 1988b. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol 8: 2964–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JM, Laz TM, Sherman F 1988. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol Cell Biol 8: 4533–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Cigan AM 1988. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol 8: 2955–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Cigan AM, Pabich EK, Valavicius BC 1988. Mutations at a Zn(II) finger motif in the yeast eIF-2 β gene alter ribosomal start-site selection during the scanning process. Cell 54: 621–632 [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol 21: 6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J 24: 3588–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon H, Clarke DD, Gershon M 1991. Evidence of steric factors in the fungitoxic mechanisms of 8-quinolinol and its 5- and 7-halogenated analogues. J Pharm Sci 80: 542–544 [DOI] [PubMed] [Google Scholar]
- Hamilton R, Watanabe CK, de Boer HA 1987. Compilation and comparison of the sequence context around the AUG start codons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res 15: 3581–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci 90: 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K 2003. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol 23: 5431–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Huang HK, Yoon H, Hannig EM, Donahue TF 1997. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev 11: 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE 1997. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol Cell Biol 17: 4146–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Takacs JE, Lorsch JR 2009. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15: 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 [DOI] [PubMed] [Google Scholar]
- Kozak M 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C, Kenski DM, Dickerson SH, Howson RW, Kuyper LF, Madhani HD, Shokat KM 2005. Chemical genomic profiling to identify intracellular targets of a multiplex kinase inhibitor. Proc Natl Acad Sci 102: 3587–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J, Dever T 2010. Molecular view of 43S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZX, Quazi NH, Deady LW, Polya GM 1996. Selective inhibition of cyclic AMP-dependent protein kinase by isoquinoline derivatives. Biol Chem Hoppe Seyler 377: 373–384 [DOI] [PubMed] [Google Scholar]
- McNabb DS, Reed R, Marciniak RA 2005. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot Cell 4: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Thomas AA 2002. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem J 367: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Lorsch JR 2008. Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition. J Biol Chem 283: 27345–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol 394: 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG 2003. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J 22: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG 2010. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev 24: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ 2004. Comparative analysis of orthologous eukaryotic mRNAs: Potential hidden functional signals. Nucleic Acids Res 32: 1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, Yeh LS, Chen NK, Ripmaster T, Schimmel P, Wang CC 2004. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J Biol Chem 279: 49656–49663 [DOI] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol 24: 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. 2007. PTC124 targets genetic disorders caused by nonsense mutations. Nature 447: 87–91 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Donahue TF 1992. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol 12: 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4: 67–73 [DOI] [PubMed] [Google Scholar]
- Zitomer RS, Walthall DA, Rymond BC, Hollenberg CP 1984. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol 4: 1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]