Abstract
The “RNA world” hypothesis rests on the assumption that RNA polymerase ribozymes can replicate RNA without the use of protein. In the laboratory, in vitro selection has been used to create primitive versions of such polymerases. The best variant to date is a ribozyme called B6.61 that can extend a RNA primer template by 20 nucleotides (nt). This polymerase has two domains: the recently crystallized Class I ligase core, responsible for phosphodiester bond formation, and the poorly characterized accessory domain that makes polymerization possible. Here we find that the accessory domain is specified by a 37-nt bulged stem–loop structure. The accessory domain is positioned by a tertiary interaction between the terminal AL4 loop of the accessory and the J3/4 triloop found within the ligase core. This docking interaction is associated with an unwinding of the A3 and A4 helixes that appear to facilitate the correct positioning of an essential 8-nt purine bulge found between the two helices. This, together with other constraints inferred from tethering the accessory domain to a range of sites on the ligase core, indicates that the accessory domain is draped over the vertex of the ligase core tripod structure. This geometry suggests how the purine bulge in the polymerase replaces the P2 helix in the Class I ligase with a new structure that may facilitate the stabilization of incoming nucleotide triphosphates.
Keywords: ribozyme, polymerase, accessory domain, RNA structure, RNA world
INTRODUCTION
The “RNA world” hypothesis suggests that prior to protein-dominated biology, RNA played a central role as both information carrier and catalyst (Orgel 1968; Gilbert 1986; Joyce 2002; Deamer and Szostak 2010). The characterization of a broad range of artificially selected ribozymes together with a growing list of natural riboswitch regulatory elements provides evidence that ribozyme-mediated metabolism in a RNA world could well have been feasible (Chen et al. 2007). The recent demonstration of a RNA-only system that can replicate indefinitely in the laboratory by the self-sustained ligation further supports this RNA early evolutionary model (Lincoln and Joyce 2009). However, a central tenet of the RNA world hypothesis, namely that a RNA polymerase ribozyme can mediate the replication of a RNA-centered organism, has yet to be demonstrated in the laboratory and would provide unique insight into the evolution of biological systems (Bartel and Unrau 1999).
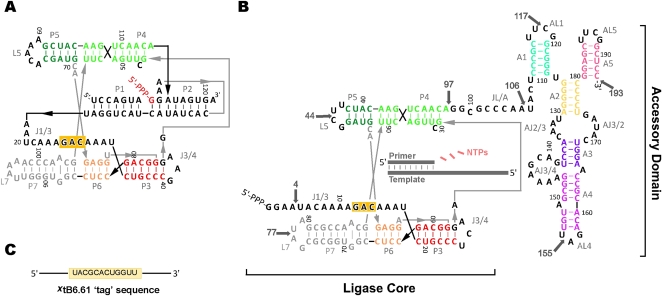
While a number of strategies to create such a polymerase have been explored, none has been as successful as those making use of the Class I ligase ribozyme as a core catalytic component (Bartel et al. 1991; Bartel and Szostak 1993; Johnston et al. 2001; McGinness et al. 2002; Lawrence and Bartel 2005). This ribozyme, originally isolated from a high diversity random sequence pool of RNA, is one of the fastest known ligase ribozymes (Bergman et al. 2000). The ribozyme promotes the formation of a 5′–3′ phosphodiester linkage between the 3′ terminus of a short RNA primer and the 5′ end of the ribozyme (Fig. 1A). Structurally the ligase resembles a tripod, containing three legs built out of the P1, P5, and P7 stems (Shechner et al. 2009). The P1 and P6 stems are positioned by a unique series of A-platforms found in the J1/3 joining sequence. The P2 helix, which is located at the vertex of the tripod, plays a major role in correctly positioning the 5′ terminal guanine triphosphate of the ribozyme with respect to the attacking 3′ hydroxyl of the primer and critical catalytic residues of the ribozyme (Bergman et al. 2004; Shechner et al. 2009). Remarkably, minor modifications to the Class I ligase ribozyme allow this ribozyme to polymerize 3–6 nucleotide (nt) triphosphates using a template sequence stabilized by hybridization to the 3′ arm of the P2 stem (Ekland and Bartel 1996). This suggested that the Class I ligase could serve as the starting point for the evolution of a true RNA polymerase ribozyme.
FIGURE 1.
The Class I ligase and the B6.61 polymerase. (A) The Class I ligase secondary structure drawn to take into account recent crystallographic data (Shechner et al. 2009). (B) Proposed secondary structure of the RNA polymerase ribozyme B6.61 based on this study. Insertion sites for tag hybridization are noted with arrows, using the notation XtB6.61, where X is the nucleotide immediately before the site of insertion. (C) The hybridization sequence inserted at each tag site.
The best RNA polymerase ribozyme to date is called B6.61 (Fig. 1B; Zaher and Unrau 2007). This polymerase, which can extend a primer-template (PT) by 20 nt, is the result of two sequential in vitro selections (Johnston et al. 2001; Zaher and Unrau 2007). The first selection detached the P1 helix from the Class I ligase and replaced it with a free PT. This change resulted in the loss of the 5′ arm of the P2 helix and converted the J1/3 region of the ligase into the 5′ terminus of the ligase core. The 3′ end of the ligase core was then attached to a high diversity RNA pool and selected for polymerase activity. The best isolate from this selection called the “Round-18” polymerase could extend a trans PT in a template-directed manner by up to 14 nt in 24 h (Johnston et al. 2001). Further evolution using in vitro encapsulated selection and a novel trans PT system, led to the emergence of B6.61 (Zaher and Unrau 2007). Differing from the Round-18 ribozyme only at its 5′ end and a residue within the accessory domain, B6.61 is uniformly two- to threefold faster than the Round 18 ribozyme under all conditions tested. Here we determine the essential residues of the accessory domain and demonstrate how this domain interacts with the ligase core.
RESULTS AND DISCUSSION
Tethering PT to the P5 and P7 legs of B6.61 results in increased polymerization efficiency
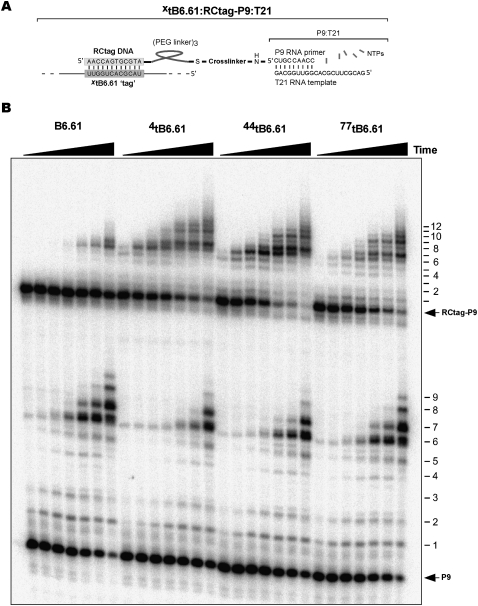
In order to better characterize PT extension by the B6.61 polymerase, we decided to localize the PT by tethering it to a set of sites on the polymerase using flexible polyethylene glycol linkers. A 12-nt “tag” sequence (henceforth tag) was built into the B6.61 polymerase at a total of eight tethering sites (Fig. 1B,C). This tag allows a 12-nt DNA “RCtag” sequence to hybridize to the polymerase. In turn this DNA was linked to the P9 RNA primer via either a 18C- or 56C-long PEG linker (Fig. 2A; Supplemental Fig. S1). The activity of the tethered PT extension was only weakly correlated with tether length. The longer linker always showed marginally slower extension kinetics, suggesting that the tethered PT concentration was well above the Km required for PT recognition by the polymerase. This is consistent with previous estimates of the PT binding affinity which are in the millimolar range (Lawrence and Bartel 2003).
FIGURE 2.
Tethered primer templates enhance the polymerization rate when hybridized to particular XtB6.61 ribozyme constructs. (A) RCtag-P9 covalently attaches the P9 RNA primer via a PEG linker to a DNA oligonucleotide able to hybridize to the ribozyme tag sequence. The P9 primer is in turn hybridized to the T21 RNA template. (B) Simultaneous time courses of untethered primer P9 extension (bottom) and tethered RCtag-P9 extension (top) by ribozymes with tags inserted at positions 4, 44, and 77. Time points: 5, 15, 30, 60, 180, 360, and 1440 min.
A number of tethering sites resulted in considerable increases in the polymerization rate. As insertion of an anchor sequence could well inhibit polymerase function, the rate of tethered and untethered primer extension was measured simultaneously using an excess of ribozyme over untethered and tethered PT. The 44tB6.61 tethered construct showed the largest polymerization enhancement, extending RCtag-P9:T21 10–20 times faster than untethered P9:T21 (Fig. 2B). The 4tB6.61 and 77tB6.61 tethered variants showed a more modest approximately fivefold rate enhancement. All three of these constructs were able to extend the tethered PT to the end of the T21 template (12 nt incorporations) (Fig. 2A). In contrast, unmodified B6.61 only extended P9:T21 by 7–8 nt. Tethering at 106tB6.61, 117tB6.61, and 193tB6.61 did not show marked changes in polymerization rate between tethered or untethered PT, while tethering at 97tB6.61 and 155tB6.61 inhibited polymerization for either PT construct (Supplemental Fig. S2). These inactive tethering sites were subsequently found to be within essential domains of the polymerase and are not discussed further.
Satisfyingly, the increased polymerization rate and efficiency of extension observed for the 4tB6.61, 44tB6.61, and 77tB6.61 constructs is consistent with the known crystal structure of the Class I ligase domain. The 44tB6.61 and 77tB6.61 tag sites are at the base of the P5 and P7 helical legs and, as such, provide a geometry that would allow the J1/3 linker region of the ligase core to correctly position the PT into the polymerase active site (Fig. 1A,B; Shechner et al. 2009). Likewise the 4tB6.61 construct appears likely to be flexible enough to allow a similar positioning of the PT. In contrast, the neutral anchor sites: 106tB6.61, 117tB6.61 and 193tB6.61, would be predicted to be on the “top” of the Class I ligase tripod, making it difficult for a tethered PT to be correctly positioned into the polymerase active site. These findings, which constrain the orientation of the PT, are consistent with an independent study in which the PT was co-localized nonspecifically to the Round 18 RNA polymerase ribozyme via hydrophobic anchors and where a three- to 20-fold increase in catalytic rate was observed (Muller and Bartel 2008).
The Class I ligase core and the accessory domain sustain polymerization in trans
The top of the Class I ligase tripod is defined by the P2 helix and the J3/4 loop (Fig. 1A; Shechner et al. 2009). Interestingly, the original polymerase selection resulted in a polymerase having an U100C mutation at the center of the residual arm of the P2 helix (Fig. 1B; Johnston et al. 2001). This is notable as throughout the selection a short RNA had been added in an attempt to reform the P2 helix and this mutation would be expected to completely destabilize such a stem. Moreover, the efficiency and fidelity of the B6.61 polymerase were improved when the short RNA was absent, further indicating that the P2 helix as defined in the Class I ligase was no longer required for polymerization (Zaher and Unrau 2007). Since the B6.61 ribozyme must have a structure analogous to the P2 stem in order to stabilize incoming nucleotide triphosphates during polymerization, we decided to study how the accessory domain might interact with the Class I ligase core in trans.
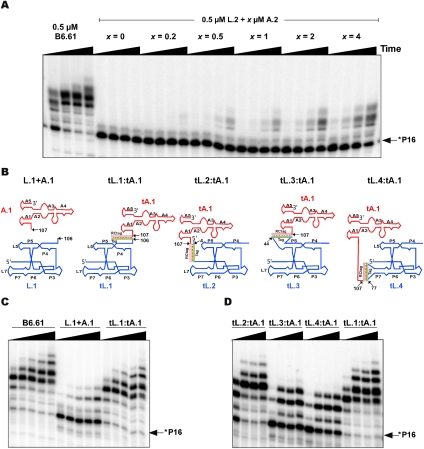
The B6.61 polymerase was first divided into two separate RNA molecules at nucleotide position 110 (Supplemental Fig. S3B, L.2 and A.2 constructs). With L.2 at 0.5 μM, the accessory domain sequence was found to promote polymerization in direct proportion to its concentration (Fig. 3A). The converse experiment, where the ligase core was titrated with respect to a fixed 0.5 μM concentration of the accessory domain A.2 sequence, produced identical results (data not shown). Since polymerization was not observed when either domain was present alone, these results imply that the two domains must cooperate with each other so as to allow polymerization. The fact that polymerization activity was in direct proportion to the concentration of the two domains and could not be made to saturate suggested that these two domains interact via a weak set of tertiary interactions that appeared likely to be enhanced by the JL/A sequence joining the two domains.
FIGURE 3.
The accessory domain of the B6.61 polymerase works either in trans or when hybridized to the ligase core. (A) Polymerization occurs in trans, and is much slower than the unimolecular B6.61 construct. Time points: 10, 50, 250, and 1500 min. (B) Trans and hybridized ligase core and accessory domain constructs showing activity. (C) Extension profiles for construct tL.1:tA.1 relative to B6.61 and trans construct L.1+A.1. Time points: 10, 60, 180, 540, and 1380 min. (D) The primer extension profile for the four hybridized constructs shown in B. Time points: 1, 16, 25, and 40 h. All RNA ribozyme constructs were at 2 μM using P16:T21 primer template pair, unless otherwise noted.
The trans domain system allowed us to systematically explore the importance of residues found at the 3′ terminus of the ligase domain. Ligase domain constructs longer than A106 all produced identical extension patterns when tested in trans with the A.1 accessory domain construct (Supplemental Fig. S3). Activity dropped precipitously for shorter constructs (Supplemental Fig. S3). This truncation series demonstrates that residues up to G101 of the ligase domain are key for polymerization to occur and contain the U100C mutation previously observed in the P2 arm of the Round 18 polymerase (Johnston et al. 2001).
The ability of the B6.61 polymerase to function in trans suggested that polymerase domain function might be explored by hybridizing the two domains together using the tag:RCtag anchoring system developed to study primer-template localization. The 12-nt tag sequence was therefore appended to the 3′ end of L.1 ligase core domain (tL.1), while the RCtag sequence was added to the 5′ end of A.1 as RNA (tA.1) (Figs. 1C, 3B). While untethered trans acting L.1 + A.1 was ∼100-fold slower than B6.61, the bimolecular tL.1:tA.1 construct appeared to fully restore activity (Fig. 3C; Supplemental Fig. S4A). Equally unexpected was the finding that moving the anchor tag sequence from the 3′ terminus to the 5′ terminus of the ligase core (tL.2) also had high levels of activity when hybridized to tA.1 (Fig. 3). This suggests that the 5′ end of the ligase core in the B6.61 polymerase is flexible enough to allow the tethered accessory domain to approach the top of the ligase tripod in the tL.2:tA.1 context, and for the tethered PT complex to approach the bottom of the tripod in the 4tB6.61 context. Consistent with this idea, tethering the accessory domain to the P5 and P7 loops did not promote efficient polymerase activity (Fig. 3B,D), in agreement with the expectation that the accessory domain needs to be at the vertex of the ligase core tripod for polymerase function. Together with our PT tethering data this indicated a general model, where the PT docks to the polymerase in the same orientation as the P1 stem in the Class I ligase structure (Fig. 1B), while the accessory domain is located at the vertex of the Class I ligase tripod structure so as to correctly mediate polymerization.
Determining the core motif of the accessory domain
In order to study the functional contribution of each structural element in the tA.1 construct, we progressively deleted sequence and simultaneously identified secondary structure elements by covariational experiments. The resulting constructs were then hybridized to tL.1 and tested for their polymerization ability as summarized in Supplemental Figure S4 (n.b. all nucleotide position henceforth are relative to the numbering shown in Fig. 1B). Disrupting helix A3 by a C136G or G168C point mutation in the tA.1 context lowered activity by 20- or threefold, respectively. A double mutation, which restored pairing, had near wild-type activity implicating A3 in the functional motif. A series of deletion constructs rapidly led to the discovery that the A1 and A5 helixes (Fig. 1B) were not essential for polymerization and that the A2 helix appeared to stabilize the previously unsuspected A3 stem via the AJ2/3 and AJ3/2 joining regions. Consistent with this, removing the 5′ arm of the A2 helix and replacing the sequence 3′ of the A3 helix, “CAUAGU…,” with “CAUACC” resulted in a construct that became functional only when combined with a C134G mutation able to increase the stability of the A3 stem by one base pair (tA.11). This insight resulted in the tD.1 construct, which increases the length of the A3 helix from four to five base pairs and only contains the A3 and A4 helices (Supplemental Fig. S4). The activity of the trans bimolecular tL.1:tD.1 construct was almost identical to that of tL.1:tA.1 when extending the P16:T21 primer template. This conclusion remained when other PTs were tested for extension (Supplemental Fig. S5). The tL.1:tD.1 construct was at most approximately twofold slower than either tL.1:tA.1 or the B6.61 unimolecular polymerase indicating that the tD.1 construct captures nearly all the functional aspects of the full accessory domain.
Further characterization found the secondary structure of the tD.1 construct to be as expected. Stem A3 when destabilized by a double G134C and C136G mutation ablated all function. Similarly a G168C and C170G double mutation resulted in very low levels of activity. Making all four point mutations simultaneously rescued nearly full levels of activity (Supplemental Fig. S4). The A4 bulged stem–loop was explored in a similar manner. Introducing a G153C mutation at the distal end of the helix considerably lowered activity that could in turn be rescued by a compensatory C159G mutation. Notably the 5′ arm of the A4 helix immediately proximal to the proposed AL4 loop (Fig. 1B) was more sensitive to mutation than the 3′ arm, suggesting a possible interaction on this side of the A4 helix with the ligase core.
The importance of the AL4 loop, the A4 bulged helix, and the AJ3/4 loop
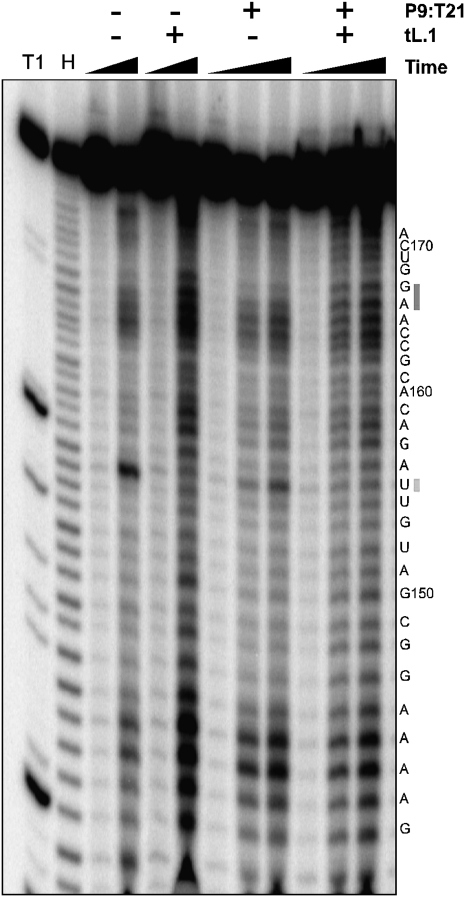
Fe(II)-EDTA radical-induced cleavage was then used to judge the solution accessibility of the residues found in the tD.1 motif. One dominant pattern of cleavage was located within the four adenosine residues found in the AJ3/4 region (notably A144 and A145) and was correlated with a set of cleavages spanning the unpaired A on the strand opposite the AJ3/4 sequence. Isolated from this location was a strong Fe(II)-EDTA radical-induced cleavage found within the AL4 loop at U155 (Fig. 4). Significantly, U155 became protected only upon addition of the tL.1 ligase domain. At the same time the pattern of cleavage ending at A165 extended to include A166 and G167 in the A3 helix. This change was correlated with a mild increase in damage at residues located just outside the AL4 triloop (G157, A158, and C159). Together these damage patterns suggest that the AL4 loop interacts with the ligase core and in the process twists the A4 and A3 stems so as to simultaneously increase solvent accessibility at both the distal end of the A4 stem and at the junction between the A3 and A4 stems.
FIGURE 4.
The ligase core affects Fe(II)-EDTA cleavage patterns in the tD.1 accessory domain. Fe(II)-EDTA cleavage patterns in the presence or absence of the tL.1 domain. Cleavage was not significantly modified by the presence or absence of PT (cleavage time points: −P9:T21; 0 and 20 min. For +P9:T21; 0, 5, and 20 min). A T1 and hydrolysis (H) ladder is shown on the left.
Consistent with this interpretation, the AL4 loop sequence had a marked effect on polymerase activity. Notably a U155C mutation and a set of constructs replacing the triloop with GUGA, AUAC, or even UACG (Cheong et al. 1990; Heus and Pardi 1991) all had minimal activity, indicating that the loop sequence and context plays a vital role in accessory domain function. The importance of the A:U base pair that potentially closes the AL4 loop was tested by a U154C mutation. This mutation showed only a small decrease in activity suggesting that either the A:U pair does not fully form or that it is involved in a nonstandard base pair when docked to the ligase core.
The unwinding/docking process is facilitated by the A151 bulge in helix A4. This bulge is not sequence-specific as an A151U mutation is well tolerated, implicating the backbone of the bulge as the functional element. Supporting this idea, N7 of A151 became increasingly modified by DEPC as a function of increasing magnesium (Supplemental Fig. S6), while the backbone at this site appears not to be strongly exposed to solvent based on our Fe(II)-EDTA results. Removing the bulge completely decreased activity by ∼20-fold, while inserting a uridine across from the bulged A151 residue nearly completely ablated activity (Supplemental Fig. S4). Together these data suggest that the A4 bulge dramatically increases the ability of the AL4 loop to correctly dock with the ligase core.
Point mutation analysis indicated that the AJ3/4 purine-rich loop (residues G139–A146) was critical for polymerase activity. When any one of these residues were deleted or mutated, polymerase activity dropped substantially relative to the tD.1 parent, suggesting that the AJ3/4 loop plays a key role in mediating efficient polymerization (Supplemental Fig. S4). In contrast, A165, which separates helix A3 from A4, was less critical. The residue could be mutated to any other base without any decrease in polymerization rate and its removal was weakly tolerated (Supplemental Fig. S4). This adenosine residue therefore appears to be primarily responsible for correctly spacing the A3 and A4 helices, which when docked to the ligase core via the AL4 loop can be unwound so as to correctly position the AJ3/4 loop structure with respect to the ligase core.
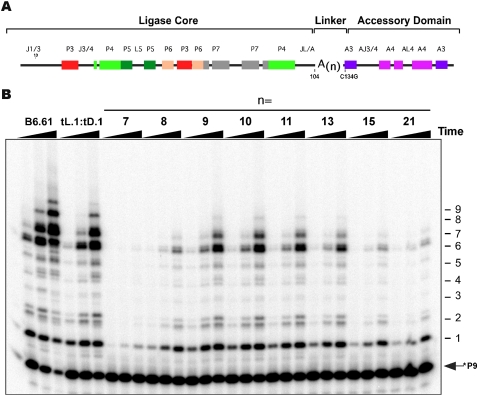
A minimal unimolecular polymerase requires an optimal JL/A linker length
Motivated by the fact that we could create a truncated, yet highly functional tL.1:tD.1 ribozyme from a system able to work only weakly in trans (L.1 + A.1), we wanted to understand how the linkage between the ligase core and the minimal accessory domain influenced polymerization behavior. We therefore synthesized a range of unimolecular constructs containing a variable adenosine spacer region inserted into the JL/A linker region (Fig. 5A). Maximal P9:T21 extension was observed for n = 10, although this rate was still two to three times slower relative to that of the tL.1:tD.1 construct and approximately five times slower than that of the unimolecular B6.61. Decreasing this optimal linker length rapidly destroyed activity, while increasing it resulted in a steady diminishment in activity (Fig. 5B).
FIGURE 5.
Systematically varying linker length between the tL.1 ligase core and the minimal tD.1 accessory domain reveals an optimal linker length. (A) The unimolecular construct tested showing the location of the variable poly(A)n linker. (B) Template extension for n-values from 7 to 21, showing extension of the P9:T21 primer template. Time points: 1, 5, and 24 h.
These findings are generally consistent with a model where the ligase core and accessory domain must be correctly spaced via the linker region so as to generate maximal polymerization efficiency. Since the two domains function in trans, the discovery of an optimal linker length reinforces our basic hypothesis that a weak set of tertiary interactions can be enhanced by the correct JL/A linker sequence and can be disrupted by linkers that are either too short or too long. We note that the optimal poly(A) linker length of n = 10 could allow a kink between the P4 helix of the ligase and the A3 helix of the accessory domain that would be similar to the tL:1:tD:1 complex in overall geometry. Shortening the linker would prevent such a kink, while too long of a linker would be too floppy for optimal activity.
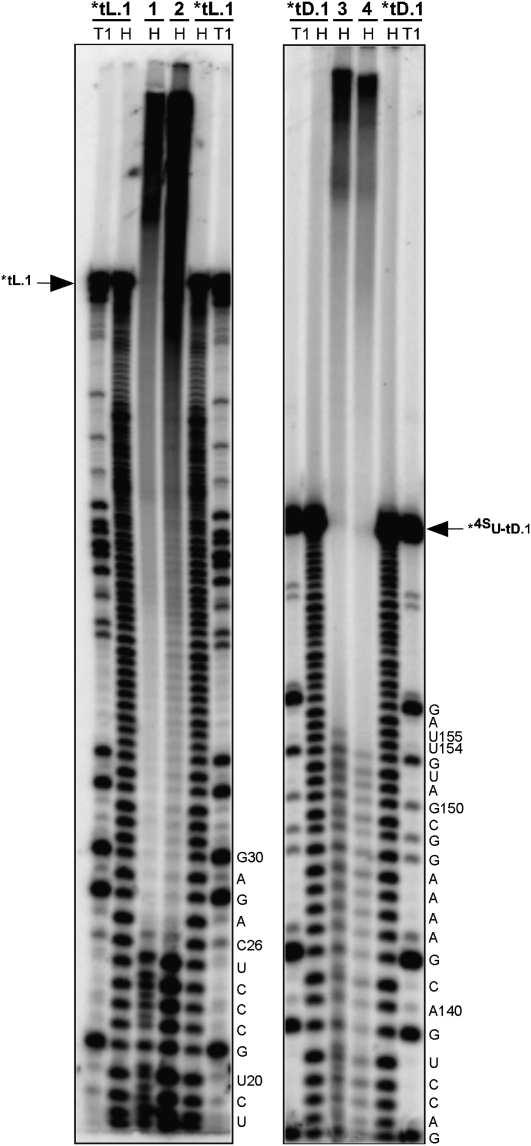
Interdomain crosslinking indicates that AL4 and J3/4 interact
Exploiting the hybridization-based system developed earlier, we developed a simple radiolabeling/4SU modification strategy to determine tertiary contacts between tA.1 and tD.1. One of the two RNA molecules was transcribed using 4SUTP instead of UTP, while the other RNA was transcribed normally. Upon hybridization of the two domains, UV irradiation at 356 nm was used to generate covalent crosslinks (Favre et al. 1998) between 4SU residues on the one molecule with adjacent residues found on the other. By radiolabeling either of the two strands in separate experiments (Supplemental Fig. S7), two dominant crosslinks could be mapped by partial alkaline hydrolysis (Fig. 6).
FIGURE 6.
Tertiary interactions mapped by UV crosslinking. Mapping of the tL.1: 4SU-tD.1 crosslinks. Each crosslinked species (1, 2, 3, and 4) was gel-purified and partially alkaline hydrolyzed (see Supplemental Fig. S7). The crosslinked residues were then mapped using a PAGE sequencing gel with T1 and hydrolysis ladders as references.
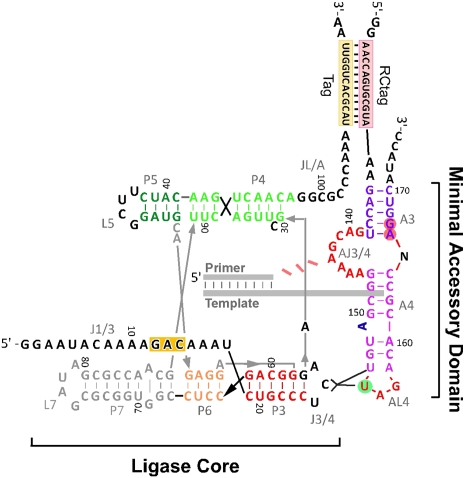
Satisfyingly, one crosslink mapped to C26 in the ligase core and 4SU154 in the accessory domain, while the second mapped to C26 in tL.1 and 4SU155 in the accessory domain. As U155 had previously been strongly implicated in an interaction with the ligase core based on Fe(II)-EDTA probing data (Fig. 4) and point mutation data, the location of these crosslinks was particularly striking. An AL4 loop interaction with the ligase J3/4 triloop would satisfy our previous constraints that place the accessory domain at the vertex of the ligase core and more importantly would serve to position the critical AJ3/4 residues close to the catalytic site of the polymerase (Fig. 7).
FIGURE 7.
Docking of the accessory domain. Crosslinking interactions between J3/4 C26 and AL4 U154 and U155 are summarized by black lines. Blue A151 bulge is required for efficient polymerization activity. Red residues in accessory domain are essential for activity. (Red circles) Increased Fe(II)-EDTA damage upon docking; (green circles) decreased Fe-EDTA(II) protection upon docking.
Proposed model and future directions
Using a range of chemical probing, mutagenesis analysis, and crosslinking studies, we propose the following model for the B6.61 polymerase. At the core, weak tertiary interactions serve to precisely position the accessory domain with respect to the ligase core. The AL4 and J3/4 docking interaction precisely positions and possibly activates the accessory domain by twisting the A3 and A4 helices with respect to each other. This likely plays a role in correctly positioning the essential AJ3/4 purine-rich bulge. The torque required for such an unwinding process necessarily implies a second, presently uncharacterized, interaction between the two domains. While the accessory domain tether might induce this torque, it appears possible that ligase residues C100 and G101 could pair directly with C141 and G142 in the accessory domain. Such an interaction would explain the importance of the vestigial 3′ P2 arm in the ligase core and explain the observed U110C mutation previously observed. Such potential interactions were investigated but not found, possibly due to the extreme sensitivity of the AJ3/4 bulge to mutation, making such point mutagenesis experiments difficult to interpret. Given the crystal structure of the ligase core and the findings presented here, we can certainly conclude that the accessory domain is draped over the top of the ligase core (Fig. 7), placing the AJ3/4 bulge in close proximity to the polymerase active site.
How this complex mediates the addition of nucleotide triphosphates is still unknown. Our PT tethering results strongly suggest that the PT transiently docks with the ligase core in an orientation similar to the P1 stem in the Class I ligase (Shechner et al. 2009). Since the first unpaired residue of the template strand in the PT must form a base pair with the next incoming NTP, critical residues found in the AJ3/4 bulge of the accessory domain in conjunction with 3′ residues of the ligase core appear ideally situated to stabilize an incoming NTP with respect to the extending template. This structure could conceivably stack an incoming NTP against G(99) so as to form a structure similar to that required to stabilize the first two nucleotides of the Class I ligase [pppG(1)pG(2)] by the P2 stem in the Class I ligase (Shechner et al. 2009).
The insight gained from this study can be used to further improve this interesting RNA polymerase enzyme. Selections based on this work can be performed that might reasonably be expected to further stabilize both the accessory domain—ligase core interaction and the affinity for PT. Both outcomes would have an important impact on improving the overall processivity and fidelity of this polymerase (Lawrence and Bartel 2003; Zaher and Unrau 2007).
MATERIALS AND METHODS
Tethering primer-template complexes to tagged polymerase ribozyme constructs
The RCtag DNA oligonucleotide construct was synthesized by coupling one or three Spacer Phosphoramidite C18 (Glen Research) units to a 3′-Thiol-Modifier C3 S-S CPG column (Glen Research), and was followed by the synthesis of the RCtag DNA sequence: 5′-AACCAGTGCGTA onto the 5′ end of the construct. All steps used standard phosphoramidite chemistry on an ABI 392 DNA/RNA synthesizer. After deprotection, these constructs where coupled to RNA primers containing a 2′-amine at their 5′- termini (Dharmacon) using the bifunctional linker Sulfo-GMBS (N-[γ-maleimidobutyryloxy]sulfosuccinimide ester [Pierce]). One nanomole amine modified RNA primer was dissolved in water, mixed with 20 nmol of sulfo-GMBS dissolved in DMF (N,N-dimethylformamide) and lyophilized to dryness. The resulting pellet was then resuspended in 2 μL phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) at pH 7.2 and incubated at room temperature for 30 min. A C18 SPICE column (Analtech) was used to purify the product, which was eluted in 20% acetonitrile, 50 mM ammonium acetate at pH 7.0. The eluted material was lyophilized to dryness and 2 nmol of RCtag-C18-3′ thiol DNA oligonucleotide was added to 25 μL of 20 mM HEPES buffer, 0.5 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-HCl) at pH 7.0 was added and allowed to react at room temperature overnight in the dark. Crosslinked RCtag DNA-C18–RNA primer construct was gel purified and stored at −20°C prior to use.
Sequence insertion and modification
The 12-nt RNA tag sequence was 5′-UACGCACUGGUU-3′. The end-tagged constructs 4tB6.61 and 193tB6.61 were constructed directly by PCR using a DNA primer containing the tag sequence, followed by transcription. Internally tagged constructs 44tB6.61, 77tB6.61, 97tB6.61 and 106tB6.61, 117tB6.61, and 155tB6.61 were constructed by three successive PCR steps. First, B6.61 DNA was PCR amplified into two DNA fragments: The first consisting of the B6.61 sequence from the 5′ end up to the insertion site followed by the 12-nt tag sequence (TACGCACTGGTT) and the second DNA piece consisting of the RCtag sequence at its 5′end followed by the rest of the B6.61 sequence starting from the “x + 1”th residue. After QIAquick (QIAGEN) purification, the two fragments, ∼10 nM each, were mixed in the absence of primers and PCR thermocycled for 30 cycles using a lower annealing temperature at 45°C for 1 min 50 s. Primers corresponding to the 5′ and 3′ terminus were then added, and PCR amplification under an annealing temperature of 50°C resulted in the internal-tagged ribozyme constructs ready for transcription. A similar approach was used to construct all other mutants used in this study.
Primer templates
The following RNA primer sequences were used in this study: P9: 5′ CUGCCAACC, P12: 5′ CUGCCAACCGUG, P16: 5′ d(cac g) CUGCCAACCGUG. Templates where as follows: T21: 5′ GAGGCUUCGCACGGUUGGCAG, T32: 5′ CGGAUCGUACCAGUCAAGCGCACGGUUGGCAG (see also Supplemental Table S1).
Kinetic assays
32P radiolabeled RNA primer at 0.1 μM was annealed to a 1-μM template in the presence of 100 mM Tris-HCl at pH 8.5. Gel-purified ribozyme components were then added to the mixture at final concentration of 2 μM, or as otherwise indicated. Extension of the primer template was initiated by the addition of 200 mM MgCl2 and 4 mM of each NTP. Reactions were incubated at 22°C and stopped by the addition of a fivefold excess of stop-mix containing 80% formamide, 40 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene cyanol, together with an approximate fourfold of a competitor RNA oligonucleotide able to hybridize to the template RNA. Samples were heated at 95°C so as to denature the PT prior to loading onto a 20% polyacrylamide sequencing gel. A Storm 820 PhosphorImager (Amersham Biosciences) and Fuji phosphorimager plates were used to expose the resulting gels.
First-order rate constants for the first nucleotide incorporation were determined by a two-parameter fit to: [F] = [F]max (1 − e−k1t), where t equals time, k1 is the fitted rate constant for extension by at least 1 nt, [F] is the fraction of the radiolabeled primer converted to extended primer of any sort, and [Fmax] is the maximum value of [F] as determined by the fit.
Solution accessibility mapped by Fe(II)-EDTA radical cleavage
Fe(II)-EDTA radical cleavage reactions were performed using 10-μL samples. 32P-5′ end-labeled RNA (0.1 μM) was heated to 80°C for 3 min and slowly cooled to 4°C over 15 min before adding buffer containing 100 mM EPPS (Sigma) at pH 8.5, and 5 mM ascorbate (Sigma). MgCl2 was added to give the concentration of free Mg2+ as indicated. The solution was incubated with 2 mM (NH4)2Fe(SO4)2 (Sigma) and 4 mM EDTA in a volume of 9 μL at 22°C for 5 min. Then, 0.5 μL 3% H2O2 (Anachemia) was added to initiate radical cleavage. After 15 min of incubation at 22°C, the reaction was stopped by the addition of 0.1 volume of a solution containing 200 mM thiourea (Sigma). Three hundred mM NaCl and 70% cold ethanol were then added to precipitate the RNA, which were later resolved on either 12% or 15% polyacrylamide sequencing gels.
DEPC probing
A 5′-end 32P radiolabeled ribozyme construct at 1 μM concentration was incubated at 22°C in 20 μL volumes with 5% by volume DEPC (Sigma), 50 mM sodium cacodylate (Sigma) pH 7.5, 1 mM EDTA, and MgCl2 at the indicated concentrations. Four volumes of stop-buffer containing 200 mM Tris-acetate pH 7.5, 375 mM sodium acetate, and 0.12 mM EDTA were then added after incubation at 22°C for 1 h in the dark. After ethanol precipitation, the pellet was dried using speed vacuum. Fifty μL of 1 M aniline (Sigma) in 15% acetic acid was used to dissolve the pellet. The mix was kept at 60°C for 20 min in the dark and dried to a pellet using the speed vacuum. After three iterations of drying and dissolving, the samples was dissolved in 30 μL loading dye containing 2 mM EDTA and run on a PAGE sequencing gel together with a hydrolysis and a T1 ladder.
4SU-mediated UV crosslinking
The DNA template of tD.1 or tA.1 were transcribed at 37°C for 1.5 h under following conditions: 40 mM Tris pH 7.9 at 20°C, 2.5 mM spermidine (Sigma), 26 mM MgCl2, 0.01% Triton X-100 (Sigma) 8 mM GTP (Amersham), 5 mM ATP (Amersham), 5 mM CTP (Amersham), 1 mM 4SUTP (Sigma), and T7 RNA polymerase. After gel purification of RNA with and without 4SUTP, the appropriate strands were then end-labeled with polynucleotide kinase. Bimolecular ribozyme constructs were then assembled in equimolar amounts as described. Reaction buffer (100 mM Tris-HCl, pH 8.5, 50 mM MgCl2) was added (final ribozyme concentration 1.0 μM) and pipetted into a polystyrene Costar 96 well plate (Corning), placed on ice and irradiated from the top with a UV transilluminator. After irradiation at 0°C for 10–20 min, three volumes of stop-solution (97% formamide, 20 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene cyanol) were added, and crosslinks resolved using a 10% or 12% PAGE.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
The work was supported by the following grants agencies: Natural Sciences and Engineering Research Council of Canada (NSERC) and the Michael Smith Foundation for Health Research (MSFHR). Graduate fellowships to L.K.L.C. (NSERC) and Q.S.W. (MSFHR) and a Senior Scholars award to P.J.U. (MSFHR) are also recognized.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2495011.
REFERENCES
- Bartel DP, Szostak JW 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418 [DOI] [PubMed] [Google Scholar]
- Bartel DP, Unrau PJ 1999. Constructing an RNA world. Trends Cell Biol 9: M9–M13 [PubMed] [Google Scholar]
- Bartel DP, Doudna JA, Usman N, Szostak JW 1991. Template-directed primer extension catalyzed by the Tetrahymena ribozyme. Mol Cell Biol 11: 3390–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH, Johnston WK, Bartel DP 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123 [DOI] [PubMed] [Google Scholar]
- Bergman NH, Lau NC, Lehnert V, Westhof E, Bartel DP 2004. The three-dimensional architecture of the class I ligase ribozyme. RNA 10: 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li N, Ellington AD 2007. Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers 4: 633–655 [DOI] [PubMed] [Google Scholar]
- Cheong C, Varani G, Tinoco I Jr 1990. Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature 346: 680–682 [DOI] [PubMed] [Google Scholar]
- Deamer D, Szostak JW, ed. 2010. The Origins of Life, Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- Ekland EH, Bartel DP 1996. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382: 373–376 [DOI] [PubMed] [Google Scholar]
- Favre A, Saintomé C, Fourrey JL, Clivio P, Laugâa P 1998. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J Photochem Photobiol B 42: 109–124 [DOI] [PubMed] [Google Scholar]
- Gilbert SD 1986. The RNA world. Nature 319: 618 [Google Scholar]
- Heus HA, Pardi A 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194 [DOI] [PubMed] [Google Scholar]
- Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP 2001. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292: 1319–1325 [DOI] [PubMed] [Google Scholar]
- Joyce GF 2002. The antiquity of RNA-based evolution. Nature 418: 214–221 [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Bartel DP 2003. Processivity of ribozyme-catalyzed RNA polymerization. Biochemistry 42: 8748–8755 [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Bartel DP 2005. New ligase-derived RNA polymerase ribozymes. RNA 11: 1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF 2009. Self-sustained replication of an RNA enzyme. Science 323: 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Wright MC, Joyce GF 2002. Continuous in vitro evolution of a ribozyme that catalyzes three successive nucleotidyl addition reactions. Chem Biol 9: 585–596 [DOI] [PubMed] [Google Scholar]
- Muller UF, Bartel DP 2008. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA 14: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE 1968. Evolution of the genetic apparatus. J Mol Biol 38: 381–393 [DOI] [PubMed] [Google Scholar]
- Shechner DM, Grant RA, Bagby SC, Koldobskaya Y, Piccirilli JA, Bartel DP 2009. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 326: 1271–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Unrau P 2007. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]