Abstract
We present a rapid experimental strategy for inferring base pairs in structured RNAs via an information-rich extension of classic chemical mapping approaches. The mutate-and-map method, previously applied to a DNA/RNA helix, systematically searches for single mutations that enhance the chemical accessibility of base-pairing partners distant in sequence. To test this strategy for structured RNAs, we have carried out mutate-and-map measurements for a 35-nt hairpin, called the MedLoop RNA, embedded within an 80-nt sequence. We demonstrate the synthesis of all 105 single mutants of the MedLoop RNA sequence and present high-throughput DMS, CMCT, and SHAPE modification measurements for this library at single-nucleotide resolution. The resulting two-dimensional data reveal visually clear, punctate features corresponding to RNA base pair interactions as well as more complex features; these signals can be qualitatively rationalized by comparison to secondary structure predictions. Finally, we present an automated, sequence-blind analysis that permits the confident identification of nine of the 10 MedLoop RNA base pairs at single-nucleotide resolution, while discriminating against all 1460 false-positive base pairs. These results establish the accuracy and information content of the mutate-and-map strategy and support its feasibility for rapidly characterizing the base-pairing patterns of larger and more complex RNA systems.
Keywords: capillary electrophoresis, footprinting, chemical mapping, RNA folding, RNA structure, high throughput
INTRODUCTION
Functional RNAs are critically involved in fundamental biological processes throughout viruses and living cells (Gesteland et al. 2006). Many RNA molecules self-assemble into specific base-pairing structures and rearrange their structures in response to other nucleic acids, proteins, and small molecules (see, e.g., Gallego and Varani 2001; Barrick et al. 2004; Pollard et al. 2006; Amaral et al. 2008; Ramakrishnan 2008). Despite continuing advances and applications of crystallography, spectroscopy, microscopy, and phylogenetic co-variance methods, the structural characterization of RNAs, particularly in large multi-state complexes like the spliceosome, remains a major challenge (see, e.g., Staley and Guthrie 1998; Noller 2005; Cruz and Westhof 2009). To address this challenge, we are pursuing a novel information-rich extension of classic chemical approaches that we call the “mutate-and-map” strategy (Kladwang and Das 2010).
Chemical mapping experiments, also known as “structure mapping” or “footprinting” methods, have been used for 30 years to probe the structures, folding kinetics, and interactions of nucleic acids in vitro and in vivo (Peattie and Gilbert 1980; Krol and Carbon 1989; Tullius 1991; Schroeder et al. 2002; Adilakshmi et al. 2006; Tijerina et al. 2007; Mitra et al. 2008). These methods permit the characterization of systems as large as the ribosome (Merryman et al. 1999; Culver and Noller 2000; Lancaster et al. 2002) or entire viral genomes (Wilkinson et al. 2008; Watts et al. 2009). The resulting nucleotide-resolution data report on RNA bases that are protected from chemical modification and are therefore likely involved in base pairs. These data are not sufficient on their own to determine nucleic acid secondary or tertiary structures, but can guide computational modeling algorithms that generate structural hypotheses (Mathews et al. 2004; Deigan et al. 2009; Quarrier et al. 2010). While these hybrid chemical/computational methods can be widely applied, they are not fully reliable, especially for non-Watson-Crick interactions, complex topologies such as pseudoknots, or protein components, which are not accurately modeled in current computational algorithms.
More experimental information is necessary for confident structure inference. The most valuable information beyond a “one-dimensional” list of residues that are protected would be a “two-dimensional” list of pairs of residues that are interacting. The desire for such pairing data motivates several approaches to RNA structure inference, including phylogenetic covariance (Levitt 1969; Gutell et al. 1992; Lehnert et al. 1996), NOE spectroscopy (Clore and Gronenborn 1985; Wuthrich 2003; Tzakos et al. 2006), and biochemical methods based on interference/suppression of modifications (Szewczak et al. 1998; Waldsich 2008), tethered cleavage (Han and Dervan 1994; Culver and Noller 2000; Joseph et al. 2000; Lancaster et al. 2002; Das et al. 2008), and various molecular rulers (Gohlke et al. 1994; Mathew-Fenn et al. 2008). Nevertheless, each of these methods is inapplicable, too arduous, and/or too low in resolution to enable rapid determination of any RNA's base-pairing patterns with nucleotide-level precision.
We recently proposed that two-dimensional (2D) residue pairing information might be attainable by augmenting the one-dimensional (1D) chemical mapping method with high-throughput mutagenesis (Kladwang and Das 2010). In this mutate-and-map approach, each single mutant of the nucleic acid system is separately synthesized and probed by chemical accessibility measurements. The mutations at a base-paired residue may release its base-pairing partner, and we hypothesize that this effect will lead to detectable changes in the partner's modification by chemical reagents. Some sequence changes may be too conservative, preserving a base-pairing interaction and leaving a residue's pairing partner protected from chemical modification. Other sequence changes may lead to more dramatic effects such as unfolding of entire helices. Nevertheless, if even a subset of mutations gives specific release of interacting residue pairs, the mutate-and-map strategy would enable the rapid and systematic determination of RNA base pairs.
The mutate-and-map approach has not yet been tested in its ability to infer full RNA base-pairing patterns. While there are important precedents, including inference of the A302/-3u contact in the Tetrahymena ribozyme (Pyle et al. 1992) and of a P7.1/P9.1 helix in the bi3 group I intron (Duncan and Weeks 2008), these prior efforts have been limited to verification of individual, previously hypothesized interactions. To establish whether mutate-and-map experiments will be more generally useful for structure inference, we are carrying out a series of proof-of-concept experiments on RNA, DNA, and ribonucleoprotein systems with known or designed structure. We recently reported our first results from this series of experiments, on a 20-bp RNA/DNA helix (Kladwang and Das 2010). In response to all possible single mutations and deletions of the DNA strand, dimethyl sulfate alkylation measurements of the A and C residues of the RNA strand gave strong, localized features. We observed unambiguous, nucleotide-resolution signals for 15 of the 17 base pairs with A or C on the RNA strand.
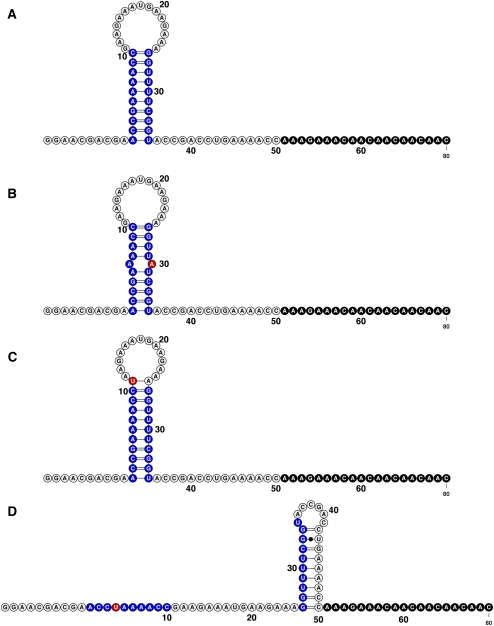
While encouraging, the prior DNA/RNA helix study did not demonstrate several remaining steps critical for inferring structures of complex RNAs: the synthesis of entire single-mutant libraries of RNA; the readout of G and U bases in addition to A and C; and the discrimination of precise base-pair “release” signals from larger-scale conformational changes induced by mutations. To address these remaining issues, we wished to apply the mutate-and-map approach to an RNA model system with at least 10 base-pairing features, an equal number of A-U and C-G base pairs, and a length small enough to still permit visual consideration of all the collected data (less than 100,000 features). We therefore designed a 35-nt system that we called the MedLoop RNA, which is expected to form a stable base-pair stem with five A-U and five C-G base pairs, closed by a 15-residue loop (Fig. 1). It provides a reasonable number of potential base-pairing features (60, counting each possible mutant in the 20 residues of the stem) to test the method. The final data set, including measurements from three chemical probes and controls, is large (total of ∼30,000 features) but still allows for visual inspection of this proof-of-concept data set. Finally, we embedded this RNA into an 80-nt sequence that is susceptible to a global conformational rearrangement upon certain mutations. The system thus provides a stringent test of the mutate-and-map method to discriminate single base pairs from large-scale changes.
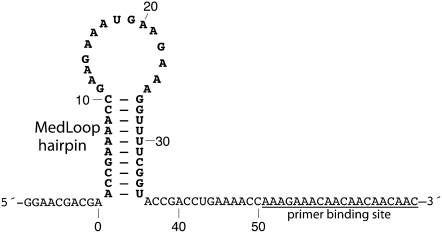
FIGURE 1.
Model system for establishing the mutate-and-map methodology for RNA structure inference. The 80-residue MedLoop RNA was designed as a 10-bp hairpin with a 15-nt internal loop (residues 1 to 35), a 10-residue 5′-flanking sequence (residues −9 to 0), and a 35-residue 3′-flanking sequence (residues 36 to 70) containing the primer binding site.
Using the MedLoop RNA model system, we report that modern molecular biology tools permit the rapid preparation and purification of a complete RNA single-mutant library. Furthermore, entire mutate-and-map data sets, with thousands of bands, can be readily measured and quantitated for three chemical probes (DMS, CMCT, and SHAPE). To rationalize strong features in the resulting data, we make a qualitative comparison to computational models of the single-mutant secondary structure ensembles. Finally, we described an automated analysis that enables the confident and accurate extraction of base-pairing signals from these information-rich measurements without using sequence information or secondary structure prediction algorithms. This study establishes a proof-of-concept of the mutate-and-map strategy for interrogating RNA structure, provides benchmark data for signal analysis, and highlights the promise of a rapid, general, and accurate approach to RNA base-pair inference.
RESULTS
Applying the mutate-and-map approach (Kladwang and Das 2010) to infer the base pairs of a target RNA requires several steps: (1) synthesizing a complete library of single-residue mutants of the target RNA, (2) chemical accessibility mapping of these RNAs, (3) understanding the features that arise in this extensive data set, and (4) automated data analysis to extract nucleotide-resolution base-pairing signals. We have developed straightforward methods for carrying out each of these steps and describe here the results for our first RNA proof-of-concept system, the MedLoop RNA sequence (Fig. 1).
An extensive library of RNA mutants
Because preparation of large mutant libraries is still uncommon in RNA biophysical studies, we first summarize the strategy, yields, and time investment associated with the preparation and purification of 120 constructs. This library included all 105 single mutants of the MedLoop RNA sequence and replicates of the unmutated (“wild type”) sequence and the first three mutants. Decreasing costs of DNA oligonucleotide synthesis and the wide availability of high-throughput purification technologies enabled inexpensive and rapid preparation of the entire library.
Our strategy was to transcribe each 80-nt RNA variant from a 100-bp DNA template that included the promoter for T7 RNA polymerase. The DNA template was prepared by annealing two commercially synthesized 60-nt DNA oligos (with 20-bp overlap), extending with a high-fidelity DNA polymerase, and then purifying with magnetic beads optimized for binding to double-stranded DNA. The resulting samples could be rapidly assayed for concentration and purity by UV absorbance measurements on an eight-channel Nanodrop and 96-well agarose gel systems. Fifty-microliter reactions with 200 pmol of each single-stranded DNA yielded 50–150 pmol of double-stranded DNA template.
RNA synthesis from these templates used standard in vitro transcription conditions (Sampson and Uhlenbeck 1988; Hartmann et al. 2005), commercial T7 RNA polymerase, and commercially available magnetic bead purification methods. Sample concentrations and purity were again checked by UV absorption and agarose gels. Parallel 40-μL transcriptions with 8 pmol of DNA template yielded 90–180 pmol of RNA after purification. The observed efficiency was similar to larger volume transcriptions purified by phenol/chloroform extraction or polyacrylamide gel electrophoresis in our laboratory. Furthermore, compared to these alternative purification strategies, the magnetic-bead-purified transcripts gave no additional impurities detectable by our reverse transcription and sequencing readouts, which are sensitive to products with populations of 0.1% of the full-length RNA (W Kladwang and R Das, in prep.).
Because samples were prepared in parallel, using 96-well plates and multi-channel pipettors, the overall process of DNA template extension, DNA purification, RNA transcription, and RNA purification of 120 samples was efficient. The synthesis time was similar to the time required for preparing single RNA samples, approximately 1 to 2 days after receiving the starting DNA oligonucleotides from a commercial source.
Mutate-and-map data reveal visually clear signatures for sites of mutation and their partner base pairs
The second step of the mutate-and-map approach requires precise measurements of the chemical accessibility of each MedLoop RNA variant at single-nucleotide resolution. In previous papers (Das et al. 2010; Kladwang and Das 2010), we described an efficient protocol for high-throughput readout of dimethyl sulfate (DMS) modification of the Watson-Crick faces of adenine and cytosine residues (at the N1 and N3 positions, respectively). As in methods published by other labs (Mitra et al. 2008; Vasa et al. 2008; Wilkinson et al. 2008), the procedure makes use of reverse transcription by fluorescent primers and multi-capillary sequencers; we further accelerated the method through the use of 96-well plate formats and oligo(dT) magnetic-bead purification steps. In addition, to achieve a more comprehensive view of the RNA's chemical reactivity, the protocol has been extended herein to two additional modification chemistries beyond DMS alkylation (Peattie and Gilbert 1980; Tijerina et al. 2007). We probed the accessibility of Watson-Crick faces of guanosine and uracil based on modification of the N1 and N3 positions, respectively, by 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT) (see, e.g., Walczak et al. 1996). We also tested the SHAPE strategy, in which the reactivity of 2′-OH groups to N-methyl isaotic anhydride (NMIA) acylation correlates with nucleotide backbone flexibility (Wilkinson et al. 2006, 2008).
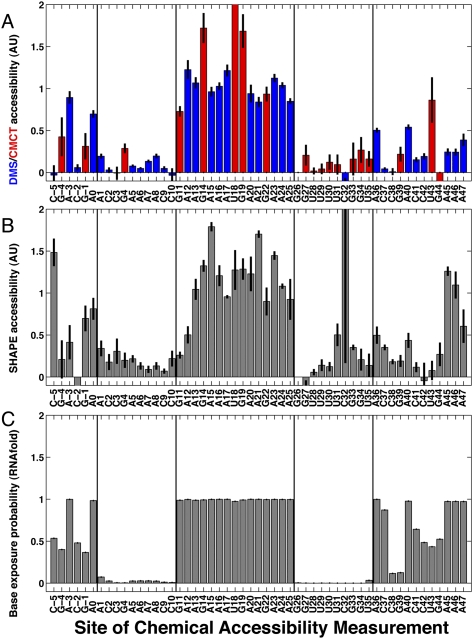
Figure 2A,B shows DMS, CMCT, and SHAPE chemical accessibilities for the starting (“wild-type”) MedLoop RNA construct without mutations. Experimental accessibilities were derived from band quantification, background subtraction, a small correction for over-modification (Vasa et al. 2008), and data averaging over 12 to 18 replicates. These data were consistent with the intended topology of the MedLoop RNA hairpin. In particular, residues 11–25 were expected to be unpaired, and these bases were, indeed, strongly modified, at levels similar or higher to the modification rates of residues outside the hairpin sequence. Residues 1–10 and 26–35 were designed to form base pairs and, as expected, show modification rates by DMS, CMCT, or NMIA that were weak or consistent with background measurements. Residues in flanking sequences (residues −9 to 0 and 36 to 50) gave higher chemical accessibilities with some modulations (see below).
FIGURE 2.
Measured and predicted accessibilities of wild-type MedLoop RNA, compared at nucleotide resolution. In all panels, vertical lines bracket the two 10-nt segments that were designed to base-pair in the MedLoop RNA. Data are quantitated from multiple independent measurements and background subtracted; error bars depict standard errors on averaged values, derived from variance within each experiment's measurements and error propagation. (A) DMS (blue at A's and C's) and CMCT (red at G's and U's) accessibilities averaged over 18 and 12 measurements, respectively; measurements were made on four different days and three independent preparations. (B) SHAPE (NMIA) accessibilities averaged over 14 measurements on three different days and two independent preparations. (C) Predicted base exposure probability from the RNAfold algorithm.
After this basic consistency check on the wild-type RNA, we measured complete mutate-and-map data sets for the MedLoop RNA. Figure 3 shows aligned DMS and CMCT electropherograms for the entire library of 120 MedLoop RNA constructs. Because the DMS and CMCT signals occur at different bases (A/C and G/U, respectively), overlaying the two signals in blue and red, respectively, allows for convenient visualization of the entire modification pattern. Supplemental Figure S1 displays the DMS and CMCT electropherograms separately, along with replicate measurements on samples independently prepared and probed by two different investigators, SHAPE electropherograms, and background measurements on unmodified samples.
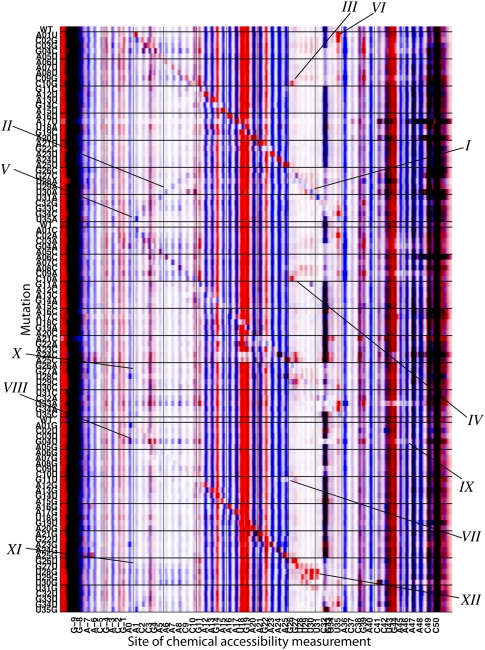
FIGURE 3.
Mutate-and-map data set for the MedLoop RNA. Chemical accessibility data for dimethyl sulfate alkylation (DMS, blue) at A's and C's and for carbodiimide modification (CMCT, red) at G's and U's are overlaid. Chemical modifications were read out by high-throughput reverse transcription with fluorescently labeled primers and capillary electrophoresis, with faster electrophoretic mobility products on the right. Raw fluorescence data (arbitrary units) are shown after automated alignment of traces and normalization to mean intensity. Ten features are marked on the data: (I) The main diagonal stripe showing localized perturbations in the U30A variant; (II–VI) punctate, off-diagonal signals corresponding to the MedLoop RNA base pairs revealed at nucleotide resolution for U30A, C10G, C10A, U35A, and A1U; (VII) protection within the 15-nt loop for G11U; (VIII) more extensive perturbations due to specific mutations, here G4U; (IX) protection of flanking regions correlated with large conformational changes in G4U; (X–XI) sites protected in experimental measurements that are predicted to be exposed by computational secondary structure prediction; (XII) unexplained exposure of multiple U's near mutation site in U28G.
Using the non-mutated MedLoop RNA sequence as a reference (“WT” in Fig. 3), several visually distinct patterns are apparent in the chemical modification profiles of the entire library of constructs. First, the mutants show chemical accessibility profiles that are qualitatively similar to the non-mutated reference, suggesting that the fold of this model system is largely robust to mutations. For example, all constructs give strong modification at residues 11–25, indicating that this loop remains unpaired as mutations are made throughout the RNA. Most of the constructs also retain strong protection at residues in the hairpin base pairs (1–10, 26–35). A few exceptions are apparent, indicating large rearrangements in secondary structure; these effects are described in the next section. In addition, there is a variable band at C32 that shows strong changes in intensity across constructs. The presence of this band in background measurements (Supplemental Fig. S1) and its weakening upon mutations of the MedLoop RNA stem suggest that it involves a reverse transcriptase stopping/pausing event mediated by stable secondary structure.
Second, as expected, clear features corresponding to local perturbations at and near the sites of mutation are apparent in the modification patterns. In Figure 3, the constructs are grouped into three libraries based on which of the three possible mutations can be made at each position; for example, the first group involves mutations of each base to its complement. Within each of the three groups, the constructs are ordered by the sequence positions of the mutation. This ordering reveals three “diagonal” stripes (labeled I in Fig. 3), corresponding to exposure of each base that has been mutated. In constructs that change an exposed base's identity from A or C to G or U (or vice versa), the pattern in Figure 3 also changes color from blue to red (or vice versa). Such effects accentuate the diagonal stripe and, in many cases, verify the desired sequence change (e.g., for U30A, labeled I in Fig. 3). In most constructs, exposure of residues neighboring in sequence is either weak or limited to immediately adjacent residues, further accentuating the diagonal feature. More “delocalized” effects are discussed in the next section.
Third, and most important, the mutate-and-map data exhibit visually clear signatures for the base pairs in the MedLoop RNA hairpin. These effects are manifest as punctate features that lie off the diagonal described above, i.e., they are due to exposure of residues caused by mutation of base-pairing partners that are distant in sequence. The most striking of these features are DMS signals at residues A5, A6, A7, and A8, exposed upon the mutations U31A, U30A, U29A, and U28A, respectively (labeled II in Fig. 3). As in our previous study on a DNA/RNA helix (Kladwang and Das 2010), the resulting internal A/A mismatches, this time within an RNA/RNA helix, appear to leave the Watson-Crick edges exposed at a level approaching half that of fully unpaired A bases. The absence of a corresponding set of SHAPE signals (Supplemental Fig. S1C) suggests that the A/A mismatch forms a well-defined noncanonical structure or set of structures rather than a generally unstructured or bulged ensemble that would be responsive to NMIA acylation.
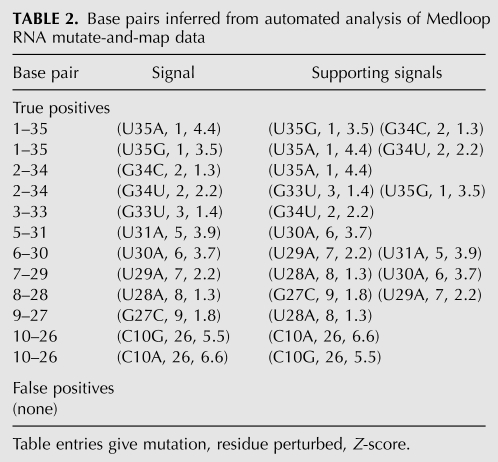
There are numerous additional sharp features corresponding to other base pairs. These include the strong reactivity of the edge base G26 to CMCT and NMIA modification upon the mutations C10G and C10A (III and IV in Fig. 3); and the “symmetric” signals for chemical modifications at A1 and U35 that arise upon mutations U35A and A1U, respectively (V and VI in Fig. 3). A more comprehensive description, based on automated signal analysis, is given below.
Rationalizing features of the mutate-and-map data from computational secondary structure prediction
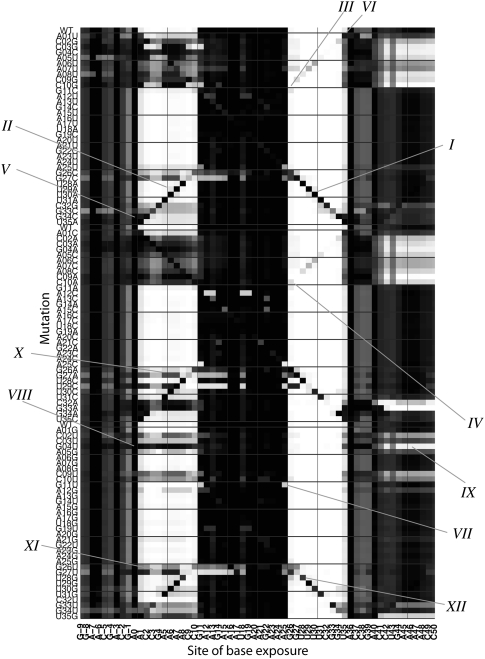
The mutate-and-map data set provides a rich source of information on how RNA structure responds to mutation. In particular, the experimental data suggest the occurrence of phenomena beyond the simple “release” of single bases upon their mutation or the mutation of their base-pairing partners. Certain mutations induced partial protections inside the long 15-residue hairpin loop (VII in Fig. 3), exposure of 10-nt strings of bases (VIII in Fig. 3), or the partial protection of sequences flanking the hairpin (IX in Fig. 3). These effects are reproducible in independent experimental replicates (Supplemental Fig. S1). We found that computational predictions of secondary structure can give a qualitative understanding of many but not all of these additional effects, which we summarize for 12 specific examples (labeled I–XII in Figs. 3, 4) in this section.
FIGURE 4.
Computational predictions for mutate-and-map features. Base exposure probabilities are shown in grayscale (white to black indicates 100% base-paired to 0% base-paired), calculated through Boltzmann ensemble enumeration by the RNAfold software for each tested mutant. To aid comparison with Figure 3, the same features I–X are marked.
For a given RNA sequence, the RNAfold module of the Vienna RNA package predicts several properties of a Boltzmann ensemble of secondary structures in addition to the minimum free energy secondary structure. To mimic our experimental data, we have carried out RNAfold calculations on each MedLoop RNA variant and plotted the predicted base accessibility in a format analogous to the mutate-and-map measurements (Figs. 2C, 4). Similar calculations with the RNAstructure and UNAfold packages are given in Supplemental Figure S2. These calculations have limitations: They are based on modeling non-pseudoknotted configurations of base pairs; the assumed energies of single mismatches and long internal loops are based on an incomplete database of thermodynamic measurements (Kierzek et al. 1999; Mathews et al. 1999; Schroeder et al. 1999; Hofacker 2004; Davis and Znosko 2007, 2008, 2010; Davis et al. 2010); and the salt conditions for the database (1 M NaCl) are different from the current experimental conditions (50 mM Na-HEPES at pH 8.0). Furthermore, uncertainties in predicting chemical modification rates from secondary structure models (as discussed in Quarrier et al. 2010) preclude a fine, quantitative comparison. Nevertheless, several features of the predicted accessibility profiles recapitulate and help rationalize patterns in the measured data.
As expected, the RNAfold minimum free energy structure of the non-mutated MedLoop RNA is predicted to exhibit the designed hairpin (Figs. 2C, 5A; cf. Fig. 1). In addition, the RNAfold calculations suggested that a diverse ensemble of base pairs between parts of the flanking sequences should provide transient protections to G −1, C37, and C38. Indeed, the DMS, CMCT, and SHAPE measurements show modulations in chemical accessibility in the flanking sequences, for example, with lower DMS accessibility of C38 relative to C41 (Fig. 2A,B). There are, however, unexplained differences, including a low experimental DMS accessibility at C37 that is not predicted by RNAfold; these discrepancies may be due to electrostatic and stacking effects that affect DMS modification rates (Tijerina et al. 2007) but are difficult to model.
FIGURE 5.
Predicted lowest free energy secondary structures by RNAfold for four MedLoop RNA variants: (A) the wild-type RNA, (B) U30A, (C) G11U, and (D) G4U. In each panel: the MedLoop RNA base-pairing regions (blue); site of mutation (red); and primer binding site (black). The figure was generated in VARNA (Darty et al. 2009).
The RNAfold calculations also recapitulate the most visually distinct signals in the mutate-and-map data. Diagonal features, marking perturbations localized to the site of mutation, are clearly visible in the simulated base accessibilities (I in Figs. 3, 4). Furthermore, the simulations recapitulate the release of single bases as their partners are mutated, including the mismatch features described above (cf. II, III, IV, V, and VI in Figs. 3, 4; see Fig. 5B for secondary structure).
Several non-trivial features of the chemical accessibility measurements were also rationalized by the calculations. Experimental data for the variant G11U showed protection of residue A25 (VII in Fig. 3), which was exposed as part of other variants' 15-nt loops. This protection is present in the simulated base accessibilities (VII in Fig. 4) and is due to an additional Watson-Crick base-pairing (Fig. 5C).
Larger-scale changes in experimental chemical accessibility of several variants involved the partial exposure of strings of several residues within the 5′ strand of the hairpin (positions 1–10). The most dramatic of these perturbations occurs upon the G4U mutation (labeled VIII in Fig. 3) but is also clearly seen in C3G, G4C, G33C, and several other variants. This exposure of the 5′ strand was typically correlated with partial protections in the 3′ flanking sequence (positions 42–48) (see, e.g., IX in Fig. 3). These large-scale changes could be rationalized by the RNAfold calculations. Each of the perturbed constructs was calculated to undergo a major conformational change in which the MedLoop RNA hairpin is disrupted: a large fraction of the RNA population is predicted to have positions 25–35 base-pair with the 3′ flanking sequence rather than positions 1–10 (Fig. 5D; VIII and IX in Fig. 4).
While the RNAfold calculations helped rationalize many features of the experimental mutate-and-map data set, we found that the agreement was not complete. Several constructs that were predicted to undergo large changes in base accessibility patterns showed no such perturbations in the experimental measurements; see, for example, all mutations at positions 26 and 27 (X and XI in Figs. 3, 4). In addition to “over-predicting” the perturbative effects of some changes, the RNAfold calculations did not provide a clear explanation for a small number of cases in which two or three residues were exposed upon mutation, such as the exposure of U29–U31 in the U28G construct (XII in Figs. 3, 4). These differences indicate that secondary structure modeling algorithms, while useful for post hoc rationalization of features, are not yet accurate enough to quantitatively predict mutate-and-map patterns.
Inferring RNA base pairs from mutate-and-map data
The task of extracting base-pair information from the resulting nucleotide-resolution data is a novel analysis challenge. In particular, the current experiment offers 10 true base pairs within the 35-nt MedLoop RNA hairpin, but there is a much larger number of potential pairs [(35 × 34)/2 − 10 = 585] for which the data could give false positives. Including the additional 35 × 25 = 875 pairings between 35 MedLoop RNA residues and 25 residues in the flanking sequence renders the problem of discriminating true base pairs even more difficult. On one hand, matching of Watson-Crick base pairs based on sequence would make this problem straightforward for the current model system. On the other hand, we desired an analysis procedure that would be generally applicable to future efforts to map non-Watson-Crick base pairs and that would also be independent of the inaccuracies and limitations of current secondary structure modeling methods. We therefore explored whether an analysis procedure could solve the problem without making use of sequence information.
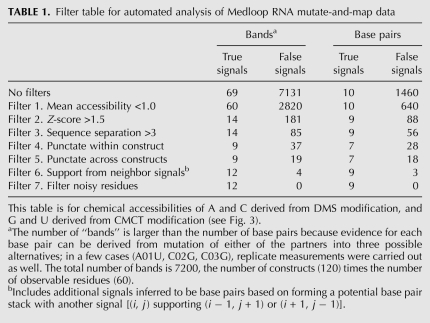
We discovered that a set of seven sequence-blind criteria, or “filters,” reproduced our visual analysis of the punctate base-pairing signals. The filters are (1) an upper bound on mean accessibility, (2) a lower bound on Z-score, (3) a minimal sequence separation, (4) a “punctate” pattern within the construct, (5) a “punctate” pattern across constructs, (6) the presence of supporting signals, and (7) a final filter for noisy residues. The effects of these progressively applied filters on the number of true-positive and false-positive signals is summarized in Table 1. A complete description of each filter, including a graphical representation of its effect on the data signals, is given in the Supplemental Material (“Description of Data Analysis Filters”; Supplemental Fig. S3). After these seven filters, the automated analysis identified nine of the 10 base pairs—each with at least one independent “supporting signal”—and 0 false positives out of 1460 possible. The final list of base pairs, Z-scores, and support information are given in Table 2. The analysis appears robust. Changing parameters (e.g., changing the Z-score cutoff for a strong signal from 1.5 to 1.0) gave identical final results. Furthermore, we applied the same automated analysis procedure to a somewhat noisier data set replicate (Supplemental Fig. S1C–D). While yielding fewer base pairs (6 of 10), this analysis of an independent replicate again gave no false positives (Supplemental Table S1).
TABLE 1.
Filter table for automated analysis of Medloop RNA mutate-and-map data
TABLE 2.
Base pairs inferred from automated analysis of Medloop RNA mutate-and-map data
DISCUSSION
Synthesizing complete single-mutant libraries for RNA
Systematic mutagenesis is a powerful approach for dissecting the biophysical and biochemical properties of macromolecular systems. For example, comprehensive alanine scanning has enabled detailed portraits of protein folding events and protein–protein interactions (see, e.g., Cunningham and Wells 1989; Weiss et al. 2000; Grantcharova et al. 2001). Such systematic mutagenesis has been rare in RNA systems but turns out to be neither difficult nor expensive with modern tools. Due to the demands of genomics, high-throughput screen assays, and general molecular biology, the cost of DNA synthesis is low and continues to decrease. Ninety-six-well magnetic bead purification methods for double-stranded nucleic acids and RNAs are commercially available. The costs of these reagents, of other PCR and transcription components, and of the time required to prepare 96 sequence variants (vs. one to two constructs) are small or comparable to present salary costs for bench scientists. Experimental interrogation of the resulting constructs, in this case of chemical accessibility profiles, can be carried out on sequencer equipment available to most labs, often in shared core facilities. Quantification of the resulting data requires computational resources and analysis skills (e.g., MATLAB) that are now available in many labs and that are accelerated by the sharing of code among labs (including herein; see Materials and Methods). We therefore propose that systematic mutagenesis studies could and should become as common in RNA biochemical analyses as they are in protein studies. We hope that the present work both demystifies the process of creating complete single-mutant libraries and demonstrates the promise of the resulting information-rich data sets.
RNA contacts revealed at nucleotide resolution by the mutate-and-map approach
The mutate-and-map strategy attempts to directly infer RNA base-pairing patterns by adding a second dimension—mutagenesis—to classic chemical mapping approaches. The power of this method is demonstrated in the expansion of information from the one dimension of Figure 2A,B to the full 2D data set of Figure 3. First, the initial 1D chemical mapping profiles show strong accessibilities to 10 loop residues (11–25) and weaker signals in residues outside this segment, but this information is not sufficient to infer the RNA's structure. The observed protections and modulations may be due to long-range base pairs, local structure (e.g., base-stacking between neighbors), low intrinsic reactivity to different chemical probes, electrostatic effects, uncertainties in signal normalization or background subtraction, or different read-through rates during reverse transcription of the RNA. These factors currently preclude a fully quantitative correlation of chemical modification rates to features of the RNA structural ensemble. Even ignoring these uncertainties, classic chemical mapping on a single RNA sequence reports on whether a residue is forming stable interactions but does not directly yield the critical information necessary for structure modeling: which other residues might be its interaction partners.
Our proposed approach attempts to uncover these interaction partners experimentally by monitoring perturbations of the chemical accessibility profiles induced by systematic single-residue mutagenesis. The resulting mutate-and-map data (Fig. 3) are experimental analogs to “contact maps” or “diagonal maps” used frequently in biomolecule structure analysis and modeling (see, e.g., Richardson 1981; Vendruscolo et al. 1997). As might be expected, the mutation of single sites leads to perturbation of chemical accessibility profiles near these sites (I in Fig. 3), corresponding to the diagonal stripes of Figure 3. The most powerful information, however, derives from features well off the diagonals, corresponding to interactions between residues distant in sequence. The MedLoop RNA was designed to contain 10 bp. Punctate off-diagonal features throughout the 2D map (e.g., II, III, IV, V, and VI) are visually clear; in combination with the diagonal stripes, these features trace out X shapes that we also expect to see as hallmarks of hairpins in data sets for other larger RNAs (W Kladwang and R Das, unpubl.). The off-diagonal features provide experimental evidence for the majority of the MedLoop RNA base pairs, and, in concert with the automated analysis described below, establish a first proof-of-concept of the mutate-and-map strategy for determining RNA base-pairing patterns.
Comparison to known structures of single RNA mismatches
Before turning to the base-pair extraction analysis, we discuss how our measurements on the MedLoop RNA system highlight both the explanatory power and limitations of our current knowledge of RNA behavior. First, the observation of the specific base-pairing signals relies on bases within single mismatches exposing their Watson-Crick edges to solvent (for DMS and CMCT accessibility) or permitting excursion of the backbone into conformations amenable to 2′-OH acylation (for SHAPE). While there is a growing literature on the structural and thermodynamic characterization of mismatches (Kierzek et al. 1999; Schroeder et al. 1999; Davis and Znosko 2007, 2010; Davis et al. 2010), we found this body of work to be only partially explanatory of our measurements. For example, the most stable non-Watson-Crick mismatch, G-G, has been extensively studied (Burkard and Turner 2000; Rypniewski et al. 2008); we expected these mismatches to form syn/anti base pairs that present Watson-Crick edges to solvent 50% of the time and permit modification by CMCT. While three such mismatches (induced by C2G, C3G, and C10G) gave CMCT signals at both G's, one did not (C32G); it is possible that steric factors due to the location of this mismatch in the interior of the helix reduced the CMCT signal.
Conversely, the most informative string of mutations (U28A, U29A, U30A, and U31A) in our mutate-and-map data sets corresponded to A/A mismatches. This was unexpected. While the solution structures of A/A mismatches have not been extensively characterized, we found a single solution structure (Richards et al. 2006) indicating the adenosine Watson-Crick edges buried within the helix, in contrast to the readily measured DMS modification rates in our measurements. Crystallographic models show alternative A/A arrangements with, e.g., Hoogsteen/sugar-edge base pairs that would explain the DMS modification rates, but the frequency of these conformations in solution remains unknown. Further understanding of chemical modification rates of mismatches would assist future mutate-and-map efforts. In lieu of fully quantitative predictions from prior structural work, we are generating an empirical table based on mutate-and-map studies of the MedLoop RNA and larger RNAs.
Comparison to computational predictions of mutant secondary structures
The full mutate-and-map data present some perturbations that are clearly different from the desired pinpointed release of a single base and its partner, and understanding the origin of these features is important for estimating the systematic errors of the mutate-and-map method. To gain insight into these features, we compared our experimental measurements to base-pairing probabilities predicted by the RNAfold algorithm for each single mutant (Figs. 4, 5). On one hand, these comparisons were helpful in qualitatively rationalizing mutation-induced protections (VII) and release of strings of bases (VIII) due to large-scale conformational rearrangements (Fig. 5A,D). On the other hand, the explanatory power of RNA secondary structure prediction is not quantitative. Several features of the data, including the robustness to mutations at positions 26 and 27 and the delocalized effects of mutations U28G and U29G, are not present in the RNAfold predictions. These inaccuracies may be due to a number of factors, including (1) the still limited database of the thermodynamic estimates of mismatch penalties; (2) the presence of non-nearest-neighbor effects in RNA structure (Mathews 2006); and (3) imprecise thermodynamic characterization of loop penalties for long loop lengths or unusual sequences (the MedLoop RNA has a highly purine-rich 15-residue loop). It will be informative to compare our data to calculations from future versions of secondary structure inference methods that implement recent experimental measurements. Nevertheless, the present disagreement underscores the need for RNA base-pair determination methods that are independent of current secondary structure modeling algorithms, motivating our sequence-blind analysis of mutate-and-map data, discussed next.
Automated inference of base pairs
General application and wide adoption of the mutate-and-map method will require an automated analysis workflow to extract residue–residue pairing signals and to assign confidence to these inferences. While the first analysis steps of band annotation and quantification are now largely automated and rapid (Das et al. 2005; Mitra et al. 2008; Vasa et al. 2008; Kladwang and Das 2010; S Yoon, J Kim, R Das, in prep.), the task of extracting base pairs from these quantitated data and discriminating them from false signals presented a novel analysis challenge. Based on considerations from our visual analysis of the data, we were able to find well-defined criteria (Table 1) that enabled the automatic detection of nine of the 10 base pairs and the complete elimination of false-positive base pairs. The analysis also succeeded on an independent, noisier replicate (Supporting Table S1). Unlike prior chemical/computational approaches to infer RNA structure, the data features that support each base pair can be explicitly delineated (Table 2) instead of relying on secondary structure prediction algorithms to fit the data. Importantly, the criteria are sequence-independent—e.g., they do not filter for solely A-U, G-C, or G-U base-pairings—and thus may be useful in future studies to identify noncanonical base pairs.
The best test of our criteria will be their application on mutate-and-map data sets on new RNAs. We expect that this automated analysis will be more challenging as the number of residues N increases; while the number of true base pairs grows as O(N), the number of false positives grows more rapidly, as O(N2). We are optimistic that this challenge can be surmounted even as N increases to the thousands of residues involved in full-length RNA messages or viral genomes. Several analysis strategies that were not used herein may be implemented as we proceed to such larger RNAs. These strategies include constraints based on the expected symmetry of the mutate-and-map signals, use of sequence information, and our accumulating knowledge of the modification rates of different mismatches. Furthermore, use of measured chemical mapping data as pseudo-energy terms in secondary structure inference is expected to be useful (see, e.g., Mathews et al. 2004; Deigan et al. 2009) but may require extension to conformational ensembles describing multiple sequences. We are making freely available the quantitated band intensities (see Materials and Methods) to encourage other groups to revise and innovate computational analyses of mutate-and-map data.
Prospect of applying the mutate-and-map strategy to larger RNAs
Building on our prior proof-of-concept on a DNA/RNA helix, we have presented the first demonstration of a mutate-and-map strategy for RNA base-pair inference, using a 35-nt hairpin within an 80-nt model RNA system. We have found this 2D extension of chemical mapping to be systematic and accurate. An automated analysis infers the majority (nine of 10) of the RNA's designed base pairs, gives direct experimental support for each interaction without reliance on models of pairing energetics or phylogenetic analysis, and discriminates true signals from large-scale rearrangements with no false positives. The success of the mutate-and-map method on this model RNA raises the prospect of rapid and confident base-pair determination for structured and partially structured RNAs that are difficult for or intractable to conventional structural approaches. High-throughput mutagenesis and chemical mapping of RNA sequences up to several hundred nucleotides in length appear feasible with our current protocols without modification. Such data may reveal base interactions beyond Watson-Crick pairs such as long-range tertiary contacts. The experiments could also be accelerated by generating single rather than all three mutants per position or by using one rather than three chemical mapping strategies. To evaluate these prospects, we are currently carrying out extensive tests of the mutate-and-map strategy on several riboswitches, ribozymes, and other non-coding RNAs with known and unknown tertiary folds.
MATERIALS AND METHODS
Preparation of DNA templates
The DNA templates for the MedLoop RNA and desired variants included the 20-nt T7 RNA polymerase promoter sequence (TTCTAATACGACTCACTATA) followed by the desired sequence. Double-stranded templates were prepared by extension of 60-nt DNA oligomers (IDT, Integrated DNA Technologies) with Phusion DNA polymerase (Finnzymes), using the following thermocycler protocol: denaturation for 2 min at 98°C, ramp to 64°C at 1°C/sec; annealing for 1 min at 64°C; extension for 10 min at 72°C; and cooling to 4°C. DNA samples were purified with AMPure magnetic beads (Agencourt, Beckman Coulter) following manufacturer's instructions. Sample concentrations were estimated based on UV absorbance at 260 nm measured on Nanodrop 100 or 8000 spectrophotometers. Verification of template length was accomplished by electrophoresis of all samples and 10-bp and 20-bp ladder length standards (Fermentas) in 4% agarose gels (containing 0.5 μg/mL ethidium bromide) and 1× TBE (100 mM Tris, 83 mM boric acid, 1 mM disodium EDTA). All sample manipulations, including following steps, were carried out in 96-well V-shaped polypropylene microplates (Greiner).
Preparation of RNA templates
In vitro transcription reactions were carried out in 40-μL volumes with 10 pmol of DNA template; 20 units of T7 RNA polymerase (New England Biolabs); 40 mM Tris-HCl (pH 8.1); 25 mM MgCl2; 2 μM spermidine; 1 mM each ATP, CTP, GTP, and UTP; 4% polyethylene glycol 1200; and 0.01% Triton X-100. Reactions were incubated for 4 h at 37°C. Transcriptions were monitored by electrophoresis of all samples along with 100–1000-nt RNA length standards (RiboRuler; Fermentas) in 4% denaturing agarose gels (1.1% formaldehyde; run in 1× TAE, 40 mM Tris, 20 mM acetic acid, 1 mM disodium EDTA), stained with SYBR Green II RNA gel stain (Invitrogen) following manufacturer's instructions. RNA samples were purified with MagMax magnetic beads (Ambion), following manufacturer's instructions; 11 mL of Lysis/Binding Solution Concentrate was supplemented with 20 mL of isopropanol, based on manufacturer's recommendations to enhance binding of small RNAs. Concentrations were measured by absorbance at 260 nm on Nanodrop 100 or 8000 spectrophotometers.
High-throughput chemical accessibility measurements
Chemical modification reactions consisted of 1.2 pmol of RNA, 66.6 mM Na-HEPES (pH 8.0) in 15-μL volumes. After incubation for 10 min at 24°C, 5 μL of modification reagent was added into the RNA mixture. There were three types of modification reagents: (1) dimethyl sulfate (DMS), freshly diluted 1 to 10 into ethanol, and again 1 to 10 into water; (2) 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT; 42 mg/mL) freshly prepared from solid stock into water; and (3) 24 mg/mL N-methyl isatoic anhydride (NMIA, for the SHAPE reaction) in anhydrous DMSO. The reactions were incubated for 15 min (DMS and CMCT) or 60 min (SHAPE) at 24°C. In control reactions (for background measurements), 5 μL of deionized water was added instead of modification reagent, and incubated for 60 min.
Different quench solutions were used for the three modification reaction types. DMS reactions were quenched with a premixed solution of 5 μL of 2-mercaptoethanol and the following components to allow for rapid purification: 3 μL of 5 M NaCl, 1.5 μL of oligo(dT) beads [poly(A) purist; Ambion], 0.25 μL of 0.5 μM 5′-rhodamine-green labeled primer (AAAAAAAAAAAAAAAAAAAAGTTGTTGTTGTTGTTTCTTT) complementary to the 3′ end of the MedLoop RNA (also used in our previous study; Kladwang and Das 2010), and 0.05 μL of a 0.5 μM Alexa-555-labeled oligonucleotide (used to verify normalization). CMCT and SHAPE (and control) reactions were quenched with the same premixed solution with 5 μL of 0.5 M Na-MES (pH 6.0), substituted for 2-mercaptoethanol. The reactions were purified by magnetic separation, rinsed with 40 μL of 70% ethanol twice, and allowed to air-dry for 10 min while remaining on a 96-post magnetic stand. The magnetic-bead mixtures were resuspended in 2.5 μL of deionized water.
The resulting mixtures of modified RNAs and primers bound to magnetic beads were reverse-transcribed by the addition of a pre-mixed solution containing 0.2 μL of SuperScript III (Invitrogen), 1.0 μL of 5× SuperScript First Strand buffer (Invitrogen), 0.4 μL of 10 mM each dnTP (dATP, dCTP, dTTP, and dITP) (Mills and Kramer 1979), 0.25 μL of 0.1 M DTT, and 0.6 μL of water. The reactions (5 μL total) were incubated for 30 min at 42°C. RNA was degraded by the addition of 5 μL of 0.4 M NaOH and incubation for 3 min at 90°C. The solutions were neutralized by the addition of 5 μL of an acid quench (2 volumes of 5 M NaCl, 2 volumes of 2 M HCl, and 3 volumes of 3 M Na-acetate). Fluorescent DNA products were purified by magnetic bead separation, rinsed with 40 μL of 70% ethanol, and air-dried for 5 min. The reverse transcription products, along with magnetic beads, were resuspended in 10 μL of a solution containing 0.125 mM Na-EDTA (pH 8.0) and a Texas-Red-labeled reference ladder (whose fluorescence is spectrally separated from the rhodamine-green-labeled products). The products were separated by capillary electrophoresis on an ABI3100 DNA sequencer. Reference ladders were created using an analogous protocol without chemical modification and the addition of, e.g., 2′-3′-dideoxy-TTP in an amount equimolar to dTTP in the reverse transcriptase reaction.
Specialized versions of the SAFA analysis scripts (Das et al. 2005; S Yoon, J Kim, R Das, in prep.) were used to analyze the ABI data. Traces were aligned by automatically shifting and scaling the time coordinate, based on cross-correlation of the Texas Red reference ladder co-loaded with all samples. Sequence assignments to bands, verified by comparison to sequencing ladders, permitted the automated peak-fitting of the traces to Gaussians. Further automated analysis of quantitated band intensities, as described in the Results section, was carried out in MATLAB.
Availability of data and code
Both the quantitated band data and MATLAB analysis scripts are being made freely available at the authors' website: http://www.stanford.edu/∼rhiju/data.html.
Prediction of RNA secondary structures and mean base accessibilities
The pf_fold() routine of the ViennaRNA package (version 1.8.4; equivalent to the “RNAfold -p” command-line) (Hofacker 2004) was used for predicting the statistical mechanics of base-pairing probabilities of the probed RNA sequences. Calculations were facilitated through Python bindings available through the software's convenient SWIG (Simplified Wrapper and Interface Generator) interface. Base-pairing probabilities were computed by summing the pairwise probability matrix pr for each residue. Additional calculations were carried out in both RNAstructure (Mathews and Turner 2006) and UNAfold (Mathews et al. 1999) and gave similar results (see Supplemental Fig. S2). These algorithms do not yet predict thermodynamics at salt concentrations lower than 1 M NaCl, but our experimental measurements were carried out in 50 mM Na-HEPES (pH 8.0). The lower salt concentration is expected to be destabilizing for RNA, and to estimate this systematic error, we repeated calculations at temperatures of 24°C, 37°C, and 50°C. While the predicted patterns were qualitatively similar at different temperatures, predictions at higher temperatures reduced the contrast between protected and exposed residues and agreed best with experimental measurements; therefore, measurements presented in the main text assumed a simulation temperature of 50°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Das lab for comments on the manuscript. Computational work was supported by a Burroughs-Wellcome Foundation Career Award at the Scientific Interface (to R.D.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2516311.
REFERENCES
- Adilakshmi T, Lease RA, Woodson SA 2006. Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic Acids Res 34: e64 doi: 10.1093/nar/gkl291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS 2008. The eukaryotic genome as an RNA machine. Science 319: 1787–1789 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard ME, Turner DH 2000. NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry 39: 11748–11762 [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM 1985. Probing the three-dimensional structures of DNA and RNA oligonucleotides in solution by nuclear Overhauser enhancement measurements. FEBS Lett 179: 187–198 [DOI] [PubMed] [Google Scholar]
- Cruz JA, Westhof E 2009. The dynamic landscapes of RNA architecture. Cell 136: 604–609 [DOI] [PubMed] [Google Scholar]
- Culver GM, Noller HF 2000. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol 318: 446–460 [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA 1989. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244: 1081–1085 [DOI] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB 2005. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Kudaravalli M, Jonikas M, Laederach A, Fong R, Schwans JP, Baker D, Piccirilli JA, Altman RB, Herschlag D 2008. Structural inference of native and partially folded RNA by high-throughput contact mapping. Proc Natl Acad Sci 105: 4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Karanicolas J, Baker D 2010. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods 7: 291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Znosko BM 2007. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry 46: 13425–13436 [DOI] [PubMed] [Google Scholar]
- Davis AR, Znosko BM 2008. Thermodynamic characterization of naturally occurring RNA single mismatches with G-U nearest neighbors. Biochemistry 47: 10178–10187 [DOI] [PubMed] [Google Scholar]
- Davis AR, Znosko BM 2010. Positional and neighboring base pair effects on the thermodynamic stability of RNA single mismatches. Biochemistry 49: 8669–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Kirkpatrick CC, Znosko BM 2010. Structural characterization of naturally occurring RNA single mismatches. Nucleic Acids Res. doi: 10.1093/nar/gkq793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CD, Weeks KM 2008. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry 47: 8504–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego J, Varani G 2001. Targeting RNA with small-molecule drugs: Therapeutic promise and chemical challenges. Acc Chem Res 34: 836–843 [DOI] [PubMed] [Google Scholar]
- Gesteland RF, Cech TR, Atkins JF, eds. 2006. The RNA world, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Gohlke C, Murchie AI, Lilley DM, Clegg RM 1994. Kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc Natl Acad Sci 91: 11660–11664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantcharova V, Alm EJ, Baker D, Horwich AL 2001. Mechanisms of protein folding. Curr Opin Struct Biol 11: 70–82 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Power A, Hertz GZ, Putz EJ, Stormo GD 1992. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res 20: 5785–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Dervan PB 1994. Visualization of RNA tertiary structure by RNA-EDTA·Fe(II) autocleavage: Analysis of tRNAPhe with uridine-EDTA·Fe(II) at position 47. Proc Natl Acad Sci 91: 4955–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann RK, Bindereif A, Schön A, Westhof E, eds. 2005. Handbook of RNA biochemistry. Wiley-VCH, Morlenbach, Germany [Google Scholar]
- Hofacker IL 2004. RNA secondary structure analysis using the Vienna RNA package. Curr Protoc Bioinformatics Chapter 12: Unit 12.2. [DOI] [PubMed] [Google Scholar]
- Joseph S, Whirl ML, Kondo D, Noller HF, Altman RB 2000. Calculation of the relative geometry of tRNAs in the ribosome from directed hydroxyl-radical probing data. RNA 6: 220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek R, Burkard ME, Turner DH 1999. Thermodynamics of single mismatches in RNA duplexes. Biochemistry 38: 14214–14223 [DOI] [PubMed] [Google Scholar]
- Kladwang W, Das R 2010. A mutate-and-map strategy for inferring base pairs in structured nucleic acids: Proof of concept on a DNA/RNA helix. Biochemistry 49: 7414–7416 [DOI] [PubMed] [Google Scholar]
- Krol A, Carbon P 1989. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol 180: 212–227 [DOI] [PubMed] [Google Scholar]
- Lancaster L, Kiel MC, Kaji A, Noller HF 2002. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell 111: 129–140 [DOI] [PubMed] [Google Scholar]
- Lehnert V, Jaeger L, Michel F, Westhof E 1996. New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: A complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol 3: 993–1009 [DOI] [PubMed] [Google Scholar]
- Levitt M 1969. Detailed molecular model for transfer ribonucleic acid. Nature 224: 759–763 [DOI] [PubMed] [Google Scholar]
- Mathew-Fenn RS, Das R, Silverman JA, Walker PA, Harbury PA 2008. A molecular ruler for measuring quantitative distance distributions. PLoS ONE 3: e3229 doi: 10.1371/journal.pone.0003229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH 2006. Revolutions in RNA secondary structure prediction. J Mol Biol 359: 526–532 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Turner DH 2006. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol 16: 270–278 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci 101: 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryman C, Moazed D, McWhirter J, Noller HF 1999. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J Mol Biol 285: 97–105 [DOI] [PubMed] [Google Scholar]
- Mills DR, Kramer FR 1979. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci 76: 2232–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A 2008. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res 36: e63 doi: 10.1093/nar/gkn267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF 2005. RNA structure: Reading the ribosome. Science 309: 1508–1514 [DOI] [PubMed] [Google Scholar]
- Peattie DA, Gilbert W 1980. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci 77: 4679–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443: 167–172 [DOI] [PubMed] [Google Scholar]
- Pyle AM, Murphy FL, Cech TR 1992. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature 358: 123–128 [DOI] [PubMed] [Google Scholar]
- Quarrier S, Martin JS, Davis-Neulander L, Beauregard A, Laederach A 2010. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA 16: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V 2008. What we have learned from ribosome structures. Biochem Soc Trans 36: 567–574 [DOI] [PubMed] [Google Scholar]
- Richards RJ, Wu H, Trantirek L, O'Connor CM, Collins K, Feigon J 2006. Structural study of elements of Tetrahymena telomerase RNA stem–loop IV domain important for function. RNA 12: 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS 1981. The anatomy and taxonomy of protein structure. Adv Protein Chem 34: 167–339 [DOI] [PubMed] [Google Scholar]
- Rypniewski W, Adamiak DA, Milecki J, Adamiak RW 2008. Noncanonical G(syn)–G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex. RNA 14: 1845–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, Uhlenbeck OC 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci 85: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SJ, Burkard ME, Turner DH 1999. The energetics of small internal loops in RNA. Biopolymers 52: 157–167 [DOI] [PubMed] [Google Scholar]
- Schroeder R, Grossberger R, Pichler A, Waldsich C 2002. RNA folding in vivo. Curr Opin Struct Biol 12: 296–300 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Szewczak AA, Ortoleva-Donnelly L, Ryder SP, Moncoeur E, Strobel SA 1998. A minor groove RNA triple helix within the catalytic core of a group I intron. Nat Struct Biol 5: 1037–1042 [DOI] [PubMed] [Google Scholar]
- Tijerina P, Mohr S, Russell R 2007. DMS footprinting of structured RNAs and RNA–protein complexes. Nat Protoc 2: 2608–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius TD 1991. DNA footprinting with the hydroxyl radical. Free Radic Res Commun 12–13: 521–529 [DOI] [PubMed] [Google Scholar]
- Tzakos AG, Grace CR, Lukavsky PJ, Riek R 2006. NMR techniques for very large proteins and rnas in solution. Annu Rev Biophys Biomol Struct 35: 319–342 [DOI] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC 2008. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M, Kussell E, Domany E 1997. Recovery of protein structure from contact maps. Fold Des 2: 295–306 [DOI] [PubMed] [Google Scholar]
- Walczak R, Westhof E, Carbon P, Krol A 1996. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA 2: 367–379 [PMC free article] [PubMed] [Google Scholar]
- Waldsich C 2008. Dissecting RNA folding by nucleotide analog interference mapping (NAIM). Nat Protoc 3: 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, Weeks KM 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS 2000. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci 97: 8950–8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96 doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich K 2003. NMR studies of structure and function of biological macromolecules (Nobel lecture). Angew Chem Int Ed Engl 42: 3340–3363 [DOI] [PubMed] [Google Scholar]