Abstract
The degradation of messenger RNA is a critical node of gene regulation. A major pathway of mRNA decay is initiated by shortening of the poly(A) tail, followed by removal of the 5′ cap structure (decapping) and subsequent degradation. Decapping is an important determinate in the destruction of many transcripts. Detailed kinetic analysis of in vivo decapping rates is necessary to understand how this step is regulated. Importantly, the product of decapping is recalcitrant for investigation, in part due to its transient nature. As such, little in vivo kinetic information is available. Here we report the development of an assay that measures decapping of mRNAs by combining splinted ligation and quantitative RT-PCR (qSL-RT-PCR). We apply this method to determine the decapping rate constant for a natural mRNA in vivo for the first time. The qSL-RT-PCR assay may be adapted for use on any mRNA, providing a new tool to study regulation of mRNA decay.
Keywords: decapping, mRNA stability, mRNA degradation, quantitative RT-PCR, splint-ligation, enzyme kinetics
INTRODUCTION
Eukaryotic mRNAs are capped by a 7-methyl guanosine linked to the mRNA by a 5′ to 5′ triphosphate—the 5′ cap. This structure promotes translation and protects mRNAs from degradation by exoribonucleases (Furuichi et al. 1977; Green et al. 1983; Schwer et al. 1998). Enzymatic removal of the cap, referred to as decapping, results in reduced translation and rapid degradation of the mRNA (Coller and Parker 2004).
mRNAs are normally degraded by two competing pathways (Parker and Song 2004; Garneau et al. 2007). In the 5′ pathway, decapping follows deadenylation and is catalyzed by the Dcp2 enzyme to produce a 7-methyl-GDP product (Fig. 1; Dunckley and Parker 1999; Wang et al. 2002; Steiger et al. 2003). The remaining mRNA, with a 5′ monophosphate, is rapidly destroyed in a 5′ to 3′ direction by the exoribonuclease Xrn1 (Fig. 1; Hsu and Stevens 1993; Muhlrad et al. 1994, 1995). In the 3′ decay pathway, deadenylation is followed by exonucleolytic degradation from the 3′ end (Muhlrad et al. 1995; Anderson and Parker 1998; Brown et al. 2000; Araki et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002). The DcpS “scavenger” decapping enzyme degrades the remaining capped species, yielding 7-methyl-GMP product (Liu et al. 2002, 2004).
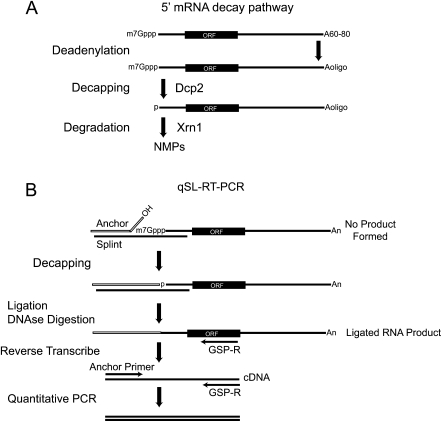
FIGURE 1.
The 5′ mRNA decay pathway. (A) mRNAs possess a 5′ 7-methyl guanosine cap (7mGppp), open reading frame (ORF) and a 3′ poly-adenosine tail, averaging 60–80 nt in yeast. mRNA decay via the 5′ pathway initiates with deadenylation of the poly-Adenosine tail to a short oligo-adenylated form (Aoligo). Dcp2 decapping enzyme subsequently removes the cap. The decapped mRNA, with a 5′ monophosphate, is destroyed by Xrn1, producing monophosphorylated nucleotides (NMP). (B) Schematic of the quantitative splinted-ligation reverse transcriptase polymerase chain reaction assay (qSL-RT-PCR). First, Anchor RNA and complementary DNA Splint oligonucleotides are annealed to the 5′ end of the target mRNA. mRNAs containing a 5′ cap cannot be ligated, as the cap prevents ligation. Decapped RNAs have a 5′ monophosphate that is ligated to the Anchor RNA 3′ hydroxyl by T4 DNA ligase. After ligation, the splint is destroyed by DNase I. The ligated RNA is converted to cDNA by reverse transcription with a reverse gene-specific primer (GSP-R). The resulting cDNA is then detected by quantitative PCR using GSP-R and a forward primer that anneals to the anchor (Anchor Primer). An internal control qPCR is performed on the same cDNA samples using gene-specific primers that amplify within the coding sequence of the mRNA.
Decapping is highly regulated (Franks and Lykke-Andersen 2008). Trans-acting proteins modulate decapping activity. Decapping of specific mRNAs is influenced by cis-acting sequences and the sequence-specific RNA-binding factors that recognize them. Small RNAs also affect decapping (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006).
Assays that measure the decapped products are essential to study regulation of decapping. The ideal assay should be sensitive, rapid, reproducible, and have a broad linear range. The reagents should be readily available, and the assay should be feasible for any mRNA. In this report, we describe a new quantitative assay for detecting decapped mRNAs. First, discussion of previous assays is informative.
Detection of decapped mRNA produced in vivo has been a longstanding challenge. Several methods have been utilized, including: selective binding to anticap antibody (Muhlrad et al. 1994, 1995; Beelman et al. 1996; Dunckley and Parker 1999; He and Jacobson 2001), selective degradation by Xrn1 (Hsu and Stevens 1993; Fischer and Weis 2002), and by trapping mRNA decay intermediates with a strong secondary structure (Decker and Parker 1993; Poole and Stevens 1997). Primer extension has been used to detect capped and uncapped RNA (Hsu and Stevens 1993; Muhlrad et al. 1995; Schwer et al. 1998; Coller et al. 2001; Hu et al. 2009, 2010). Ligation mediated RT-PCR was also used (Fromont-Racine et al. 1993; Couttet et al. 1997; Couttet and Grange 2004). Recently, a modified ligation method was developed to detect decapped mRNAs by RT-PCR (Hu et al. 2009, 2010). A bridging DNA oligonucleotide—a “splint”—mediates specific and efficient ligation of a 5′ anchor RNA oligonucleotide to the mRNA of interest. This splinted ligation method has been used to create hybrid RNAs in vitro (Moore and Query 2000) and for detection of small RNAs (Maroney et al. 2007, 2008) and 5′ ends of bacterial mRNAs (Celesnik et al. 2007, 2008).
In this report, we advance the splinted ligation method to create a quantitative decapping assay with broad linear response and sensitivity, hereon referred to as quantitative splinted ligation reverse transcription PCR assay, or qSL-RT-PCR. This method measures decapped mRNA levels with a detection range spanning at least four orders of magnitude from as little as 1.5 ng of total cellular RNA. We then apply qSL-RT-PCR to measure the in vivo decapping rate of the RPL41A mRNA.
RESULTS
The qSL-RT-PCR assay specifically detects and quantitates decapped mRNA
With the goal of measuring decapped mRNA, we coupled splinted ligation with reverse transcription and quantitative polymerase chain reaction. The RPL41A mRNA from Saccharomyces cerevisiae (encoded by YDL184C) was used because the 5′ end is mapped and the mRNA is abundant (50 copies per cell) (Yu and Warner 2001). RPL41A coding sequence is closely related to RPL41B, yet these mRNAs differ substantially in their UTR sequences, making it possible to develop a detection strategy specific for RPL41A (Yu and Warner 2001). An overview of the qSL-RT-PCR assay is shown in Figure 1B. First, an RNA Anchor oligonucleotide is ligated to the 5′ end of the mRNA of interest. The 5′ half of the DNA splint specifically anneals to the 5′ end of the mRNA of interest, while the 3′ half anneals to an Anchor RNA oligonucleotide. DNA ligase joins the 3′ hydroxyl of the RNA anchor to the 5′ monophosphate of the decapped mRNA. Following ligation, the DNA splint is destroyed by DNAse 1. Next, the ligated RNA is purified and analyzed by quantitative RT-PCR.
To begin the qSL-RT-PCR assay, total RNA was purified from yeast cells lacking the XRN1 gene (Fig. 1A). This genetic background stabilizes the decapped mRNA to permit efficient detection (Muhlrad et al. 1995). qSL-RT-PCR was performed to measure the cycle threshold (Ct) and melting temperature of the amplified product. Reactions were separated by agarose gel electrophoresis to detect the expected product of 224 bp (Fig. 2A, lane 1). As an internal control, the RPL41A was detected using standard qRT-PCR with specific primers to amply an internal 158-bp product (Fig. 2A, lane 2).
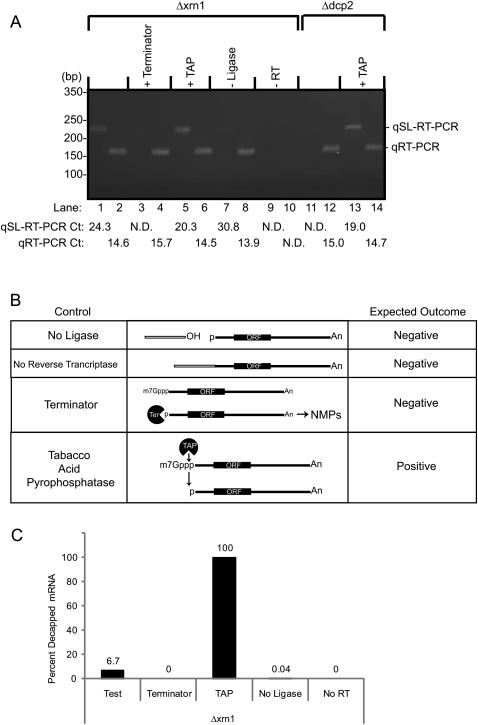
FIGURE 2.
The qSL-RT-PCR assay specifically detects decapped mRNA. (A) qSL-RT-PCR was used to detect endogenous decapped RPL41A mRNA present in 10 μg of total RNA isolated from yeast strains lacking either the XRN1 (Δxrn1) or the DCP2 (Δdcp2) genes (odd-numbered lanes). Standard qRT-PCR was used to detect total RPL41A RNA (even-numbered lanes). In control reactions, total RNA was treated as indicated at the top of the gel, including Terminator (lanes 3,4) or Tobacco Acid Pyrophosphatase (+TAP) treatment (lanes 5,6,13,14). In lanes 7 and 8, DNA ligase was omitted (−Ligase). Reverse transcriptase was omitted from reactions in lanes 9 and 10 (–RT). Cycle thresholds measured for each sample are indicated at the bottom of the figure. Reactions that did not yield a detectable cycle threshold are labeled “N.D.” (Not Detected). (B) Critical controls for the qSL-RT-PCR assay demonstrate specificity for decapped mRNA. Control reactions are indicated on the left. In the middle, a diagram of each control is depicted. On the right, the expected outcome of qSL-RT-PCR is indicated for each control reaction. (Terminator) The Terminator enzyme specifically destroys uncapped mRNA with a 5′ monophosphate but does not degrade capped mRNA, thereby demonstrating specificity of the assay for decapped RNA. Tobacco Acid Phosphatase removes 5′ cap, leaving a 5′ monophosphate that is detected by the qSL-RT-PCR assay, thereby serving as a positive control. (No ligase) Omission of DNA ligase prevents ligation of anchor to mRNA, thereby preventing amplification of product. (No Reverse Transcriptase) In the absence of reverse transcriptase, no product should be generated, thereby demonstrating dependence on cDNA conversion of mRNA. (C) Relative amounts of decapped RPL41A mRNA in A were determined for the indicated reactions from the strain lacking Xrn1. Calculations are described in the Materials and Methods.
Control reactions are necessary to demonstrate specificity of the assay (Fig. 2B). First, RNA was treated with the exoribonuclease, Terminator, which selectively degrades uncapped mRNA, but not capped mRNA. Terminator abolished detection of decapped mRNA (Fig. 2A, lane 3). As a positive control, mRNA was decapped with Tobacco Acid Pyrophosphatase (TAP), which increased the amount of decapped RPL41A mRNA detected (Fig. 2A, lane 5). Total RPL41A was detected by qRT-PCR in these samples (Fig. 2A, lanes 4,6). We note that TAP treatment did not decrease the Ct of qSL-RT-PCR to equivalent value of qRT-PCR. Several considerations account for this: First, the amplicons and forward primer differ, and therefore parameters of reverse transcription and quantitative PCR, differ. Second, the qSL-RT-PCR signal depends on the efficiency of Anchor ligation and TAP. Importantly, the TAP control demonstrates that qSL-RT-PCR detects an increase in decapped RPL41A mRNA in Δxrn1 cells (Fig. 2A, cf. lanes 1 and 5) and Δdcp2 cells (Fig. 2A, cf. lanes 11 and 13).
Ligation of the anchor RNA to the decapped mRNA is requisite for detection by qSL-RT-PCR (Fig. 2B). Therefore, omission of DNA ligase is an important negative control that resulted in loss of signal from the qSL-RT-PCR assay (Fig. 2A, lane 7), but does not affect the internal qRT-PCR (Fig. 2A, lane 8). This control is crucial to exclude artifactual amplification by residual DNA splint. To control for DNA contamination, mock reverse transcription reactions were performed wherein reverse transcriptase is omitted (Fig. 2B). As expected, no product was detected by either assay (Fig. 2A, lanes 9,10).
To further validate the detection of decapped mRNA, RNA was purified from cells lacking decapping enzyme, Dcp2 (Wang et al. 2002; Parker and Song 2004). In this strain, no decapped RPL41A was detected by qSL-RT-PCR (Fig. 2A, lane 11), yet RPL41A mRNA was present (Fig. 2A, lane 12). This result demonstrates that the decapped RPL41A is generated by Dcp2. When this RNA sample was treated with TAP, decapped RNA was readily detected by qSL-RT-PCR (Fig. 2A, lane 13). We conclude that the qSL-RT-PCR specifically detects decapped mRNA.
To compare the amounts of decapped mRNAs, we used the comparative Ct method (Livak and Schmittgen 2001; Schmittgen and Livak 2008). Ct values (Fig. 2A) from the qSL-RT-PCR assay were normalized to the total amount of RPL41A using Ct values from qRT-PCR for that sample. The amount of decapped RPL41A was then calculated relative to the TAP-treated sample (Fig. 2A, lanes 5,6), which was set to 100% for detected decapped RPL41A mRNA (Fig. 2C). In the test sample, 6.7% of the RPL41A mRNA was decapped and detectable by qSL-RT-PCR. Terminator treatment completely destroyed decapped mRNA. When DNA ligase was omitted, the relative amount of decapped mRNA was 0.04%, or 147-fold below the test sample; thus, background signal was low. Taken together, we conclude that the qSL-RT-PCR assay is highly specific for decapped mRNA and can measure differences in the amount of decapped mRNA.
The qSL-RT-PCR assay has a broad linear detection range and high sensitivity
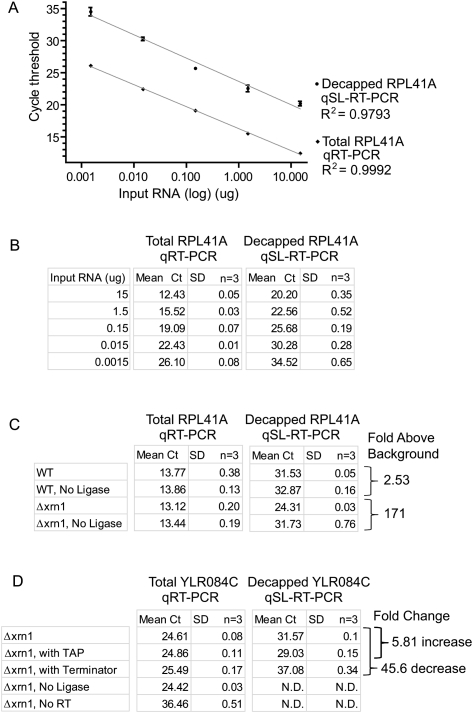
To determine the detection range and sensitivity of the qSL-RT-PCR, the amount of input RNA was varied. Total RNA was purified from yeast cells (Δxrn1) and a series of 10-fold dilutions were made. Three replicate dilution series were made to assess variability. qSL-RT-PCR was performed on each sample, and the resulting mean Ct values were plotted against the amount of input RNA (Fig. 3A). Both decapped and total RPL41A mRNA were detected over the entire range of RNA concentrations. Signal of the decapped mRNA was proportional to the input RNA. Linearity of the response was excellent: Nonlinear regression analysis yielded correlation coefficients of R2 = 0.9793 for qSL-RT-PCR and R2 = 0.9992 for qRT-PCR (Fig. 3A). Strikingly, decapped RPL41A was measured in as little as 1.5 ng of input RNA. These results were reproducible, as shown in Figure 3A, in which plots for the mean Ct values for three replicates are graphed with standard deviations for each measurement. Because standard deviations are small, the values are also listed in Figure 3B. These results demonstrate that the qSL-RT-PCR assay provides a sensitive means to measure decapped mRNA with a broad, linear detection range and excellent reproducibility.
FIGURE 3.
The qSL-RT-PCR assay has a broad, linear dynamic range for sensitive detection of decapped RPL41A RNA. (A) Five 10-fold serial dilutions of total RNA from Δxrn1 cells were analyzed using qSL-RT-PCR and qRT-PCR assays to detect decapped and total RPL41A mRNA, respectively. Triplicate samples were analyzed and the mean cycle threshold (Ct) values are plotted against input RNA amount (15, 1.5, 0.15, 0.015, and 0.0015 μg) on a logarithmic scale. Standard deviation is indicated above and below each data point. (B) Mean Ct values and standard deviations (SD) from A are shown in the table. Nonlinear regression analysis was used to determine correlation coefficients (R2) for each curve. (C) qRT-PCR and qSL-RT-PCR assays were performed on 10 μg of total RNA to measure RPL41A mRNA in wild-type BY4742 cells (WT) and Δxrn1. Mean Ct values and standard deviations are indicated as determined from triplicate samples. Fold increase above background was calculated relative to control reactions lacking T4 DNA ligase for each strain, determined from ΔCt of qSL-RT-PCR. (D) qRT-PCR and qSL-RT-PCR assays were performed on 7.5 μg of total RNA to measure YLR084C mRNA in Δxrn1 cells. Control samples were treated with TAP or Terminator. T4 DNA ligase or Reverse Transcriptase were omitted from control reactions as indicated. Mean Ct values and standard deviations are indicated and were determined from triplicate samples. Fold change in decapped mRNA level was calculated from the ΔCt of qSL-RT-PCR values. Reactions that did not yield a detectable cycle threshold are labeled “N.D.” (Not Detected).
Based on the observed sensitivity, we tested the ability of qSL-RT-PCR to detect decapped RPL41A mRNA in wild-type cells. Equal mass of RNA (10 μg), purified from wild-type or Δxrn1 yeast cells, was analyzed by qRT-PCR and qSL-RT-PCR. The mean Ct values were determined for each assay from three replicates (Fig. 3C). As a control for false-positive background signal, DNA ligase was ommitted from control reactions. The amount of RPL41A mRNA in each strain was measured by qRT-PCR (Fig. 3C). In Δxrn1 cells, decapped mRNA was detected 171-fold above background (ΔCt = 7.42, P = 0.000074), whereas in wild-type cells decapped RNA was 2.53-fold above background (ΔCt = 1.43, P = 0.00018). Therefore, decapped RPL41A mRNA was detectable in wild-type cells, though the signal is close to background under these conditions. It is noteworthy that Hu and coworkers detected decapped mRNA in wild-type cells using splinted ligation and RT-PCR (Hu et al. 2009, 2010). As expected, decapped mRNA is stabilized by deletion of XRN1 and, therefore, is readily detectable above background.
To further demonstrate the sensitivity of qSL-RT-PCR, we tested its ability to detect a low abundance, cell cycle-regulated mRNA encoded by the YLR084C gene. YLR084C mRNA was measured by qRT-PCR from replicate RNA purifications from the Δxrn1 strain and detected at a mean Ct of 24.61 (Fig. 3D), which is ∼1000-fold below that of RPL41A (Mean Ct ∼14) Fig. 2A,C). To detect decapped YLR084C mRNA, we first mapped the 5′ end of this mRNA using 5′ rapid amplification of cDNA ends. A specific splint DNA oligonucleotide was created and used in the qSL-RT-PCR assay to detect decapped YLR084C mRNA (Mean Ct = 31.57) (Fig. 3D). Treatment of the RNA with TAP increased the decapped YLR084C by 5.81-fold, whereas Terminator treatment decreased it by 45.6-fold (Fig. 3D). Omission of ligase completely abolished detection of decapped YLR084C mRNA but had no effect on detection by qRT-PCR (Fig. 3D). Omission of reverse transcriptase resulted in significant loss of detection of both total and decapped YLR084C mRNA (Fig. 3D). Taken together, the results presented here demonstrate that qSL-RT-PCR detects abundant (RPL41A) and rare (YLR084C) decapped mRNAs.
Measurement of in vivo decapping rate
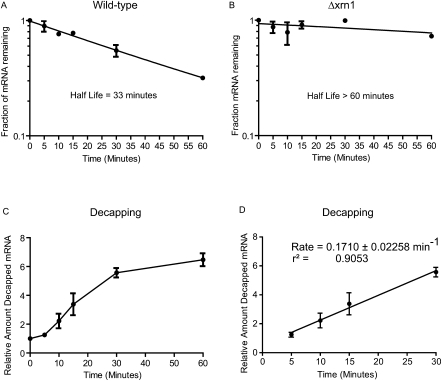
To determine the rate of decapping of RPL41A mRNA in vivo, transcription was inhibited with Thiolutin, samples were collected at time points, and accumulation of decapped RPL41A was measured. The half-life of RPL41A is 33 min in wild-type yeast cells (Fig. 4A). To measure the accumulation of decapped mRNA, degradation was blocked by deletion of the XRN1 gene, which greatly stabilized RPL41A, with over 70% of the mRNA remaining after 60 min (Fig. 4B). Thus, RPL41A mRNA is predominantly degraded by the 5′ pathway. qSL-RTP-CR was then performed on each time point. The resulting Ct values were normalized to total RPL41A detected in that sample. Next, the amount of decapped RPL41A at each time point was determined using the comparative Ct method (Schmittgen and Livak 2008). As expected, decapped RPL41A increased over the time course (Fig. 4C). Mean Ct values from biological replicates were plotted with standard error for each measurement (Fig. 4). The resulting curve demonstrated a brief lag phase during the first 5 min, followed by a linear phase for 30 min (Fig. 4C). The curve then plateaus between 30 and 60 min. The linear phase between 5 and 30 min was analyzed using linear regression analysis. The slope of the resulting line measures the rate constant of 0.171 +/− 0.0226 min−1. Therefore, qSL-RT-PCR detected accumulation of decapped endogenous mRNA and measured the decapping rate constant.
FIGURE 4.
The in vivo decapping rate of RPL41A mRNA was determined using qSL-RT-PCR assay. The half-life of RPL41A was measured in wild-type yeast (A) and in a strain wherein the XRN1 gene is deleted (Δxrn1) (B). (C) Decapped RPL41A mRNA was measured using the qSL-RT-PCR assay in the Δxrn1 strain following transcription shutoff. Relative amount of decapped RNA was calculated after normalization to total RPL41A at each time-point, relative to time = 0. (D) The rate of decapping was determined by linear regression analysis of the data in the graph in C with linear reaction kinetics between 5 and 30 min. The slope of the line, the reaction rate, is shown along with error and correlation coefficient. In all graphs, the mean value of the replicates is plotted and standard error is indicated above and below the data points.
DISCUSSION
In this report, we develop a quantitative assay, qSL-RT-PCR, that measures decapped mRNA produced in vivo by natural mRNA decay. The assay is versatile; any endogenous mRNA may be analyzed using qSL-RT-PCR. The assay has a broad dynamic range with a linear response over four orders of magnitude of input RNA (Fig. 3). The sensitivity of qSL-RT-PCR far exceeds Northern blotting. Indeed, decapped mRNA can be detected from as little as 1.5 ng of total cellular RNA (Fig. 3). The measurements are highly reproducible (Figs. 3,4). qSL-RT-PCR is fast and all of the reagents are readily available.
The specificity of qSL-RT-PCR is a major strength. By design, qSL-RT-PCR measures only one species—decapped mRNA. Specificity is imparted by splinted ligation; T4 DNA ligase has strong preference for perfect base pairing on each side of the nick to be ligated (Engler 1982; Wu and Wallace 1989). A 3′ hydroxyl and 5′ monophosphate are essential for ligation; therefore, RNAs that do not receive a 5′ cap (with a 5′ triphosphate) or degradation products from RNase A (with a 5′ hydroxyl) are not detected. Specificity also imposes restrictions. First, the mRNA's 5′ end must be mapped with single nucleotide resolution. Second, decapped products that are dephosphorylated or trimmed will not be detected. Third, competing decay pathways may diminish the magnitude of decapped RNA (He and Jacobson 2001).
When performing qSL-RT-PCR, we recommend optimization of several parameters. First, the critical controls described here should be performed. To optimize splinted ligation, the length of the DNA splint oligonucleotide is important—longer splints work better. The effect of splint length on ligation efficiency was systematically investigated: >40 bp of complementarity worked best (Kurschat et al. 2005). Titration of the splint can improve the assay and reduce background. Efficient DNase I digestion of the splint after ligation is crucial. PCR conditions should be optimized, specifically primer design and amplification efficiencies.
Measurement of decapping rates in vivo has proven difficult, yet meeting this challenge is essential to study regulation of decapping. We have demonstrated the ability of qSL-RT-PCR to measure the decapping rate constant of a natural mRNA. To stabilize the decapped mRNA, 5′ decay must be inhibited by inactivation of Xrn1. As no specific inhibitors of Xrn1 have been identified, Xrn1 was inactivated genetically. In other eukaryotes, genetic inactivation or RNAi-mediated depletion of Xrn1 may block decay of decapped mRNAs, thereby permitting determination of decapping rates.
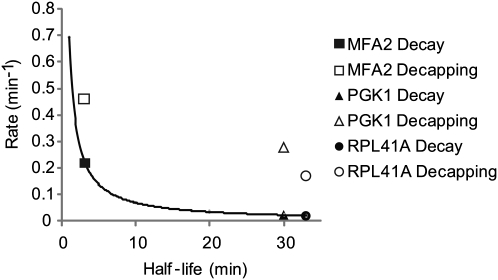
Comparison of the RPL41A decapping rate with the previously estimated rates for MFA2 and PGK1 mRNAs is informative. MFA2 and PGK1 mRNAs, modified with a secondary structure, were monitored by Northern blot (Muhlrad and Parker 1992; Muhlrad et al. 1995; Cao and Parker 2001). The half-life of oligo-adenylated mRNA intermediate was measured for each mRNA and used to calculate respective decapping rate constants (Cao and Parker 2001). In Figure 5, we compare the decapping rates of all three mRNAs. For reference, the first-order decay rates of each mRNA, calculated from their half-lives, are plotted on the same graph (Fig. 5). From this data, several observations emerge. First, the decapping rate constants measured by the two methods are similar in magnitude (MFA2 = 0.462 min−1, PGK1 = 0.2772 min−1, RPL41A = 0.171 min−1). Second, the mRNAs with short half-lives are decapped faster than those with longer half-lives (Fig. 5). Third, decapping rates are faster than the decay rate, indicating that decapping is not the rate-determining step. Indeed, deadenylation is often the rate-limiting step of mRNA decay (Shyu et al. 1991; Muhlrad and Parker 1992; Decker and Parker 1993). Interestingly, the in vivo decapping rates are slower than those measured in vitro with purified enzyme and substrate (Jones et al. 2008). This discrepancy likely reflects competitive binding of other proteins to the cap in vivo, such as eIF4E, or perhaps limiting concentration of decapping enzyme.
FIGURE 5.
Comparison of mRNA half-life, decay, and decapping reaction rates. The reaction rates of decay (in black) and decapping (in white) are compared with mRNA half-lives of three mRNAs, MFA2, PGK1, and RPL41A. Decay rates were derived from each mRNA's half-life. The trend-line curve plots the decay rate for mRNAs with half-lives ranging from 1 to 33 min. Decapping rates and mRNA half-lives for PGK1 and MFA2 were measured previously by Northern blot analysis of the poly(G) tract containing RNA (Muhlrad and Parker 1992; Muhlrad et al. 1995; Cao and Parker 2001). RPL41A decapping rate and half-life were measured by qSL-RT-PCR and qRT-PCR, respectively, as shown in Figure 4.
We anticipate that qSL-RT-PCR assay will prove useful for analysis of other RNAs and processes; for instance, other processing events such as endonucleolytic cleavage (Gatfield and Izaurralde 2004; Doma and Parker 2006; Eberle et al. 2009; Tomecki and Dziembowski 2010). Lastly, the ability to monitor decapping rates of natural mRNAs in vivo will greatly facilitate study of the features and factors that regulate decapping to control decay and translation.
MATERIALS AND METHODS
Yeast strains
AGY81: BY4742 MATalpha his3delta1 leu2delta0 lys2delta0 ura3delta0 Open Biosystems
AGY95: BY4742 MATalphaa xrn1∷KANr
AGY39: yRP1358 MATa his4-539 leu2-3112 lys2-201 trp1 ura3-52 dcp2∷TRP1
Oligonucleotides
Synthetic oligonucleotides were purchased from Integrated DNA technologies.
NB33: Anchor RNA oligonucleotide
5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUG-3′
NB34: RPL41A splint
5′-GCTCTCATTTCGATTGAATCGATGTGGTCTCATCAAAGCCAGCAAACGCAGTGTTCATTC
ATCGCCATCAGC-3′
NB35: Anchor forward PCR primer
5′-GCTGATGGCGATGAATGAACACTGC-3′
NB39: RPL41A reverse PCR primer
5′-CTAACATGTTAATTCATCAATGACATTACGATACTCTTG-3′
NB47: RPL41A forward PCR primer
5′-GAGAGCCAAGTGGAGAAAGAAGAGAACTAGA-3′
NB62: YLR084C splint
5′-GGGATGATTAAAGCGCACACTTTCCTTCCATCAAAGCCAGCAAACGCAGTGTTCATTCA
TCGCCATCAGC-3′
NB56: YLR084C reverse transcription primer
5′-GAGTGTCCAGAGACGATGAACAAAC-3′
NB58: YLR084C reverse PCR primer
5′- GTTTTCTCTTGCGTTTCTTCGTTTCCCG-3′
NB63: YLR084C forward PCR primer
5′-GCGCTTTAATCATCCCCATACTAGACTTTG-3′
RNA purification
Yeast cultures were grown in YPAD media at 30°C to an OD600 nm of 0.8. Cells were harvested at 4000g for 10 min, washed once with H2O, and suspended in 400 μL of TES (10 mM Tris-HCl at pH 6.8, 10 mM EDTA, 0.2% w/v SDS). Next, the RNA was extracted using acid phenol (pH 4.3) at 65°C, extracted twice with chloroform, and precipitated with ethanol and 0.3 M sodium acetate (pH 5.2) at −20°C. RNA was pelleted by centrifugation and washed with 70% ethanol. Purified RNA was resuspended in RNAse-free H2O and analyzed using a Nanodrop spectrophotometer (Thermo Scientific) and by formaldehyde gel electrophoresis.
Transcription shut-off
To measure mRNA decay rates, thiolutin, provided by Pfizer, was used to inhibit transcription (Jimenez et al. 1973; Passos and Parker 2008). Cultures of yeast (35 mL) were grown in YPAD to an OD600 nm of 0.8. Cells were treated with 5 μg/mL thiolutin and 5-mL samples were collected at the indicated times. Cells were pelleted at 13,000g for 1 min and rapidly frozen in a dry-ice–ethanol bath. Total RNA was then extracted from each sample.
Splinted-ligation
Total RNA (10 μg, unless otherwise indicated) was mixed with 20 picomoles of RPL41A splint oligonucleotide NB34 and 30 picomoles of RNA anchor NB33. For YLR084C, 7.5 μg of total RNA was mixed with 20 picomoles of splint NB62 and 30 picomoles RNA anchor NB33. To anneal oligos, samples were sequentially incubated for 5 min from 70°C to 60°C to 42°C, and finally to 25°C. Next, 20 U RNase Inhibitor Plus (Promega), 20 U High Concentration T4 DNA ligase (Promega), and T4 DNA ligase buffer were added to each sample and incubated overnight at 15°C. To degrade the splint and genomic DNA, each sample was treated with 10 U of RQ1 DNase (Promega) or Turbo DNase (Ambion) in the supplied 1 x DNase buffer for up to 3 h at 37°C. RNA was then extracted using acid-phenol:chloroform and then chloroform. The RNA was precipitated with ethanol and 0.3 M sodium acetate (pH 5.2) and 20 μg of glycoblue coprecipitant (Ambion). After washing with 70% ethanol, the RNA was suspended in 13.3 μL of RNase-free H2O.
Reverse transcription
Prior to reverse transcription, RNA samples were treated with DNase 1 to destroy any contaminating genomic DNA. For splinted ligation samples, this step is incorporated into the Splinted Ligation method described above. One unit of RQ DNase 1 (Promega) was used per microgram of total RNA. DNase treatment was performed in supplied DNase buffer for 1 h at 37°C. Reactions were then heat inactivated for 15 min at 65°C in 1 x DNase stop buffer. Two micrograms, unless otherwise noted, of DNase-treated RNA were added to reverse transcriptase assays. Reverse transcription reactions contained 20 picomoles of gene-specific reverse primer (NB39 for RPL41A or NB56 for YLR084C), 0.5 mM each dNTP, 1.2 mM MgCl2, 1 x GoScript buffer, and 1 μL of GoScript (Promega) in a 20-μL reaction and incubated 1 h at 42°C, then heat inactivated for 15 min at 65°C.
Quantitative PCR
Amplification of PCR products was measured using GoTaq qPCR master mix (Promega) with 200 nM of each primer in 50-μL reactions. For detection of splinted ligation products, the anchor forward PCR primer NB35 and RPL41A reverse PCR primer NB39 were used with 2 μL of template cDNA from splinted ligation reactions. For detection of total RPL41A mRNA, primers NB47 and NB39 were used. A splinted ligation product of YLR084C was detected using NB35 and NB58. Total YLR084C mRNA was detected using NB58 and NB63. A Bio-Rad CFX 96 C1000 real-time PCR instrument was used for all assays. Cycling parameters were: Steps (1) 95°C for 2 min, (2) 95°C for 30 sec, (3) 57°C for 30 sec, (4) 72°C for 40 sec, with steps 2–4 repeated 40 cycles. Reactions were analyzed by thermal melting curve and by gel electrophoresis. PCR amplification efficiencies of each primer set were determined (Pfaffl 2001). Primers NB35 and NB39 amplification efficiency was 102.7% and primers NB47 and NB39 were 102.9%.
Tobacco acid pyrophosphatase treatment
Ten μg total RNA was decapped by treatment with 20 U of RNase Inhibitor Plus (Promega) and 5 U Tabacco Acid Pyrophosphatase (Epicentre Biosystems) in the supplied 1 x TAP buffer for 30 min at 37°C.
Terminator treatment
Ten μg of total RNA was incubated with 20 U of RNase Inhibitor Plus and 10 U of Terminator (Epicentre Biosystems) in the supplied 1 x Terminator buffer for 30 min at 37°C.
Data analysis
Quantitative PCR assays were analyzed and cycle threshold (Ct) values determined using CFX Manager software (Bio-Rad). Calculations and graphs were created using Graphpad Prism version 5.0. Students t-test was used to measure significance. Relative changes were calculated using the comparative Ct method, ΔΔCt (Livak and Schmittgen 2001; Schmittgen and Livak 2008). First, the Ct value from qSL-RT-PCR assay for each timepoint sample (“target”) was normalized to the Ct of total RPL41A detected by qRT-PCR in that same sample (“reference”) by calculating ΔCttest = Cttarget test – Ctreference test. For time-course experiments, the ΔCt of decapped RPL41A RNA at time = 0 min (“calibrator”) was calculated using the equation ΔCtcalibrator = Cttarget calilbrator – Ctreference calibrator. The ΔΔCt for each timepoint was then calculated from the equation ΔΔCt = ΔCttest – ΔCtcalibrator. Relative expression for each timepoint was then calculated using the equation Ratio = 2-ΔΔCt (Livak and Schmittgen 2001; Schmittgen and Livak 2008). The relative amount of decapped RNA detected, relative to time = 0 min, was then plotted against the time the sample was harvested following transcription shutoff. The decapping rate was then determined by linear regression curve fitting using GraphPad Prism. The measured rate was 0.1710 +/− 0.0228 min−1 with a correlation coefficient of 0.9053 between 5 and 30 min determined from two biological replicates (Fig. 3).
To determine the percentage of decapped RNA (Fig. 2), relative expression was determined using the ΔΔCt method (Livak and Schmittgen 2001; Schmittgen and Livak 2008). To do so, ΔΔCt = ΔCttest was calculated for each sample (including test, terminator treated, TAP treated, no DNA ligase, and no reverse transcriptase controls), and −ΔCtcalibrator was calculated using the TAP treated sample as the “calibrator”. Next, the relative ratio was calculated and the TAP treated sample (ratio = 1) was set to 100%.
RPL41A mRNA half-lives were determined from the qRT-PCR analysis of time-course samples following addition of Thiolutin transcription inhibitor (Fig. 4A,B). The amounts of mRNA remaining at each timepoint relative to time = 0 min were determined from Ct values by calculating 2ΔCt where ΔCt = (Ct [calibrator t = 0] −Ct [test timepoint]). Half-life of RPL41A in wild-type cells is based on six biological replicates, and half-life in Δxrn1 cells is determined from two biological replicates. The resulting data was then plotted against time, and nonlinear regression analysis of biological replicate samples were fitted using first order decay kinetics (GraphPad Prism 5) (Fig. 4A,B). mRNA decay rates for RPL41A, PGK1, and MFA2 (Fig. 5) were calculated from the measured mRNA half-lives (Muhlrad and Parker 1992; Muhlrad et al. 1995) using the equation Rate = ln(2)/(t1/2).
ACKNOWLEDGMENTS
We thank members of the Goldstrohm lab and Dr. Trista Schagat for helpful discussions. We thank Pfizer for providing Thiolutin. In addition, we thank Dr. Wenqian Hu for his intellectual contributions. N.B. was supported by the Cellular and Molecular Biology Training Grant NIH NIGMS 5T32GM007315-34. J.C. is supported by a grant from the National Institutes of Health (GM080465).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2436411.
REFERENCES
- Anderson JS, Parker RP 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J 20: 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JT, Bai X, Johnson AW 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7: 1192–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell 27: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG 2008. PABLO analysis of RNA 5′-phosphorylation state and 5′-end mapping. Methods Enzymol 447: 83–98 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Grange T 2004. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T 1997. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci 94: 5628–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Engler MJ, Richardson CC 1982. DNA Ligases. In The Enzymes (ed. Boyer PD), pp. 3–29 Academic Press, New York [Google Scholar]
- Fischer N, Weis K 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J 21: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Bertrand E, Pictet R, Grange T 1993. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res 21: 1683–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, LaFiandra A, Shatkin AJ 1977. 5′-Terminal structure and mRNA stability. Nature 266: 235–239 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Green MR, Maniatis T, Melton DA 1983. Human β-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell 32: 681–694 [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol 21: 1515–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Stevens A 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Petzold C, Coller J, Baker KE 2010. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A, Tipper DJ, Davies J 1973. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrob Agents Chemother 3: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BN, Quang-Dang DU, Oku Y, Gross JD 2008. A kinetic assay to monitor RNA decapping under single-turnover conditions. Methods Enzymol 448: 23–40 [DOI] [PubMed] [Google Scholar]
- Kurschat WC, Muller J, Wombacher R, Helm M 2005. Optimizing splinted ligation of highly structured small RNAs. RNA 11: 1909–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rodgers ND, Jiao X, Kiledjian M 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 21: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Jiao X, Liu H, Gu M, Lima CD, Kiledjian M 2004. Functional analysis of mRNA scavenger decapping enzymes. RNA 10: 1412–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maroney PA, Chamnongpol S, Souret F, Nilsen TW 2007. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA 13: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Chamnongpol S, Souret F, Nilsen TW 2008. Direct detection of small RNAs using splinted ligation. Nat Protoc 3: 279–287 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC 2000. Joining of RNAs by splinted ligation. Methods Enzymol 317: 109–123 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev 6: 2100–2111 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol 15: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Passos DO, Parker R 2008. Analysis of cytoplasmic mRNA decay in Saccharomyces cerevisiae. Methods Enzymol 448: 409–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole TL, Stevens A 1997. Structural modifications of RNA influence the 5′ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem Biophys Res Commun 235: 799–805 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schwer B, Mao X, Shuman S 1998. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res 26: 2050–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Belasco JG, Greenberg ME 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev 5: 221–231 [DOI] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dziembowski A 2010. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA 16: 1692–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762 [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Wallace RB 1989. Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene 76: 245–254 [DOI] [PubMed] [Google Scholar]
- Yu X, Warner JR 2001. Expression of a micro-protein. J Biol Chem 276: 33821–33825 [DOI] [PubMed] [Google Scholar]