Abstract
BACKGROUND
Pluripotent stem cells have been derived from a variety of sources such as from the inner cell mass of preimplantation embryos, from primordial germ cells, from teratocarcinomas and from male germ cells. The recent development of induced pluripotent stem cells demonstrates that somatic cells can be reprogrammed to a pluripotent state in vitro.
METHODS
This review summarizes our current understanding of the origins of mouse and human pluripotent cells. We pay specific attention to transcriptional and epigenetic regulation in pluripotent cells and germ cells. Furthermore, we discuss developmental aspects in the germline that seem to be of importance for the transition of germ cells towards pluripotency. This review is based on literature from the Pubmed database, using Boolean search statements with relevant keywords on the subject.
RESULTS
There are distinct molecular mechanisms involved in the generation and maintenance of the various pluripotent cell types. Furthermore, there are important similarities and differences between the different categories of pluripotent cells in terms of phenotype and epigenetic modifications. Pluripotent cell lines from various origins differ in growth characteristics, developmental potential, transcriptional activity and epigenetic regulation. Upon derivation, pluripotent stem cells generally acquire new properties, but they often also retain a ‘footprint’ of their tissue of origin.
CONCLUSIONS
In order to further our knowledge of the mechanisms underlying self-renewal and pluripotency, a thorough comparison between different pluripotent stem cell types is required. This will progress the use of stem cells in basic biology, drug discovery and future clinical applications.
Keywords: pluripotency, embryonic stem cells, epigenetics, germ cells, iPS cells
Introduction
In mammals, most tissues are known to harbour stem cells that allow tissue maintenance and repair. The two key features that allow them to do so are: (i) self-renewal, which is the ability to produce new stem cells with equal characteristics as the original stem cell and (ii) the capacity to give rise to differentiated cell types. Stem cells are further characterized by their differentiation potential. For example, unipotent stem cells such as spermatogonial stem cells (SSCs) can give rise to just one differentiated cell type; in this case spermatozoa. Adult stem cells are multipotent and can produce multiple differentiated cell types that are, in general, within the same lineage as the original stem cell. Examples are haematopoietic stem cells, hair follicle stem cells and intestinal stem cells. Pluripotent stem cells such as embryonic stem (ES) cells are even more plastic and can give rise to cells of all three embryonic germ layers as well as germ cells. ES cells and other pluripotent stem cells can proliferate indefinitively, whereas the proliferative capacity of adult stem cells is limited. A fertilized oocyte and blastomeres of the first cleavage stages are considered totipotent, because they can still give rise to all embryonic and extraembryonic lineages. These cells are, however, not considered stem cells because they lack the ability to self-renew.
Because pluripotent stem cells have the capacity to give rise to unlimited numbers of any differentiated cell type, they are considered good candidates to regenerate damaged tissues and organs in future cell therapies. Furthermore, pluripotent stem cells are powerful research tools to study fundamental processes in development and they have considerable utility for drug screens and toxicity testing.
Methods
Mouse and human pluripotent stem cell lines have been derived from a variety of sources such as the pluripotent inner cell mass (ICM) of preimplantation embryos, from primordial germ cells (PGCs), from teratocarcinomas and from male germ cells. Early embryos and the germline seem particularly permissive to the derivation of pluripotent stem cells, but pluripotency is not restricted to these cells, because rather unexpectedly somatic cells can be reprogrammed to a pluripotent state as well. The difference between pluripotent cells of various origins could affect future use in cell therapies, research or drug development.
Here, we review the origins of mouse and human pluripotent cells, the molecular mechanisms that are involved in the generation and maintenance of these cells, and the similarities and differences between the different categories of pluripotent cells in terms of phenotype and epigenetic modifications. At the Pubmed database, Boolean search statements with relevant keywords on the subject were used to obtain relevant literature ranging from 1954 until 2010. Key words that were used include Pluripotency, Embryonic Stem cells, Epigenetics, Germ Cells, Teratocarcinoma and iPS cells.
Embryonic Stem cells
Mouse ES cell lines were first described early in the 1980s and were generated from the ICMs of blastocyst stage embryos (Evans and Kaufman, 1981; Martin, 1981). As well as using the ICMs of blastocyst stage embryos, mouse ES cell lines have also been generated from explanted or disaggregated morulae and cleavage stage embryos and even from biopsied blastomeres (Eistetter, 1989; Delhaise et al., 1996; Tesar, 2005; Chung et al., 2006; Lorthongpanich et al., 2008).
The capacity of cells to differentiate into derivatives of all three germ layers and germ cells can be assessed in various ways. In vitro differentiation can be achieved through the formation of the so-called embryoid bodies (EBs). These structures will be formed when ES cells are grown in drops hanging from the lid of a Petri-dish or upon culture of ES cells in non-adherent dishes. Aggregated ES cells will start to differentiate and recapitulate embryonic development to a limited extent, thereby forming derivatives of all three embryonic germ layers and germ cells. In addition to the EB approach, there are numerous other methods for the in vitro differentiation of ES cells to specific cell types.
Another more stringent way to determine pluripotency of ES cells is through in vivo differentiation, for example by teratoma formation. In this assay, cells are injected subcutaneously into the testis or in the kidney capsule of immunocompromised mice, after which they will induce tumour formation consisting of tissues of all three embryonic germ layers if the injected cells were indeed pluripotent. In even more rigorous tests for pluripotency, cells are aggregated with morula stage embryos or injected into the blastocoel of host embryos. Under these conditions, pluripotent cells can integrate with the cells of the host embryo and participate in its development, contributing to tissues of all three germ layers including germ cells, resulting in chimeras.
Finally, in the tetraploid complementation assay, pluripotent cells are combined with tetraploid preimplantation embryos. The tetraploid cells will preferentially develop into the trophectoderm lineage whereas the developing embryo will be exclusively produced from transplanted diploid cells. Tetraploid complementation is generally considered to be the most stringent test for pluripotency, because in the less stringent chimera assay, compensating embryonic cells from the host can mask limitations in developmental potential of the injected cells (Jaenisch and Young, 2008). Pluripotency of mouse ES cells has been validated with all of the above methods including the tetraploid complementation assay (Nagy et al., 1993).
In general, mouse ES cells are co-cultured on a supportive layer of mitotically inactive mouse embryonic fibroblasts (feeder cells) in the presence of fetal calf serum (FCS) and leukaemia inhibitory factor (LIF). The cytokine LIF promotes the self-renewing and pluripotent phenotype of mouse ES cells through activation of Stat3 signalling by binding to heterodimers of the LIF-receptor and the signal transducer Gp130 (Fig. 1; Smith et al., 1988; Williams et al., 1988; Yoshida et al., 1994). In the absence of serum, mouse ES cells require stimulation by bone morphogenetic protein-4 (BMP4) to prevent the cells from differentiating (Ying et al., 2003). BMP4 contributes to the preservation of the mouse ES cell phenotype by activating Smad transcription factors, which in turn induce the expression of members of the Id (Inhibitor of differentiation) gene family. Indeed forced expression of Id allows self-renewal of mouse ES cells in the presence of LIF and absence of BMP4 or serum. Id proteins contribute to the self-renewal of ES cells by inhibition of differentiation towards the neural lineage (Ying et al., 2003).
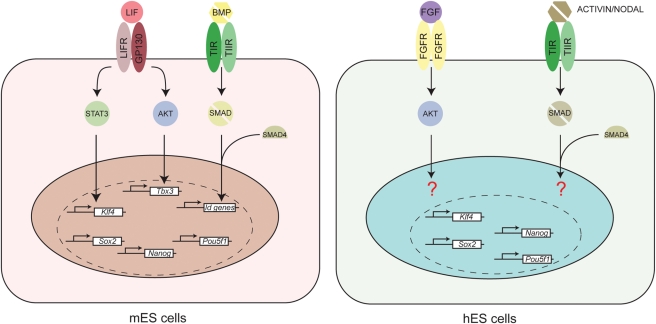
Figure 1.
Schematic representation of the cell signal transduction pathways that are involved in pluripotency of mouse and human ES cells. Mouse ES cells: LIF binds to LIFR and GP130 heterodimers, which results in the activation of STAT3 signalling and AKT signalling and subsequent activation of the downstream target genes Klf4 and Tbx3. BMP binds to heterodimers of the Type I and Type II receptors. As a result, three Smad transcription factors (one of which is Smad 4) trimerize and translocate to the nucleus where they activate the expression of Id genes. LIF and BMP signalling also results in the expression of other members of the pluripotency factor network (e.g. Sox2, Nanog and Pou5f1), but it is unclear if this is a direct or an indirect effect. Human ES cells: FGF binds to FGF-receptor homodimers, which leads to AKT signalling. In parallel, Activin or Nodal homodimers binds to heterodimers of the Type I and Type II receptors. As a result, three Smad transcription factors (one of which is Smad 4) trimerize and translocate to the nucleus. Cooperatively, FGF and Activin/Nodal maintain pluripotency of human ES cells. The question marks depict our lack of knowledge on how these cell signal transduction pathways act on the pluripotency factor network (dashed line) in human ES cells.
Human ES cell lines were first described in the late 1990s (Thomson et al., 1998). In contrast to mouse ES cells that are usually derived from freshly collected embryos, the main source for human ES cells is cryopreserved human surplus embryos that have been donated by patients undergoing in vitro fertilization (Lerou et al., 2008). Similar to mouse ES cells, human ES cell lines have also been derived from morula and cleavage stage embryos and from biopsied blastomeres (Klimanskaya et al., 2006, 2007; Chung et al., 2008; Feki et al., 2008; Lerou et al., 2008; Strelchenko et al., 2004; van de Velde et al., 2008; Geens et al., 2009; Ilic et al., 2009). The currently available arsenal for determining pluripotency of human cells is restricted to in vitro differentiation and teratoma formation. More stringent methods such as chimera formation and tetraploid complementation are not possible in humans due to the clear ethical issues, and it remains therefore unresolved whether human ES cells can participate in normal development. In an effort to determine their in vivo developmental capacity, human ES cells have been injected into mouse blastocysts that were subsequently transferred to pseudo-pregnant foster mice and allowed to develop until embryo day 8.5 (E8.5) (James et al., 2006). Although rare, human ES cells persisted up to E8.5, however, it was unresolved if the human ES cells were capable of functionally integrating into the host tissue, or if the human cells were proliferating at this stage (James et al., 2006).
Human ES cells are generally maintained on feeder cells in the presence of basic fibroblast growth factor (bFGF) and FCS or Knockout Serum Replacement. In contrast with mouse ES cells, human ES cells have a flattened epithelial morphology and their long-term culture depends on FGF/Nodal and Activin pathways (Fig. 1; Vallier et al., 2005). Furthermore, in the absence of feeders, LIF prevents mouse ES cells but not human ES cells from differentiating, despite the presence of a functional LIF signal transduction pathway in human ES cells (Thomson et al., 1998; Reubinoff et al., 2000; Humphrey et al., 2004). Another difference between human ES cells and mouse ES cells is that in vitro human ES cells can differentiate into cells that resemble trophectoderm, either spontaneously (Thomson et al., 1998; Reubinoff et al., 2000) or upon stimulation with BMP-4 (Xu et al., 2002). Thus, human ES cells have a different developmental potential than mouse ES cells even though both cell types are considered pluripotent. If future studies demonstrate that human ES cells can differentiate into all extraembryonic tissues in addition to their capacity to differentiate into all three germ layers and germ cells, then it could be argued that human ES cells are totipotent, like the zygote or early cleavage stage blastomeres.
Various studies have reported ES cell lines from non-human primates (NHP; Thomson et al., 1995, 1996; Suemori et al., 2001; Sasaki et al., 2005; Mitalipov et al., 2006). NHP ES cells share growth characteristics, colony morphology and marker expression with human ES cells and it is therefore reasonable to assume that they represent similar cell types. In contrast with human ES cells, it is possible to examine the in vivo developmental competence of NHP ES cells in chimeric embryos. However, to date, there are no reports of chimeric animals derived from blastocysts injected with NHP ES cells. Although injected NHP ES cells persist in blastocyst stage embryos (Mitalipov et al., 2006), transfer of 15 chimeric blastocysts to pseudo-pregnant females did not result in chimearic offspring. This indicates that the potential of NHP ES cells to contribute to chimeras may be low. Given the similarities between NHP ES cells and human ES cells, human ES cells may have an equally limited ability to participate in normal embryo development.
Epiblast of post-implantation mouse and rat embryos
Pluripotent stem cells that are isolated from the epiblast of post-implantation mouse and rat embryos (EpiSCs) can be maintained in defined culture conditions that also support self-renewal of human ES cells (Brons et al., 2007; Tesar et al., 2007). EpiSCs express markers of pluripotency such as POU domain, class 5, transcription factor 1 (Pou5f1, also known as Oct-4), Nanog and Sox2, and can differentiate in vitro into all three germ layers and germ cells. However, compared with mouse ES cells, EpiSCs have a flattened epithelial morphology that is more reminiscent of human ES cells (Brons et al., 2007; Tesar et al., 2007). Furthermore, similar to human ES cells (Xu et al., 2002), EpiSCs can differentiate to cells that express markers for trophectoderm upon stimulation with BMP4 (Brons et al., 2007). The similarities between mouse EpiSCs and human ES may indicate that human ES cells are derived from epithelialized epiblast cells in the human embryo (Brons et al., 2007; Tesar et al., 2007).
In comparison with mouse ES cells, EpiSCs have low potential to contribute to chimeras, and germline transmission has not been described. Nevertheless, EpiSCs can differentiate into all three germ layers as demonstrated by their capacity to form teratomas after ectopic injection into immunodeficient mice and in vitro EpiSCs can also differentiate towards PGCs (Brons et al., 2007; Tesar et al., 2007; Hayashi et al., 2009). Mouse ES cells can give rise to EpiSC-like cells when cultured under EpiSC conditions, and EpiSCs can be converted to pluripotent ES-like cells after culture under ES cell-conditions (Fig. 2; Bao et al., 2009; Guo et al., 2009).
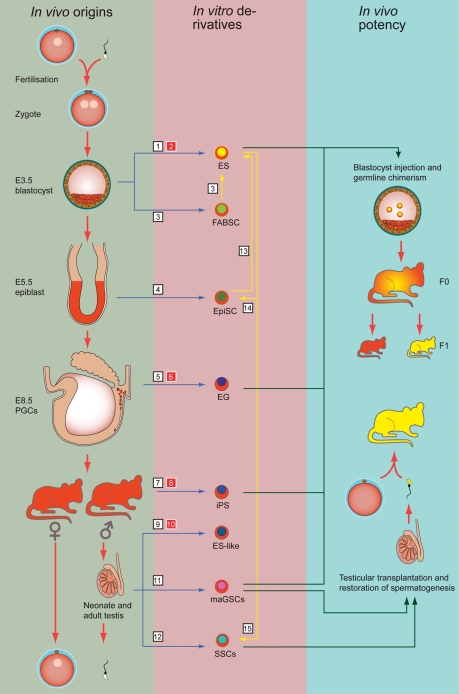
Figure 2.
Origins of mammalian pluripotent cells. In the green column on the left are the developmental stages and in vivo origins from which mammalian pluripotent stem cells have been derived. In the pink column in the middle are the stem cell types that can be propagated in vitro. In the blue column on the right is indicated which in vitro cultured mouse cells can participate in all three germ layers including germ cells as determined by the chimera assay. In the same column is indicated which in vitro cultured mouse cell types can restore spermatogenesis as determined by the testis transplantation assay. Blue arrows indicate the derivation of in vitro cell lines from in vivo origins, yellow arrows indicate in vitro transformation/differentiation of one cell type into another, green arrows indicate in vivo functionality tests of in vitro cultured cells. White numbered squares refer to studies performed in mice and red numbered squares refer to studies performed in human: 1Evans and Kaufman (1981), Martin (1981); 2Thomson et al. (1998); 3Chou et al. (2008); 4Brons et al. (2007), Tesar et al. (2007); 5Matsui et al. (1992), Resnick et al. (1992); 6Liu et al. (2004), Shamblott et al. (1998), Turnpenny et al. (2003); 7Okita et al. (2007), Takahashi and Yamanaka (2006); 8Takahashi et al. (2007); 9Kanatsu-Shinohara et al. (2004), Ko et al. (2009), Seandel et al. (2007); 10Conrad et al. (2008), Golestaneh et al. (2009), Kossack et al. (2009), Mizrak et al. (2009); 11Guan et al. (2006); 12Kanatsu-Shinohara et al. (2003); 13Bao et al. (2009), Guo et al. (2009), Silva et al. (2009); 14Guo et al. (2009); 15Nayernia et al. (2006).
Explanted ICMs from preimplantation mouse embryos can give rise to a distinct type of stem cells when they are cultured under conditions that also support growth of human ES cells and EpiSCs. These cell lines have been designated FAB-SCs (for bFGF, Activin and BIO-derived stem cells; Chou et al., 2008). In contrast to EpiSCs, FAB-SCs have global micro-RNA expression profiles similar to those of ES cells, which is suggestive of an intermediary state between regular mouse ES cells and EpiSCs (Chou et al., 2008). Despite their early embryonic origin and the expression of the pluripotency factors POU5F1, SOX2 and NANOG, FAB-SCs are not pluripotent because they fail to differentiate in common in vitro and in vivo differentiation assays. However, FAB-SCs can convert to pluripotency upon transient LIF/BMP4 stimulation, or by ectopic expression of E-cadherin, an important protein for cell–cell adhesion (Chou et al., 2008).
Under standard culture conditions, mouse ES cells are heterogeneous in their expression of the pluripotency genes Nanog, Stella and Rex1 (Chambers et al., 2007; Hayashi et al., 2008; Toyooka et al., 2008). For these genes, reporter cell lines have been designed that express a fluorescent protein when the gene of interest (for example Nanog) is transcriptionally active. These cell lines have been used to sort ES cells that express Nanog, Stella or Rex1. However, during culture a population of ES cells will emerge that is negative for the gene that was initially selected for. The converse is also true; cells that are positive for Nanog, Stella or Rex1 will emerge from populations of ES cells that initially did not express these genes (Chambers et al., 2007; Hayashi et al., 2008; Toyooka et al., 2008).These findings indicate that within ES cells there are subpopulations of cells that are in a dynamic equilibrium. On the basis of gene expression patterns and developmental potential, it has been suggested that the Stella/Rex1-positive and Stella/Rex1-negative ES cells span the ICM and epiblast phenotype, respectively (Hayashi et al., 2008; Toyooka et al., 2008). This suggests that mouse ES cells are in a continuous process of differentiation from the ICM towards the epiblast phenotype and dedifferentiation from the epiblast phenotype towards that of ICM cells.
Pluripotency and the developing embryo
In order to better understand the mechanism behind pluripotency, it is essential to understand the developmental processes that lead to the formation of blastocyst stage embryos and the establishment of the pluripotent cell population in these embryos that can give rise to ES cells or EpiSCs. Three different cell types can be recognized in blastocyst stage embryos: the trophectoderm (which gives rise to the trophoblast/placenta and the chorion), precursors of the primitive endoderm (which in rodents primarily gives rise to parietal endoderm and the visceral endoderm of the yolk sac after implantation) and precursors of the pluripotent epiblast (which gives rise to the embryo proper, the umbilical cord and the amnion). The formation of these three cell populations is established by two consecutive lineage segregations: firstly the trophectoderm and the cells of the ICM are segregated and secondly, the primitive endoderm and the epiblast are segregated within the ICM (Fig. 3).
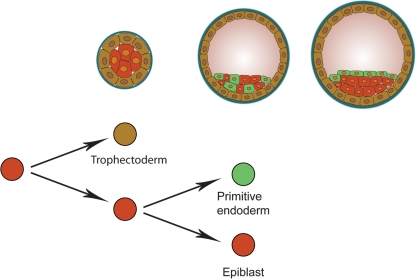
Figure 3.
The first two lineage segregation events in mammals. From left to right: three successive stages (morula, early blastocyst and late blastocyst) in mammalian development. The two consecutive lineage segregation events that occur in this developmental period are schematically depicted below these embryonic stages. The first segregation of cell lineages occurs in morula stage embryos and results in the formation of the trophectoderm (brown) and the cells of the ICM (red). In the ICM, this event is followed by a second segregation event that initially results in the formation of randomly distributed primitive endoderm precursors (green) and epiblast precursors (red). These cells are subsequently sorted out to form the primitive endoderm and the pluripotent epiblast at the late blastocyst stage.
Segregation of the trophectoderm and the ICM
The early cell fate decisions in the developing embryo are governed by key transcriptional regulators. In mouse embryos, a key player in segregation of the trophectoderm and the ICM is the transcription factor POU5F1, which is expressed in all cells up to the morula stage but becomes restricted to the ICM of blastocysts and subsequently the epiblast (Palmieri et al., 1994). Although the trophectoderm of Pou5f1 null embryos is functional and embryos implant at the blastocyst stage, Pou5f1 null embryos fail to develop an ICM or any of its derivatives, resulting in embryos that are entirely composed of trophectoderm cells instead. As a consequence, Pou5f1 null embryos are not viable and die shortly after implantation (Nichols et al., 1998).
Another key transcription factor involved in specification of the trophectoderm lineage in mouse embryos is the caudal-related homeodomain protein CDX2. Expression of CDX2 in mouse blastocysts is the inverse of POU5F1 expression and is restricted to the trophectoderm (Beck et al., 1995).
Functional studies in mouse ES cells suggest that this complementary spatial expression is established through reciprocal inhibition of CDX2 and POU5F1 (Rossant et al., 2003; Niwa et al., 2005) and it has been proposed that this mutual repression causes the specification of POU5F1-positive epiblast cells versus CDX2-positive trophectoderm cells in late blastocysts (Rossant et al., 2003; Niwa et al., 2005; Strumpf et al., 2005; Ralston et al., 2010). However, in compacting morulae, the mutual inhibitory relation between CDX2 and POU5F1 is not yet clear, because CDX2 and POU5F1 are present in nuclei of all cells at the moment of compaction (Dietrich et al., 2007). Co-expression of CDX2 and POU5F1 might reflect a certain degree of plasticity of inner and outer cells at this stage of development. Indeed, at the moment of compaction, processes involved in polarization and cell allocation are still reversible, since isolated and re-aggregated outer cells of compacted embryos (16 cells, but not 32 cells) are able to develop into blastocysts with ICM and trophectoderm, and give rise to fertile offspring (Suwinska et al., 2008).
CDX2 acts in concert with the transcription factor GATA3 to induce trophoblast gene expression (Ralston et al., 2010). The expression of GATA3 and CDX2 in preimplantation embryos is in turn regulated by the TEA DNA binding domain/transcription enhancer factor (TEAD/TEF) family transcription factor TEAD4 (Fig. 4; Yagi et al., 2007; Nishioka et al., 2008; Ralston et al., 2010). Although TEAD4 is expressed in both inside and outside cells, its activity is spatially regulated along the inside–outside axis of the embryo (Nishioka et al., 2009). In cells localized at the outside, TEAD4 acts in conjunction with the TEAD coactivator protein YAP. However, in inside cells, YAP is excluded from the nuclei by the Hippo signalling pathway component LATS (Fig. 4; Nishioka et al., 2009). TEAD4 is therefore inactive in inside cells and consequently Cdx2 and Gata3 are not expressed in these cells. To summarize, the first lineage segregation that results in the formation of the trophectoderm and the ICM is governed by Hippo signalling, which patterns the cooperative activity of YAP and TEAD4 to the outer cells of the embryo. In these cells, YAP and TEAD4 induce the expression of the trophectoderm genes Cdx2 and Gata3, which in turn leads to the induction of a trophoblast gene expression programme and the down-regulation of POU5F1. In inner cells, TEAD4 activity is suppressed by LATS-mediated reduction of nuclear YAP localization and initial POU5F1 expression is maintained.
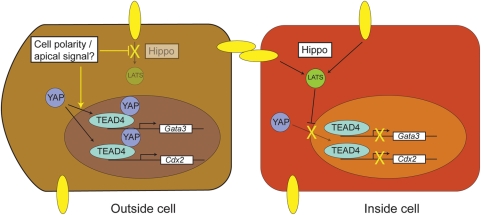
Figure 4.
The segregation of the trophectoderm from the ICM. The segregation of the trophectoderm and the ICM is governed by Hippo signalling. In the outer cells of the embryo (left). Hippo signalling is suppressed possibly as a result of a signal generated by cell polarity. YAP is translocated to the nucleas where it co-operates with TEAD4 to induce the expression of the trophectoderm genes Cdx2 and Gata3, which in turn leads to the induction of a trophoblast gene expression program and the down-regulation of POU5F1. In inner cells (right), Hippo signalling is active resulting in LATS-mediated phosphorylation of YAP. Phosphorylated YAP is sequestered in the cytoplasm. In the absence of nuclear YAP localization, TEAD4 remains inactive, the trophectoderm genes Cdx2 and Gata3 are consequently not expressed, and initial POU5F1 expression is maintained (Nishioka et al., 2009).
Segregation of the primitive endoderm and the epiblast
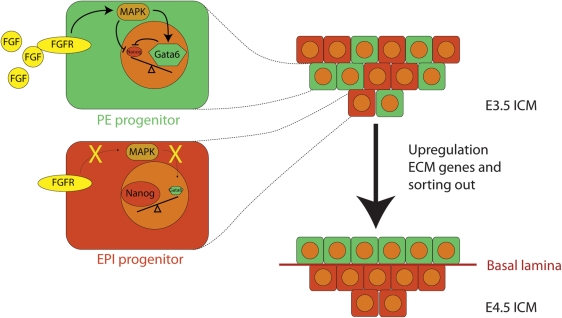
In mouse embryos, SOX2 plays an important role in the development of the pluripotent epiblast. The expression pattern of SOX2 in preimplantation embryos is comparable with that of POU5F1 and indeed Sox2 null embryos die around implantation due to a defective epiblast development (Avilion et al., 2003). The SOX2/POU5F1-positive ICM of embryonic day 3.5 (E3.5) blastocysts has a heterogeneous constitution and consists of cells that express the transcription factor GATA6 and other cells that express the transcription factor NANOG (Fig. 5). The GATA6- and NANOG-positive cell populations are the precursors of, respectively, the primitive endoderm and the pluripotent epiblast. Embryos from which Nanog has been genetically deleted lack pluripotent epiblast cells and, in agreement, ICM cells from E3.5 Nanog mutants differentiate into endoderm-like cells when cultured in vitro (Mitsui et al., 2003). In embryos that lack Gata6, primitive endoderm formation is initiated, but no functional visceral endoderm is formed in post-implantation embryos (Morrisey et al., 1998; Koutsourakis et al., 1999). Consistent with their in vivo roles POU5F1, SOX2 and NANOG are expressed in pluripotent ES cells, whereas CDX2, GATA3 and GATA6 are not. Human ES cell generation is optimal when Day 6 blastocysts are used, which coincides with restricted expression of POU5F1 to the ICM and of CDX2 to the trophectoderm (Chen et al., 2009). At this stage of development, NANOG is expressed in a subset of the ICM cells (Cauffman et al., 2009).
Figure 5.
The segregation of the primitive endoderm from the epiblast. Schematic representation of the ICM of E3.5 (top-right) and E4.5 embryos (bottom-right). In E3.5 embryos, cellular differences in FGF-signalling (enlarged cells on the left) result in a heterogeneous ICM with NANOG-positive (red) and GATA6-positive (green) cells, which are the precursors for the epiblast (EPI) and the primitive endoderm (PE) respectively. Extracellular matrix (ECM) genes are up-regulated and the cells are sorted out to form the primitive endoderm and epiblast lineages at E4.5 (Chazaud et al., 2006).
The mosaic expression of Gata6 and Nanog in mouse embryos depends on FGF-mediated GRB2-Ras-MAP kinase signalling (Chazaud et al., 2006). GRB2 activation through FGF-signalling leads to the suppression of Nanog and the activation of Gata6 (Chazaud et al., 2006; Hamazaki et al., 2006). As such, mouse Grb2−/− embryos fail to develop primitive endoderm and have a homogenous ICM of which all cells express NANOG but do not express GATA6 (Cheng et al., 1998; Chazaud et al., 2006). The Grb2−/− phenotype is similar to those of mouse embryos lacking Fgf4 or Fgfr2, which also fail to develop primitive endoderm (Feldman et al., 1995; Wilder et al., 1997; Arman et al., 1998).
The role of FGF in the development of the mouse primitive endoderm has been further confirmed by adding exogenous FGF4 or small chemical compounds that interfere with FGF-signalling to mouse embryo cultures (Nichols et al., 2009b; Yamanaka et al., 2010). Small molecule inhibition of FGF-receptor activation, or of MEK in developing mouse embryos from the 8-cell stage until the blastocyst stage, blocks primitive endoderm formation and results in fewer precursors of the primitive endoderm and more epiblast precursors than in control embryos (Nichols et al., 2009b; Yamanaka et al., 2010). Nevertheless, when inhibitors are removed from the culture before E3.75, the embryos recover and restore a mutually exclusive expression pattern of NANOG and GATA6, which subsequently segregate into the primitive endoderm and the epiblast. In a comparable regulative manner, embryos that have been initially cultured in control media up to E3.75 develop ICMs that consist entirely of NANOG-positive cells when transferred to conditions in which FGF-signalling is inhibited (Yamanaka et al., 2010). This plasticity of ICM cells to convert from epiblast progenitor to primitive endoderm progenitor and vice versa is progressively lost after E4.0 (Yamanaka et al., 2010). The plasticity of ICM cells is reminiscent of the plasticity of the various populations in ES cells that are in dynamic equilibrium, such as the NANOG-positive and NANOG-negative populations (Chambers et al., 2007).
FGF4-stimulation of embryos results in effects that are opposite to FGF-inhibition. The ICMs of preimplantation mouse embryos that have been cultured from E1.5 onwards in the presence of exogenous FGF4 develop more primitive endoderm precursors (Yamanaka et al., 2010). From E4.0 to E4.5 onwards, exogenous FGF4 does not alter the constitution of the ICM, which demonstrates that at this stage plasticity is lost and the epiblast and primitive endoderm cell lineage are committed (Yamanaka et al., 2010). Thus, modulating FGF-signalling before E4.0 can shift the fate of ICM cells to either primitive endoderm or pluripotent epiblast.
Small chemical compounds that interfere with FGF-signalling have also been used to block differentiation of ES cells. Chemical inhibition of MEK and GSK3β (negative regulator of WNT-signalling) enables feeder-free maintenance of mouse ES cells in the absence of growth factors or cytokines. It has been proposed that ES cells are intrinsically self-renewing and maintain this state if properly shielded from inductive differentiation and this condition has therefore been referred to as the ground state of ES cell self-renewal (Ying et al., 2008). Similar conditions with combinations of small molecule inhibitors have enabled the generation of germline competent ES cells from previously non-permissive strains of mice (Nichols et al., 2009a). Furthermore, these conditions allowed the derivation of the first authentic rat ES cells with germline-competence (Buehr et al., 2008; Li et al., 2008).
Since the first reports of mouse ES cells in the 1980s, ES cell lines have been derived from mouse (Evans and Kaufman, 1981; Martin, 1981), rat (Buehr et al., 2008; Li et al., 2008) and a few primate species; e.g. rhesus monkey (Thomson et al., 1995) and human (Thomson et al., 1998). Why most other mammals have been recalcitrant when it comes to ES cell derivation remains enigmatic, but may be caused by crucial differences in early embryo development between species (Kuijk et al., 2008). To better understand differences in establishment and maintenance of the pluripotent cell population in various species, it is essential to do functional studies on early development in species other than the mouse. It is to be expected that the use of small chemical compounds to block differentiation will facilitate ES cell derivation from hitherto non-permissive species.
The role of pluripotency factors in ES cells
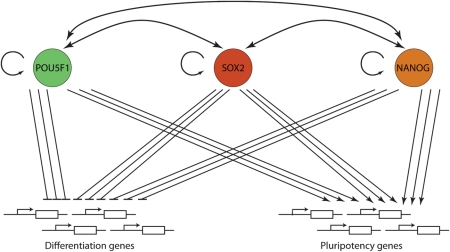
In both human and mouse ES cells, the transcription factors POU5F1, SOX2 and NANOG can bind to the same promoter regions of a large number of genes, thereby regulating transcriptional activity of these genes (Boyer et al., 2005; Loh et al., 2006; Chen et al., 2008; Sharov et al., 2008). Targets include genes that code for transcription factors, signal transduction components and epigenetic modifiers that cooperatively promote pluripotency and self-renewal. In addition, POU5F1, SOX2 and NANOG bind to the promotors of genes that are silent in ES cells, expression of which is associated with lineage commitment and differentiation (Fig. 6; Boyer et al., 2005; Loh et al., 2006; Sharov et al., 2008). These findings demonstrate that POU5F1, SOX2 and NANOG act together to control pluripotency. Indeed individual depletion of each factor leads to increased expression of genes that are implicated in developmental processes and differentiation (Niwa et al., 2000; Chambers et al., 2003; Matin et al., 2004; Boyer et al., 2005; Hyslop et al., 2005; Loh et al., 2006; Fong et al., 2008; Sharov et al., 2008). Furthermore, POU5F1, SOX2 and NANOG form an autoregulatory positive feedback loop through mutual binding to their promoter regions (Boyer et al., 2005), which is considered to facilitate stability of the pluripotent state (Masui et al., 2007; Jaenisch and Young, 2008). While LIF plays an important role in the derivation and maintenance of murine ES cells, it is not required during early murine embryonic development. In the absence of LIF or its receptors, embryos develop normally, at least until mid-gestation (Stewart et al., 1992; Yoshida et al., 1994; Li et al., 1995; Ware et al., 1995; Nakashima et al., 1999). However, mutant embryos that lack the LIF-receptor Gp130 fail to develop an epiblast if implantation is delayed by diapause; i.e. delayed implantation common to lactating females (Nichols et al., 2001). It has therefore been hypothesized that LIF signalling suppresses differentiation of epiblast progenitors to epiblast cells, which is consistent with the function of LIF signalling in diapause and in mouse ES cells. The responsiveness of mouse ES cells to LIF can therefore be attributed to the role of this cytokine in diapause (Nichols et al., 2001).
Figure 6.
A network controlling pluripotency. The transcription factors POU5F1, SOX2 and NANOG bind to the same promoter regions of a large number of genes, thereby regulating transcriptional activity of these genes (Boyer et al., 2005; Loh et al., 2006; Chen et al., 2008; Sharov et al., 2008). The cooperative binding of these factors results in the activation of genes that code for transcription factors, signal transduction components, and epigenetic modifiers that promote pluripotency and self-renewal, including Pou5f1, Sox2 and Nanog (arrows). In addition, POU5F1, SOX2 and NANOG bind to the promotors of genes that are silent in ES cells, expression of which is associated with lineage commitment and differentiation (blunted arrows).
In the absence of LIF, activation of Stat3 is sufficient to maintain ES cell pluripotency, which is achieved by activation of the transcription factors KLF4 and SOX2 (Fig. 1; Niwa et al., 1998, 2009; Matsuda et al., 1999). In addition to activating Stat3, LIF sustains self-renewal and pluripotency by activating the PI(3)K-Akt pathway, which results in the elevated transcription of Tbx3 and Nanog (Fig. 1; Niwa et al., 2009). Overexpression of Klf4, Tbx3 (Niwa et al., 2009) or Nanog (Chambers et al., 2003; Mitsui et al., 2003) supports LIF-independent self-renewal of mouse ES cells. However, upon loss of Tbx3, Sox2 or Nanog, mouse ES cells have an increased propensity to differentiate (Chambers et al., 2003, 2007; Mitsui et al., 2003; Ivanova et al., 2006).
Epigenetics and pluripotency
Lineage segregation events in early embryo development are characterized by a progressive restriction in cellular plasticity of the newly formed cells. This increasing loss in developmental potential is established through epigenetic modifications, which are heritable non-genetic alterations that influence the transcriptional activity of genes. Through the acquisition of different epigenetic programmes, daughter cells of one fertilized oocyte can give rise to phenotypically distinct cell types even though these cells are genetically identical: the best-known examples of epigenetic modifications are DNA methylation and histone modifications.
DNA methylation
DNA methylation is established by the covalent binding of a methyl group to the five position of a cytosine residue often in the context of a cytosine followed by guanine (CpG) dinucleotide. DNA methylation is performed by a group of enzymes called DNA methyltransferases (Dnmts). De novo DNA methylation is accomplished by DNMT3a and DNMT3b (Okano et al., 1999). Upon DNA replication, DNA methylation patterns are copied to the newly synthesized DNA strand by Dnmt1, which ensures that the appropriate levels and patterns of DNA methylation are maintained after each cell division (Leonhardt et al., 1992; Li et al., 1992; Lei et al., 1996). Targeted deletion of DNMT3a and DNMT3b results in embryonic lethality, which demonstrates that de novo DNA methylation is vital for normal development (Okano et al., 1999).
DNA methylation patterns are dynamically regulated throughout early development and these changes are necessary to establish DNA methylation patterns that are consistent with the pluripotent phenotype of ICM cells of blastocyst stage embryos (Farthing et al., 2008). In the mouse, there is a rapid global decrease in DNA methylation of the paternal genome shortly after fertilization, preferentially in the Line1 family of transposons (Lane et al., 2003; Farthing et al., 2008), whereas DNA methylation patterns of the maternal genome decrease more gradually during the first cleavage divisions (Mayer et al., 2000). During the following cell divisions until the blastocyst stage, global methylation levels decrease further probably because at these stages DNMT1 is actively retained in the cytoplasm, thereby preventing maintenance of DNA methylation (Carlson et al., 1992; Rougier et al., 1998; Cardoso and Leonhardt, 1999). From the morula up to the blastocyst stages there is a cell-specific wave of de novo DNA methylation, which results in extensive DNA methylation in the ICM, but less DNA methylation in the trophectoderm of blastocyst stage embryos (Dean et al., 2001; Santos et al., 2002).
Epigenetic regulation through DNA methylation seems to play an important role in pluripotency. In mouse ES cells, genes that have promoters with high CpG content are generally hypomethylated, whereas genes with relatively low CpG content are hypermethylated (Meissner et al., 2008; Mohn et al., 2008). During in vitro differentiation of mouse ES cells, the majority of the promoters maintain their methylation level, except for pluripotency genes and germline-specific genes that gain methylation levels after differentiation (Farthing et al., 2008; Mohn et al., 2008).
The majority of promoter regions that are hypomethylated in mouse ES cells have similar low levels of methylation in sperm cells (Farthing et al., 2008). These regions appear to have been epigenetically prepared to participate in the development of a totipotent embryo. A notable exception is the DNA methylation pattern at the promoter regions of Nanog, which is hypermethylated in sperm cells (Imamura et al., 2006; Farthing et al., 2008). The paternal allele of the Nanog promoter is demethylated after fertilization to allow faithful NANOG expression in blastocyst stage embryos (Farthing et al., 2008).
DNA methylation dynamics in early human embryo development seem to deviate from those of the mouse, particularly at the expanded blastocyst stage. In human zygotes, the paternal pronucleus generally has lower DNA methylation levels than the maternal pronucleus, although not to the same extent as in the mouse (Beaujean et al., 2004; Fulka et al., 2004). Similar to the mouse, human embryos have progressively weaker DNA methylation patterns at later stages in preimplantation development. However, at the expanded blastocyst stage the pattern in the human is the inverse of that in the mouse, with higher DNA methylation levels in the trophectoderm versus the ICM (Fulka et al., 2004). It would be interesting to investigate whether these differences in methylation patterns contribute to the differences between human and mouse ES cells.
Imprinting
Imprinting is an epigenetic type of gene regulation established through DNA methylation that allows mono-allelic and parent-of-origin-dependent gene expression. Imprint marks are erased and reset during gametogenesis at differentially methylated DNA regions that serve as imprinting control regions (Hemberger et al., 2009). The association of defects in erasure, establishment and maintenance of imprints in a variety of human disorders and diseases underscores the importance of appropriate imprinting (Murphy and Jirtle, 2003). Imprinting plays a critical role regulating placental growth and suckling behaviour and thereby growth through nutrient partitioning between the mother and the fetus/baby (Reik and Walter, 2001). In general, maternally expressed imprinted genes limit fetal growth while paternally expressed imprinted genes promote fetal growth (Tada et al., 1998). The dynamics of imprint erasure and establishment of new imprints are important and dynamic epigenetic processes in germ cells (discussed below).
Histone modifications
The fundamental structure of chromatin is composed of spherical particles called nucleosomes that recur every 200 ± 40 base pairs (Oudet et al., 1975; McGhee and Felsenfeld, 1980; Luger et al., 1997). They are assembled from two copies each of histone proteins H2A, H2B, H3 and H4, around which 146 base pairs of DNA are wrapped in 1.67 left-handed superhelical turns (Luger et al., 1997). Importantly, the N-terminus of histone tails protrude from this nucleosome structure and can be chemically modified at certain residues by methylation, acetylation, phosphorylation, sumoylation or ubiquitination (Goldberg et al., 2007). These modifications have great impact on DNA accessibility and transcriptional activity.
In ES cells, a large number of lineage-specific genes are acetylated at lysine residue 9 of histone 3 (H3K9) and methylated at lysine 4 of histone 3 (H3K4). In addition to these markers of open chromatin, the same genes are marked with repressive tri-methylation at lysine residue 27 of histone 3 (H3K27me3; Azuara et al., 2006; Bernstein et al., 2006; Zhao et al., 2007). These opposing epigenetic modifications are called bivalent domains and it has been hypothesized that by these epigenetic modifications genes are primed for transcription, but elongation of transcripts is repressed (Azuara et al., 2006; Bernstein et al., 2006). Thus, bivalent domains may enable pluripotency of ES cells by silencing genes encoding developmentally important transcription factors while keeping them poised for activation (Bernstein et al., 2006).
In addition to the DNA methylation patterns that seem to prepare the DNA of sperm cells to participate in the developing embryo (Farthing et al., 2008), human sperm cells have histone modifications that also seem to prepare the sperm cells for future embryo development (Hammoud et al., 2009). For example, there is significantly more, H3K27me3 AT promoters of developmental genes in sperm and there is considerable overlap with the pattern of H3K27me3 in ES cells. Furthermore, bivalent genes have been detected in sperm cells that bear both the repressive mark H3K27me3 and the active mark H3K4me3 reminiscent of patterns in ES cells (Hammoud et al., 2009).
Chromatin remodelling complexes
Chromatin structure can be further modified through the activity of ATP-dependent complexes that affect nucleosome mobility and thereby may alter gene expression (Ho and Crabtree, 2010). Four families of ATP-dependent chromatin remodelling complexes can be recognized: SWI/SNF, ISWI, CHD and INO80 complexes (Ho and Crabtree, 2010).
Particular assemblies of chromatin remodelling complexes are essential for pluripotency of cells. For instance, in mouse ES cells, a SWI/SNF complex called esBAF (ES cell-specific Brahma-associated factor) is essential for self-renewal and pluripotency (Ho et al., 2009b). The core ATPase subunit of esBAF is BRG, which has a genome-wide high degree of overlap in binding affinity to target genes that are also bound by the core pluripotency network of transcription factors POU5F1, SOX2 and NANOG (Ho et al., 2009a).
Nucleosome-remodelling and histone deacetylase (NURD) complexes belong to the CHD family of chromatin remodelers. Like esBAF complexes, NURD complexes function as transcriptional repressors. Indeed, mouse ES cells from which the NURD-component MBD3 has been genetically deleted no longer require LIF for self-renewal, but they fail to commit to developmental lineages after differentiation is initiated (Kaji et al., 2006).
The pluripotency factors NANOG and POU5F1 interact with MTA1, a core component of NURD complexes, to form a unique deacetylase complex called NODE (NANOG and OCT4 associated deacetylase) (Liang et al., 2008). The activity of NODE is functionally distinct from MBD3 containing NURD complexes. Whereas MBD3-deficient ES cells fail to differentiate properly, mouse ES cells that lack the NODE component MTA1 differentiate even in the presence of LIF (Liang et al., 2008). These studies demonstrate that ATPase-dependent chromatin remodelling complexes are important for pluripotency of ES cells.
X chromosome inactivation
An epigenetic process that is strongly associated with loss of pluripotency in mice is X chromosome inactivation. X chromosome inactivation occurs in female cells to obtain a similar dosage of X-linked gene expression levels as male cells that have just one X chromosome. In early female embryo development of the mouse, the paternally inherited X chromosome is firstly inactivated from the 4-cell stage onward (Patrat et al., 2009). At the blastocyst stage, the paternally inherited X chromosome becomes reactivated in the ICM and, consequently, there are temporarily two active X chromosomes. Shortly thereafter, one of the active X chromosomes is randomly inactivated in the ICM (Mak et al., 2004; Okamoto et al., 2004). Random X chromosome inactivation is a stochastic process that starts with counting the number of X chromosomes and subsequent selection of the future active and inactive X chromosomes (Monkhorst et al., 2008). The non-coding RNA Xist plays an important role in X-inactivation. Xist RNA coats the inactive X chromosome and attracts protein complexes that are also required for the silencing process (Brockdorff et al., 1992; Brown et al., 1992; Zhao et al., 2008).
Comparable with the ICM cells from which they have been derived, female mouse ES cell lines have two active X chromosomes, but upon differentiation, one of the X chromosomes is randomly inactivated in each cell. Recently, mechanistic insight was provided for this negative correlation between Xist activity and pluripotency of cells. NANOG, POU5F1 and SOX2 can bind to intron 1 of Xist, thereby repressing the X-inactivating activity of this gene (Navarro et al., 2008). The up-regulation of the pluripotency factors NANOG, POU5F1 and SOX2 in early mouse embryo development may therefore lead to the inhibition of Xist and consequently to the reactivation of the paternal X chromosome. This is supported by the observation that female mouse embryos lacking NANOG fail to reactivate the inactive paternal X chromosome and do not develop pluripotent epiblast cells (Silva et al., 2009). Blastocysts that have experimentally induced high numbers of NANOG-positive cells also have more cells with two active X chromosomes than control embryos (Nichols et al., 2009b).
Other types of pluripotent stem cells
In addition to ES cells and EpiSCs, pluripotent stem cells have also been derived from a variety of other sources.
Embryonal carcinoma cells
Teratocarcinomas are a subset of germline tumours that have an additional component of malignant undifferentiated stem cells called embryonal carcinoma (EC) cells (Andrews, 1988). In males of the 129 mouse strain, ∼1% spontaneously develops teratocarcinomas (Stevens and Little, 1954), but teratoma formation can also be experimentally induced by subcutaneous or testicular injection of peri-implantation embryos into host mice (Kirby, 1963; Stevens, 1964, 1970). A subset of these induced tumours will be malignant and can be progressively re-transplanted into new hosts, demonstrating that they harbour cells that have self-renewal capacity (Stevens, 1970). Mouse EC cell lines have limited differentiation potential and contribute poorly to chimeric mice probably due to genetic aberrations, although some mouse EC cell lines can give rise to germline chimeras (Mintz and Illmensee, 1975). Importantly, in 1967 it was described how mouse EC cells can be cultured in vitro without loss of pluripotency (Finch and Ephrussi, 1967).
Human EC cell lines were first cultured as permanently transplantable xenografts in hamster cheek pouches (Pierce et al., 1957), but in the 1970s in vitro cultures were established (Fogh and Trempe, 1975; Hogan et al., 1977). The developmental potential of most human EC cell lines is poor, probably due to high degrees of aneuploidy (Andrews, 2002; Yu and Thomson, 2008). Nevertheless, human EC cell lines have been described with multilineage differentiation potential (Pera et al., 1989; Teshima et al., 1988) and several human EC lines such as TERA-2 can be induced to differentiate into multiple cell types by exposure to retinoic acid or by teratoma formation in mice (Andrews, 1984; Andrews et al., 1984). The pluripotent human EC cells are rather similar to human ES cells, exemplified by for instance high levels of alkaline phosphatase activity, expression of the cell surface antigens SSEA3 and SSEA4, expression of the transcription factors POU5F1 and NANOG, and the ability to differentiate towards cells that resemble trophectoderm (Andrews, 2002).
Embryonic germ cells
PGCs can also convert to pluripotent cells, when they are cultured in vitro on feeders in the presence of stem cell factor, LIF and bFGF (Matsui et al., 1992; Resnick et al., 1992).
In mouse development, the first identifiable PGC progenitors initially have a mesodermal character (McLaren and Lawson, 2005). However, this preliminary mesodermal character is gradually replaced by an expression pattern that, through the expression of some key pluripotency genes such as Sox2 (Yabuta et al., 2006) and Nanog (Yamaguchi et al., 2005) is reminiscent of ES cells and the pluripotent epiblast. Transcriptional similarities between PGCs and ES cells are maintained between E8.5 and E13.5 (Mise et al., 2008), which coincides with the developmental period during which EG cell lines can be derived (Matsui et al., 1992; Resnick et al., 1992; Labosky et al., 1994; Stewart et al., 1994; Durcova-Hills et al., 2006). NANOG is important for proper germ cell development as illustrated by the inability of Nanog−/− germ cells to survive beyond E11.5 (Chambers et al., 2007). POU5F1 is also expressed in PGCs and germ cell-specific loss of this factor results in premature apoptosis prior to colonization of the genital ridges (Kehler et al., 2004).
In the mouse, newly emerging and migratory germ cells undergo genome-wide epigenetic reprogramming events. For example, in newly formed PGCs of female embryos the inactive X chromosome is gradually reactivated between E7.5 and E12.5 (Chuva de Sousa Lopes et al., 2008; Sugimoto and Abe, 2007). Furthermore, between E7.5 and E8.5, a shift from H3K9me2 towards H3K27me3 can be observed (Seki et al., 2005; Hajkova et al., 2008). Strikingly, EG cells have thus far only been derived from PGCs beyond E8.5, which indicates that the epigenetic changes that take place before E8.5 are essential for the establishment of EG cells.
From E10.5, when mouse PGCs colonize the gonads, germ cells are subjected to DNA demethylation of imprinted genes and extensive histone modifications, by which the epigenome of the germline is reset (Hajkova et al., 2002, 2008). This process is essential for the germline to set-up gender-specific imprinting patterns later in development. In males, imprints are prenatally established in diploid gonocytes and in females imprints are established at the diplotene stage of meiosis (Murphy and Jirtle, 2003). Accordingly, mouse EG cell lines derived from E11.5 and E12.5 germ cells lack imprints of differentially methylated regions (Labosky et al., 1994; Tada et al., 1998). Chimeras made with these imprint-free EG cells display fetal overgrowth and skeletal abnormalities (Tada et al., 1998). In contrast, EG cells derived from E8.5 to E11.5 retain imprints and can contribute to healthy chimeras (Tada et al., 1998).
Several labs have reported the generation of human EG cell lines from gonadal germ cells of 5–9-week-old human embryos (Shamblott et al., 1998; Turnpenny et al., 2003; Liu et al., 2004). The efficiency of deriving new EG cell colonies correlates well with the number of POU5F1-positive cells in the fetal testis (Kerr et al., 2008). Human EG cell lines resemble mouse EG cells in morphology and growth requirements, but they are difficult to maintain undifferentiated and form EBs spontaneously in culture. As a result, long-term culture (>30 passages) of these cells has not been demonstrated.
Interestingly, in contrast to human ES cells, human EG cells do not have the capacity to generate teratomas in nude mice, even though human EG cells express the pluripotency markers POU5F1, NANOG, SSEA4, TRA-1-61, TRA-1-80 and alkaline phosphatase (Shamblott et al., 1998; Turnpenny et al., 2003, 2005; Liu et al., 2004). However, unlike human ES and EC cells, human EG cells are positive for SSEA-1 (Shamblott et al., 1998, 2001). Another difference between human ES cells and human EG cells is that the latter do not express SOX2 (Perrett et al., 2008). It is possible that human EG cells are incapable of self-renewal and have low differentiation potential in teratoma assays because they lack critical SOX2 expression. Since these cells endogenously express POU5F1 and NANOG, it would be interesting to examine if their developmental potency and self-renewal capacities can be enhanced by the introduction of exogenous SOX2.
Analogous with mice, derivation of EG cell lines from human PGCs at developmental stages before the colonization of the developing gonads might prove more successful for the establishment of EG cell lines that display self-renewal and can differentiate into all three germ layers.
Testis-derived multipotent stem cells
The germline seems to be particularly permissive to the derivation of pluripotent stem cells, because in addition to germ cell tumors and PGCs, SSCs are another potential source of pluripotent stem cell lines. SSCs originate from gonocytes that migrate to the basal membrane of the seminiferous tubules shortly after birth (de Rooij and Russell, 2000). Although SSCs are unipotent and can only differentiate to sperm cells in vivo, several groups have independently reported that in vitro cultured SSCs derived from neonate or adult mice gave rise to pluripotent cell colonies that resemble ES cells (Kanatsu-Shinohara et al., 2004; Guan et al., 2006; Seandel et al., 2007; Kanatsu-Shinohara et al., 2008; Ko et al., 2009). These SSC-derived pluripotent cells were given various names such as ES-like cells (Kanatsu-Shinohara et al., 2004), multipotent adult germline stem cells (maGSCs; Guan et al., 2006), and germline-derived pluripotent stem cells (gPS cells; Ko et al., 2009).
After testicular transplantation, cultured SSCs can restore spermatogenesis, whereas the pluripotent cells that have been derived from SSCs give rise to teratomas after transplantation (Ko et al., 2009).
Pluripotent stem cells derived from neonate mouse testis have different epigenetic properties from those derived from adult mouse testis. The androgenetic DNA methylation patterns of H19 and Igfr2 in pluripotent stem cells that have been derived from adult SSCs indicate that imprints are unaffected by the conversion (Ko et al., 2009). In contrast, pluripotent stem cells that have been derived from neonatal mouse testis have largely erased imprinting patterns reminiscent of gonocytes (Kanatsu-Shinohara et al., 2004), suggesting that ES-like cells from neonate mouse testis originate from germ cells that had not yet been subjected to imprinting reset.
Various recent reports have independently shown that pluripotent stem cells can also be generated from biopsies of human testes (Conrad et al., 2008; Golestaneh et al., 2009; Kossack et al., 2009; Mizrak et al., 2009). These cells share characteristics with human ES cells such as the expression of various markers for pluripotency, high telomerase activity, in vitro differentiation potential and the capacity to form teratomas with derivatives of the three germ layers upon subcutaneous injection into immunocompromised mice (Conrad et al., 2008; Golestaneh et al., 2009; Kossack et al., 2009; Mizrak et al., 2009).
Importantly, it has been described that pluripotent stem cells derived from human SSCs are not capable of giving rise to teratomas at similar efficiencies as human ES cells, which could indicate that not all cells had been fully converted to pluripotency (Golestaneh et al., 2009; Kossack et al., 2009; Mizrak et al., 2009).
Reprogramming of somatic cells
In addition to the germline and ICM origins of pluripotent stem cell lines described above, various methods have been described that can be used to reprogramme somatic cells to a pluripotent state. These techniques are particularly interesting for regenerative medicine purposes, because they can potentially be used to generate pluripotent stem cell lines that are immunocompatible with the patient. Such patient-specific stem cell lines would circumvent the risk that grafted cells are rejected after transplantation. In addition, patient-specific stem cells could be useful to study the aetiology of disease and for drug development.
One approach for the derivation of patient-specific pluripotent stem cell lines is fusion of somatic cells with pluripotent cells. In the mouse, it has been demonstrated that somatic cells can be reprogrammed to a pluripotent state by fusion with EC, ES or EG cells (Miller and Ruddle, 1976, 1977; Tada et al., 1997, 2001). Similarly, human somatic cells have been reprogrammed to pluripotency through fusion with ES cells. The pluripotent developmental potential of these hybrid cell lines has been confirmed in vitro by embryoid body formation and in vivo by teratoma formation (Cowan et al., 2005; Yu et al., 2006). However, because these hybrid cells are tetraploid, they are unlikely candidates for cell therapies.
Patient-specific pluripotent stem cell lines can, in theory, also be derived through ‘therapeutic cloning’. With this technique, somatic cells from patients are used in somatic cell nuclear transfer (SCNT). The resulting embryos can subsequently be used for the derivation of ES cell lines. However, ‘therapeutic cloning’ has proved to be difficult and so far no true human ES cell lines of SCNT-origin have been successfully generated. Owing to these various ethical and practical difficulties and due to the development of a promising new technique to reprogramme somatic cells, the interest for therapeutic cloning has declined over the past few years.
The new development that is mainly responsible for the diminishing interest in therapeutic cloning is the discovery that somatic cells (e.g. fibroblasts and mature B lymphocytes) can be reprogrammed to pluripotency upon ectopic induction of the transcription factor genes c-Myc, Klf4, Pou5f1 and Sox2 to form iPS cells (Takahashi and Yamanaka, 2006; Hanna et al., 2008). Importantly, mouse iPS cells have passed all tests of pluripotency including the stringent tetraploid complementation assay (Boland et al., 2009; Kang et al., 2009; Zhao et al., 2009).
The therapeutic potential of iPS cells has been demonstrated in a humanized sickle cell anemia mouse model. Firstly, the human sickle haemoglobin allele was corrected by gene-specific targeting of iPS cells derived from this mouse model. Haematopoietic progenitors that were subsequently derived from these corrected iPS cells could reconstitute the haematopoietic system of the sickle cell anemia mice and correct the disease phenotype (Hanna et al., 2007).
Mouse iPS cells reprogrammed with c-Myc, Klf4, Pou5f1 and Sox2 exhibit a high degree of tumorigenicity in chimeras and their progeny through reactivation of the oncogenic transgene C-MYC (Okita et al., 2007). The carcinogenic potential of iPS-derived cells is a major obstacle for applications such as cell therapies. However, more recent studies have conveniently demonstrated that C-MYC is dispensable for the reprogramming process (Nakagawa et al., 2008; Wernig et al., 2008). Moreover, neural stem cells endogenously express SOX2, KLF4 and C-MYC and can be reprogrammed to pluripotency with a single exogenous factor, namely POU5F1 (Kim et al., 2009). It is questionable whether reprogramming of the somatic epigenome is uniquely dependent on these three/four factors, or that there is perhaps a larger set of factors that in various combinations can lead to nuclear reprogramming.
The ectopic expression of the four reprogramming factors resets the epigenome of somatic cells to one that highly resembles that of pluripotent stem cells (Maherali et al., 2007). For example, female mouse iPS cells have reactivated their silenced X chromosome and upon differentiation one of the two X chromosomes is again silenced (Maherali et al., 2007). Furthermore, in contrast with their somatic antecedents, the Nanog and Pou5f1-promoter regions are largely unmethylated in iPS cells (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). Moreover, iPS cells have global H3K4me3 and H3K27me3 resembling the bivalent domains of mouse ES cells (Maherali et al., 2007; Wernig et al., 2007, Yu et al., 2009).
Various other combinations of reprogramming factors have been used for the generation of iPS cell lines and certain combinations seem to yield better quality iPS cells than others in terms of their in vivo developmental potential. For example, mouse cells reprogrammed with Pou5f1, Sox2, Klf4 and Tbx3 have higher germline transmission frequencies in chimaeras than iPS cells that were reprogrammed with Pou5f1, Sox2 and Klf4 (Han et al., 2010).
Mouse ES cells are more efficient in giving rise to viable mice in tetraploid complementation assays than genetically matched iPS cells are (Stadtfeld et al., 2010). The transcriptional differences between mouse ES cells and genetically identical iPS cells mainly originate from the imprinted Dlk1–Dio3 gene cluster on chromosome 12, which is maternally expressed. In most iPS cells, this gene cluster is silenced and only in a small proportion of iPS cells the activity of genes on this cluster is similar to that of ES cells. The activity of genes on this gene cluster is predictive for the developmental potential of iPS cells in chimeras and in tetraploid complementation assays (Stadtfeld et al., 2010). It seems that in iPS cells this gene cluster is silenced during the reprogramming process, by an as yet unknown mechanism.
Human iPS cells have been generated by lentiviral transduction of the same reprogramming factors used to create mouse iPS cells (POU5F1, SOX2, KLF4 and C-MYC; Takahashi et al., 2007; Yu et al., 2007) and with a subset of these factors in combination with NANOG and LIN28 (Li et al., 2009). Human iPS cells resemble human ES cells in growth conditions, colony morphology, transcriptome, expression of cell surface antigens, and potential to differentiate into all three germ layers in in vitro differentiation and teratoma assays. The fact that the same genes can induce pluripotency in human and mouse suggests conservation between species of the transcriptional network of pluripotency even though the extrinsic factors and signals that maintain pluripotency of iPS cells are not conserved between mouse and human.
Although human iPS cells closely resemble human ES cells, iPS cells have unique gene expression signatures that distinguish them from ES cells (Chin et al., 2009). Human iPS cells have been reported to bear residual gene expression from the donor cell type used in the reprogramming process (Marchetto et al., 2009; Ghosh et al., 2010). Genes involved in the differentiated state of the donor cells are not completely turned off after reprogramming and there is an incomplete induction of genes that are associated with pluripotency of stem cells. Moreover, various imprinted genes have aberrant expression levels in some human iPS cell lines as compared with human ES cells (Pick et al., 2009).
In addition to transcriptional differences between human ES cells and human iPS cells, there are also indications of functional differences between these cell types. For example, reduced efficiencies have been reported for the differentiation of iPS cells towards retinal and hemangioblastic cell types (Meyer et al., 2009; Feng et al., 2010). Moreover, differentiated derivatives from human iPS cells have been reported to have limited growth rates, be more apoptotic, and senesce earlier when compared with differentiated derivatives from human ES cells (Feng et al., 2010).
Comparing human ES cells with genetically matched human iPS cells will shed light on the question of reported differences between human iPS cells and ES cells originating from differences in genetic background and the presence of viral transgenes, or faithfully representing fundamental differences between these pluripotent cell types. Furthermore, such an approach will elucidate whether imprinted genes are also aberrantly silenced in human iPS cells, as is the case for mouse iPS cells (Stadtfeld et al., 2010).
Indubitably, iPS cells have been and will be extremely valuable for stem cell research and our understanding of pluripotency, but the current apparent differences between human iPS cells and human ES cells make it questionable whether iPS cells are suitable for regenerative medicine purposes at this time.
Conclusion
The study of pluripotent stem cells has progressed dramatically since the first derivation and culture of mouse ES cells in 1981. It has now become clear that various tissues can give rise to pluripotent stem cells in culture. Unexpectedly, it has even become possible to reprogram differentiated somatic cells to a pluripotent state. Although the ultimate goal of using pluripotent stem cells for regenerative medicine still seems a long way off, these cells can and already are being used in toxicity screens and drug screening/discovery experiments. In particular, the use of disease-specific stem cells can be informative in these types of experiments.
Despite the similarities between the various pluripotent stem cell types, there are important discrepancies, such as differences in transcriptional activity, developmental potential and epigenetic regulation. Dependent on the application, this variance could have significant effects on the outcome, for example in drug toxicity screens, directed differentiation experiments or future regenerative medicine. An important goal for the near future is to understand the underlying causes of the differences between the various pluripotent stem cell types. It is possible that we can use this knowledge to reprogramme somatic cells to a pluripotent state analogous to that of the pluripotent cells in blastocyst stage embryos.
The degree to which human pluripotent stem cells are indeed truly pluripotent cannot be established as thoroughly as is possible for mouse pluripotent stem cells, which makes it in some aspects difficult to compare these species. Remarkably, establishing ES cells from other mammalian species has proved to be more difficult, which could result from embryonic differences between mammals (Keefer et al., 2007; Kuijk et al., 2008). It would be helpful if more knowledge could be obtained on the mechanisms that are important for self-renewal in the various stem cells types, as this could help the successful derivation of pluripotent cell types from mammals other than mice and humans. Indeed generation of iPS cells from various mammalian species might give new insights into the mechanisms responsible for self-renewal and pluripotency.
Funding
SCDSL was funded by the Nederlands Organization for Scientific Research (VENI 916.76.015 grant), N.G. was funded by the Dutch Science Organization NWO and the National Institutes of Health.
References
- Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–293. doi: 10.1016/0012-1606(84)90316-6. doi:10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- Andrews PW. Human teratocarcinomas. Biochim Biophys Acta. 1988;948:17–36. doi: 10.1016/0304-419x(88)90003-0. [DOI] [PubMed] [Google Scholar]
- Andrews PW. From teratocarcinomas to embryonic stem cells. Philos Trans R Soc Lond B Biol Sci. 2002;357:405–417. doi: 10.1098/rstb.2002.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews PW, Damjanov I, Simon D, Banting GS, Carlin C, Dracopoli NC, Fogh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. doi:10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. doi:10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. doi:10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. doi:10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, Meehan R, Young L. Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol. 2004;14:R266–R267. doi: 10.1016/j.cub.2004.03.019. doi:10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. doi:10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. doi:10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. doi:10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. doi:10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. doi:10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. doi:10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. doi:10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. doi:10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. doi:10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Cauffman G, De RM, Sermon K, Liebaers I, Van d V. Markers that define stemness in ESC are unable to identify the totipotent cells in human preimplantation embryos. Hum Reprod. 2009;24:63–70. doi: 10.1093/humrep/den351. doi:10.1093/humrep/den351. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. doi:10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. doi:10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. doi:10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. doi:10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen AE, Egli D, Niakan K, Deng J, Akutsu H, Yamaki M, Cowan C, Fitz-Gerald C, Zhang K, Melton DA, et al. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–106. doi: 10.1016/j.stem.2008.12.001. doi:10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, Tortorice CG, Cardiff RD, Cross JC, Muller WJ, et al. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95:793–803. doi: 10.1016/s0092-8674(00)81702-x. doi:10.1016/S0092-8674(00)81702-X. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. doi:10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YF, Chen HH, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, Lu J, McKay RD, Geijsen N. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. doi:10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Klimanskaya I, Becker S, Marh J, Lu SJ, Johnson J, Meisner L, Lanza R. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature. 2006;439:216–219. doi: 10.1038/nature04277. doi:10.1038/nature04277. [DOI] [PubMed] [Google Scholar]
- Chung Y, Klimanskaya I, Becker S, Li T, Maserati M, Lu SJ, Zdravkovic T, Ilic D, Genbacev O, Fisher S, et al. Human embryonic stem cell lines generated without embryo destruction. Cell Stem Cell. 2008;2:113–117. doi: 10.1016/j.stem.2007.12.013. doi:10.1016/j.stem.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes S, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008;4:e30. doi: 10.1371/journal.pgen.0040030. doi:10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S, Renninger M, Hennenlotter J, Wiesner T, Just L, Bonin M, Aicher W, Buhring HJ, Mattheus U, Mack A, et al. Generation of pluripotent stem cells from adult human testis. Nature. 2008;456:344–349. doi: 10.1038/nature07404. doi:10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. doi:10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. doi:10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaise F, Bralion V, Schuurbiers N, Dessy F. Establishment of an embryonic stem cell line from 8-cell stage mouse embryos. Eur J Morphol. 1996;34:237–243. doi: 10.1076/ejom.34.4.237.13046. doi:10.1076/ejom.34.4.237.13046. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. doi:10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G, Adams IR, Barton SC, Surani MA, McLaren A. The role of exogenous fibroblast growth factor-2 on the reprogramming of primordial germ cells into pluripotent stem cells. Stem Cells. 2006;24:1441–1449. doi: 10.1634/stemcells.2005-0424. doi:10.1634/stemcells.2005-0424. [DOI] [PubMed] [Google Scholar]
- Eistetter HR. Pluripotent embryonal stem cell lines can be established from disaggregated mouse morulae. Dev Growth Differ. 1989;31:275–282. doi: 10.1111/j.1440-169X.1989.00275.x. doi:10.1111/j.1440-169X.1989.00275.x. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. doi:10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. doi:10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feki A, Bosman A, Dubuisson JB, Irion O, Dahoun S, Pelte MF, Hovatta O, Jaconi ME. Derivation of the first Swiss human embryonic stem cell line from a single blastomere of an arrested four-cell stage embryo. Swiss Med Wkly. 2008;138:540–550. doi: 10.4414/smw.2008.12385. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. doi:10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010 doi: 10.1002/stem.321. doi:10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Finch BW, Ephrussi B. Retention of multiple developmental potentialities by cells of a mouse testicular teratocarcinoma during prolonged culture in vitro and their extinction upon hybridization with cells of permanent lines. Proc Natl Acad Sci USA. 1967;57:615–621. doi: 10.1073/pnas.57.3.615. doi:10.1073/pnas.57.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh J, Trempe G. New human tumor cell lines. In: Fogh J, editor. Human Tumor Cells In Vitro. New York: Plenum; 1975. pp. 115–159. [Google Scholar]
- Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;26:1931–1938. doi: 10.1634/stemcells.2007-1002. doi:10.1634/stemcells.2007-1002. [DOI] [PubMed] [Google Scholar]
- Fulka H, Mrazek M, Tepla O, Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. doi:10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- Geens M, Mateizel I, Sermon K, De RM, Spits C, Cauffman G, Devroey P, Tournaye H, Liebaers I, Van d V. Human embryonic stem cell lines derived from single blastomeres of two 4-cell stage embryos. Hum Reprod. 2009;24:2709–2717. doi: 10.1093/humrep/dep262. doi:10.1093/humrep/dep262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. doi:10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. doi:10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Golestaneh N, Kokkinaki M, Pant D, Jiang J, Destefano D, Fernandez-Bueno C, Rone JD, Haddad BR, Gallicano GI, Dym M. Pluripotent stem cells derived from adult human testes. Stem Cells Dev. 2009;8:1115–1126. doi: 10.1089/scd.2008.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. doi:10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. doi:10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. doi:10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. doi:10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki T, Kehoe SM, Nakano T, Terada N. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol Cell Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. doi:10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. doi:10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. doi:10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. doi:10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136:3549–3556. doi: 10.1242/dev.037747. doi:10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. doi:10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;8:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. doi:10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009a;106:5187–5191. doi: 10.1073/pnas.0812888106. doi:10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009b;106:5181–5186. doi: 10.1073/pnas.0812889106. doi:10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Fellous M, Avner P, Jacob F. Isolation of a human teratoma cell line which expresses F9 antigen. Nature. 1977;270:515–518. doi: 10.1038/270515a0. doi:10.1038/270515a0. [DOI] [PubMed] [Google Scholar]
- Humphrey RK, Beattie GM, Lopez AD, Bucay N, King CC, Firpo MT, Rose-John S, Hayek A. Maintenance of pluripotency in human embryonic stem cells is STAT3 independent. Stem Cells. 2004;22:522–530. doi: 10.1634/stemcells.22-4-522. doi:10.1634/stemcells.22-4-522. [DOI] [PubMed] [Google Scholar]
- Hyslop L, Stojkovic M, Armstrong L, Walter T, Stojkovic P, Przyborski S, Herbert M, Murdoch A, Strachan T, Lako M. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells. 2005;23:1035–1043. doi: 10.1634/stemcells.2005-0080. doi:10.1634/stemcells.2005-0080. [DOI] [PubMed] [Google Scholar]
- Ilic D, Giritharan G, Zdravkovic T, Caceres E, Genbacev O, Fisher SJ, Krtolica A. Derivation of human embryonic stem cell lines from biopsied blastomeres on human feeders with minimal exposure to xenomaterials. Stem Cells Dev. 2009;18:1343–1350. doi: 10.1089/scd.2008.0416. doi:10.1089/scd.2008.0416. [DOI] [PubMed] [Google Scholar]
- Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. doi:10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, Decoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;7102:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. doi:10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295:90–102. doi: 10.1016/j.ydbio.2006.03.026. doi:10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. doi:10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. doi:10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. doi:10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Lee J, Inoue K, Ogonuki N, Miki H, Toyokuni S, Ikawa M, Nakamura T, Ogura A, Shinohara T. Pluripotency of a single spermatogonial stem cell in mice. Biol Reprod. 2008;78:681–687. doi: 10.1095/biolreprod.107.066068. doi:10.1095/biolreprod.107.066068. [DOI] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. doi:10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Keefer CL, Pant D, Blomberg L, Talbot NC. Challenges and prospects for the establishment of embryonic stem cell lines of domesticated ungulates. Anim Reprod Sci. 2007;98:147–168. doi: 10.1016/j.anireprosci.2006.10.009. doi:10.1016/j.anireprosci.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin KJ, Scholer HR, et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. doi:10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr CL, Hill CM, Blumenthal PD, Gearhart JD. Expression of pluripotent stem cell markers in the human fetal testis. Stem Cells. 2008;26:412–421. doi: 10.1634/stemcells.2007-0605. doi:10.1634/stemcells.2007-0605. [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, rauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Kirby D. The development of mouse blastocysts transplanted to the scrotal and cryptorchid testis. J Anat. 1963;97:119–130. [PMC free article] [PubMed] [Google Scholar]
- Klimanskaya I, Chung Y, Becker S, Lu SJ, Lanza R. Human embryonic stem cell lines derived from single blastomeres. Nature. 2006;444:481–485. doi: 10.1038/nature05142. doi:10.1038/nature05142. [DOI] [PubMed] [Google Scholar]
- Klimanskaya I, Chung Y, Becker S, Lu SJ, Lanza R. Derivation of human embryonic stem cells from single blastomeres. Nat Protoc. 2007;2:1963–1972. doi: 10.1038/nprot.2007.274. doi:10.1038/nprot.2007.274. [DOI] [PubMed] [Google Scholar]
- Ko K, Tapia N, Wu G, Kim JB, Bravo MJ, Sasse P, Glaser T, Ruau D, Han DW, Greber B, et al. Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell. 2009;5:87–96. doi: 10.1016/j.stem.2009.05.025. doi:10.1016/j.stem.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Kossack N, Meneses J, Shefi S, Nguyen HN, Chavez S, Nicholas C, Gromoll J, Turek PJ, Reijo-Pera RA. Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells. 2009;27:138–149. doi: 10.1634/stemcells.2008-0439. doi:10.1634/stemcells.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Kuijk EW, Du PL, van Tol HT, Oei CH, Haagsman HP, Colenbrander B, Roelen BA. Differences in early lineage segregation between mammals. Dev Dyn. 2008;237:918–927. doi: 10.1002/dvdy.21480. doi:10.1002/dvdy.21480. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Barlow DP, Hogan BL. Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development. 1994;120:3197–3204. doi: 10.1242/dev.120.11.3197. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. doi:10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. doi:10.1016/0092-8674(92)90561-P. [DOI] [PubMed] [Google Scholar]
- Lerou PH, Yabuuchi A, Huo H, Takeuchi A, Shea J, Cimini T, Ince TA, Ginsburg E, Racowsky C, Daley GQ. Human embryonic stem cell derivation from poor-quality embryos. Nat Biotechnol. 2008;26:212–214. doi: 10.1038/nbt1378. doi:10.1038/nbt1378. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. doi:10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- Li M, Sendtner M, Smith A. Essential function of LIF receptor in motor neurons. Nature. 1995;378:724–727. doi: 10.1038/378724a0. doi:10.1038/378724a0. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. doi:10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. doi:10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. doi:10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu H, Pan Y, Tang S, Xiong J, Hui N, Wang S, Qi Z, Li L. Human embryonic germ cells isolation from early stages of post-implantation embryos. Cell Tissue Res. 2004;318:525–531. doi: 10.1007/s00441-004-0990-7. doi:10.1007/s00441-004-0990-7. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. doi:10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lorthongpanich C, Yang SH, Piotrowska-Nitsche K, Parnpai R, Chan AW. Development of single mouse blastomeres into blastocysts, outgrowths and the establishment of embryonic stem cells. Reproduction. 2008;135:805–813. doi: 10.1530/REP-07-0478. doi:10.1530/REP-07-0478. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. doi:10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de NM, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. doi:10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. doi:10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. doi:10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. doi:10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, Moore HD, Andrews PW. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–668. doi: 10.1634/stemcells.22-5-659. doi:10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. doi:10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. doi:10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. doi:10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. doi:10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McLaren A, Lawson KA. How is the mouse germ-cell lineage established? Differentiation. 2005;73:435–437. doi: 10.1111/j.1432-0436.2005.00049.x. doi:10.1111/j.1432-0436.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Shearer RL, Capowski EE, Wright LS, Wallace KA, McMillan EL, Zhang SC, Gamm DM. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Ruddle FH. Pluripotent teratocarcinoma-thymus somatic cell hybrids. Cell. 1976;9:45–55. doi: 10.1016/0092-8674(76)90051-9. doi:10.1016/0092-8674(76)90051-9. [DOI] [PubMed] [Google Scholar]
- Miller RA, Ruddle FH. Teratocarcinoma X friend erythroleukemia cell hybrids resemble their pluripotent embryonal carcinoma parent. Dev Biol. 1977;56:157–173. doi: 10.1016/0012-1606(77)90159-2. doi:10.1016/0012-1606(77)90159-2. [DOI] [PubMed] [Google Scholar]
- Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci USA. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. doi:10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise N, Fuchikami T, Sugimoto M, Kobayakawa S, Ike F, Ogawa T, Tada T, Kanaya S, Noce T, Abe K. Differences and similarities in the developmental status of embryo-derived stem cells and primordial germ cells revealed by global expression profiling. Genes Cells. 2008;13:863–877. doi: 10.1111/j.1365-2443.2008.01211.x. doi:10.1111/j.1365-2443.2008.01211.x. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. doi:10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. doi:10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Mizrak SC, Chikhovskaya JV, Sadri-Ardekani H, van Daalen S, Korver CM, Hovingh SE, Roepers-Gajadien HL, Raya A, Fluiter K, de Reijke TM, et al. Embryonic stem cell-like cells derived from adult human testis. Hum Reprod. 2009;25:158–167. doi: 10.1093/humrep/dep354. doi:10.1093/humrep/dep354. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. doi:10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. doi:10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. doi:10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Jirtle RL. Imprinting evolution and the price of silence. Bioessays. 2003;25:577–588. doi: 10.1002/bies.10277. doi:10.1002/bies.10277. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, bramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. doi:10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci. 1999;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. doi:10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. doi:10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- Nayernia K, Nolte J, Michelmann HW, Lee JH, Rathsack K, Drusenheimer N, Dev A, Wulf G, Ehrmann IE, Elliott DJ, et al. In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev Cell. 2006;11:125–132. doi: 10.1016/j.devcel.2006.05.010. doi:10.1016/j.devcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. doi:10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nichols J, Chambers I, Taga T, Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128:2333–2339. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- Nichols J, Jones K, Phillips JM, Newland SA, Roode M, Mansfield W, Smith A, Cooke A. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat Med. 2009a;15:814–818. doi: 10.1038/nm.1996. doi:10.1038/nm.1996. [DOI] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009b;19:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. doi:10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. doi:10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. doi:10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature genetics. 2000;24:372–376. doi: 10.1038/74199. doi:10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. doi:10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. doi:10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. doi:10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. doi:10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. doi:10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Oudet P, Gross-Bellard M, Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4:281–300. doi: 10.1016/0092-8674(75)90149-x. doi:10.1016/0092-8674(75)90149-X. [DOI] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. doi:10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le BP, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. doi:10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Cooper S, Mills J, Parrington JM. Isolation and characterization of a multipotent clone of human embryonal carcinoma cells. Differentiation. 1989;42:10–23. doi: 10.1111/j.1432-0436.1989.tb00602.x. doi:10.1111/j.1432-0436.1989.tb00602.x. [DOI] [PubMed] [Google Scholar]
- Perrett RM, Turnpenny L, Eckert JJ, O'Shea M, Sonne SB, Cameron IT, Wilson DI, Meyts ER, Hanley NA. The early human germ cell lineage does not express SOX2 during in vivo development or upon in vitro culture. Biol Reprod. 2008;78:852–858. doi: 10.1095/biolreprod.107.066175. doi:10.1095/biolreprod.107.066175. [DOI] [PubMed] [Google Scholar]
- Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. doi:10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- Pierce B, Verney EL, Dixon FJ. The biology of testicular cancer. I. Behavior after transplantation. Cancer Res. 1957;17:134–138. [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. doi:10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. doi:10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. doi:10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Rossant J, Chazaud C, Yamanaka Y. Lineage allocation and asymmetries in the early mouse embryo. Philos Trans R Soc Lond B Biol Sci. 2003;358:1341–1348. doi: 10.1098/rstb.2003.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. doi:10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. doi:10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Sasaki E, Hanazawa K, Kurita R, Akatsuka A, Yoshizaki T, Ishii H, Tanioka Y, Ohnishi Y, Suemizu H, Sugawara A, et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus) Stem Cells. 2005;23:1304–1313. doi: 10.1634/stemcells.2004-0366. doi:10.1634/stemcells.2004-0366. [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. doi:10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. doi:10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci USA. 1998;95:13726–13731. doi: 10.1073/pnas.95.23.13726. doi:10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamblott MJ, Axelman J, Littlefield JW, Blumenthal PD, Huggins GR, Cui Y, Cheng L, Gearhart JD. Human embryonic germ cell derivatives express a broad range of developmentally distinct markers and proliferate extensively in vitro. Proc Natl Acad Sci USA. 2001;98:113–118. doi: 10.1073/pnas.021537998. doi:10.1073/pnas.021537998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Masui S, Sharova LV, Piao Y, Aiba K, Matoba R, Xin L, Niwa H, Ko MS. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. doi:10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. doi:10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. doi:10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LC. Experimental production of testicular teratomas in mice. Proc Natl Acad Sci USA. 1964;52:654–661. doi: 10.1073/pnas.52.3.654. doi:10.1073/pnas.52.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LC. The development of transplantable teratocarcinomas from intratesticular grafts of pre- and postimplantation mouse embryos. Dev Biol. 1970;21:364–382. doi: 10.1016/0012-1606(70)90130-2. doi:10.1016/0012-1606(70)90130-2. [DOI] [PubMed] [Google Scholar]
- Stevens LC, Little CC. Spontaneous testicular teratomas in an inbred strain of mice. Proc Natl Acad Sci USA. 1954;40:1080–1087. doi: 10.1073/pnas.40.11.1080. doi:10.1073/pnas.40.11.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. doi:10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Gadi I, Bhatt H. Stem cells from primordial germ cells can reenter the germ line. Dev Biol. 1994;161:626–628. doi: 10.1006/dbio.1994.1058. [DOI] [PubMed] [Google Scholar]
- Strelchenko N, Verlinsky O, Kukharenko V, Verlinsky Y. Morula-derived human embryonic stem cells. Reprod Biomed Online. 2004;9:623–629. doi: 10.1016/s1472-6483(10)61772-5. doi:10.1016/S1472-6483(10)61772-5. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. doi:10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Suemori H, Tada T, Torii R, Hosoi Y, Kobayashi K, Imahie H, Kondo Y, Iritani A, Nakatsuji N. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev Dyn. 2001;222:273–279. doi: 10.1002/dvdy.1191. doi:10.1002/dvdy.1191. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. doi:10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol. 2008;322:133–144. doi: 10.1016/j.ydbio.2008.07.019. doi:10.1016/j.ydbio.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. doi:10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Tada M, Hilton K, Barton SC, Sado T, Takagi N, Surani MA. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol. 1998;207:551–561. doi: 10.1007/s004270050146. doi:10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. doi:10.1016/S0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. doi:10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. doi:10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tesar PJ. Derivation of germ-line-competent embryonic stem cell lines from preblastocyst mouse embryos. Proc Natl Acad Sci USA. 2005;102:8239–8244. doi: 10.1073/pnas.0503231102. doi:10.1073/pnas.0503231102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. doi:10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Teshima S, Shimosato Y, Hirohashi S, Tome Y, Hayashi I, Kanazawa H, Kakizoe T. Four new human germ cell tumor cell lines. Lab Invest. 1988;59:328–336. [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci USA. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. doi:10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Hearn JP. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod. 1996;55:254–259. doi: 10.1095/biolreprod55.2.254. doi:10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. doi:10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. doi:10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Turnpenny L, Brickwood S, Spalluto CM, Piper K, Cameron IT, Wilson DI, Hanley NA. Derivation of human embryonic germ cells: an alternative source of pluripotent stem cells. Stem Cells. 2003;21:598–609. doi: 10.1634/stemcells.21-5-598. doi:10.1634/stemcells.21-5-598. [DOI] [PubMed] [Google Scholar]
- Turnpenny L, Cameron IT, Spalluto CM, Hanley KP, Wilson DI, Hanley NA. Human embryonic germ cells for future neuronal replacement therapy. Brain Res Bull. 2005;68:76–82. doi: 10.1016/j.brainresbull.2005.08.014. doi:10.1016/j.brainresbull.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. doi:10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- van de Velde H, Cauffman G, Tournaye H, Devroey P, Liebaers I. The four blastomeres of a 4-cell stage human embryo are able to develop individually into blastocysts with inner cell mass and trophectoderm. Hum Reprod. 2008;23:1742–1747. doi: 10.1093/humrep/den190. doi:10.1093/humrep/den190. [DOI] [PubMed] [Google Scholar]
- Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, et al. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. doi:10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. doi:10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wilder PJ, Kelly D, Brigman K, Peterson CL, Nowling T, Gao QS, McComb RD, Capecchi MR, Rizzino A. Inactivation of the FGF-4 gene in embryonic stem cells alters the growth and/or the survival of their early differentiated progeny. Dev Biol. 1997;192:614–629. doi: 10.1006/dbio.1997.8777. doi:10.1006/dbio.1997.8777. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. doi:10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. doi:10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. doi:10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. doi:10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. Nanog expression in mouse germ cell development. Gene Expr Patterns. 2005;5:639–646. doi: 10.1016/j.modgep.2005.03.001. doi:10.1016/j.modgep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. doi:10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. doi:10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. doi:10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K, Shoyab M, Taga T, Kishimoto T. Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech Dev. 1994;45:163–171. doi: 10.1016/0925-4773(94)90030-2. doi:10.1016/0925-4773(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22:1987–1997. doi: 10.1101/gad.1689808. doi:10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, He P, Slukvin II, Thomson JA. Human embryonic stem cells reprogram myeloid precursors following cell–cell fusion. Stem Cells. 2006;24:168–176. doi: 10.1634/stemcells.2005-0292. doi:10.1634/stemcells.2005-0292. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, ntosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. doi:10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. doi:10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. doi:10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. doi:10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. doi:10.1038/nature08267. [DOI] [PubMed] [Google Scholar]