Abstract
Atrial fibrillation (AF) is the most common cardiac rhythm abnormality and represents a major burden, both to patients and to health-care systems. In recent years, increasing evidence from population-based studies has demonstrated that AF is a heritable condition. Although familial forms of AF have been recognized for many years, they represent a rare subtype of the arrhythmia. However, despite their limited prevalence, the identification of mutations in monogenic AF kindreds has provided valuable insights into the molecular pathways underlying the arrhythmia and a framework for investigating AF encountered in the general population. In contrast to these rare families, the typical forms of AF occurring in the community are likely to be multigenic and have significant environmental influences. Recently, genome-wide association studies have uncovered common sequence variants that confer increased susceptibility to the arrhythmia. In the future, the elucidation of the genetic substrate underlying both familial and more typical forms of AF will hopefully lead to the development of novel diagnostic tools as well as more targeted rhythm control strategies. In this article, we will focus on monogenic forms of AF and also provide an overview of case–control association studies for AF.
Keywords: Atrial fibrillation, Mutation, Family, Genetics
1. Introduction
Atrial fibrillation (AF) is the most prevalent cardiac rhythm abnormality and is a major cause of morbidity and mortality.1 Previous studies have reported a significant increase in the risk of stroke, dementia, heart failure, and mortality associated with AF.1–6 Established risk factors for AF include impaired left ventricular function, valvular heart disease, hypertension, and advancing age.6–9 In a minority of cases, AF occurs in the absence of these risk factors, a disease subtype referred to as lone AF.
Monogenic AF families have been recognized for many years. In 1943, Wolff10 described a case of three brothers with AF. In the ensuing years, larger kindreds with heritable AF have been identified and studied. Investigators have used linkage analysis to identify a number of loci for familial AF.11,12 In addition, mutations in both ion channel coding genes13–18 and non-ion channel coding genes19–21 have been reported. AF has also been described as a concomitant disease in patients with inherited arrhythmia syndromes such as the Brugada syndrome22 and long QT syndrome23,24 (LQTS) and in patients with familial cardiomyopathies such as hypertrophic cardiomyopathy and dilated cardiomyopathy (DCM).25,26
AF has traditionally been perceived as a predominantly sporadic condition with rare familial subtypes. However, in recent years, increasing evidence from population-based studies has emerged to suggest that the commonly occurring AF phenotype has a significant genetic component. Investigators from the Framingham Heart Study (FHS) have reported that a parental history of AF almost doubles the risk of future disease in offspring.27 Similar findings regarding the heritability of AF were observed among Icelanders.28 Investigators at Mayo Clinic and Massachusetts General Hospital have also reported that for individuals in whom a first-degree relative is diagnosed with lone AF, the risk of developing AF is significantly higher than that of the general population.29,30
In contrast to monogenic forms of AF, in which rare genetic mutations with high penetrance are responsible for the condition, the more common AF phenotype is likely to be caused by common genetic polymorphisms interacting with various environmental factors. In early case–control studies comparing cohorts of AF to controls, AF risk has been associated with polymorphisms in a variety of different genes.31–40 However, in many such studies, the results have not been systematically replicated and sample sizes have been relatively small. More recently, genome-wide association studies (GWAS) have identified common genetic variants or single-nucleotide polymorphisms (SNPs) that confer increased susceptibility to AF and have led to significant advances in understanding the genetic basis of AF in the general population.
Despite the fact that monogenic forms of AF are relatively rare, studies in familial AF kindreds have provided valuable insights into the molecular pathways underlying the arrhythmia. The identification of single gene mutations has also provided a framework for interrogation of genetic polymorphisms that predispose to the commonly occurring AF phenotype. Eventually, the elucidation of the genetic substrate underlying the different forms of AF may lead to the development of novel approaches for diagnosis and treatment of the arrhythmia. In the following review, we will discuss monogenic forms of AF and also highlight results from case–control association studies in cohorts with non-familial AF. GWAS in AF will be discussed separately in this edition.
2. Susceptibility loci for AF
In 1997, Brugada et al.11 described a susceptibility locus for AF in three families with autosomal dominant AF. Using linkage analysis, the locus for AF was mapped on chromosome 10q22–24; however, the causative mutation at this locus remains elusive. In 2003, a second susceptibility locus on chromosome 6q12–q16 was identified in a large family which also had autosomal dominantly inherited AF.12 Of note, both susceptibility loci overlap with loci that have previously been reported for familial DCM.41–43 It remains to be determined whether DCM and AF are linked in these families.
3. Monogenic mutations in AF
3.1. Ion channel mutations
The majority of mutations identified in monogenic AF families have been located in genes that encode ion channel subunits (summarized in Table 1 and Figure 1). Functional analyses of these mutations have revealed either gain-of-function effects or loss-of-function effects. Interestingly, electrophysiological remodelling in patients with non-familial forms of chronic AF results in similar ion channel phenotypes.44 These observations suggest that the different forms of AF share common underlying mechanisms. However, these mechanisms are presently not clearly understood.
Table 1.
Summary of monogenic mutations associated with AF
| Gene | Gene product | Family/proband characteristics | Documented familial segregation, yes/no | Functional assay performed, yes/no | Functional effect of mutation | References |
|---|---|---|---|---|---|---|
| KCNQ1 | α-subunit of IKs channel | Chinese family with autosomal dominant AF | Yes | Yes | Gain-of-function effect with increased IKs | 13 |
| KCNQ1 | α-subunit of IKs channel | Isolated case of AF detected in utero (Caucasian) | No | Yes | Gain-of-function effect with increased IKs | 55 |
| KCNQ1 | α-subunit of IKs channel | Caucasian family with autosomal dominant AF | Yes | Yes | Gain-of-function effect with increased IKs | 56 |
| KCNE2 | β-subunit of IKs channel | Two Chinese AF kindreds | Yes | Yes | Gain-of-function effect with increased IKs | 14 |
| KCNE5 | β-subunit of IKs channel | Isolated non-familial case of AF (Caucasian) | No | Yes | Gain-of-function effect with increased IKs | 58 |
| KCNJ2 | Kir 2.1 channel | Chinese AF kindred | Yes | Yes | Gain-of-function effect with increased IK1 | 16 |
| KCNA5 | Kv1.5 channel | Caucasian proband with refractory AF | Yes | Yes | Loss-of-function effect with reduced IKur | 17 |
| SCN5A | Sodium channel α-subunit | Caucasian family with variable expression of AF, DCM and impaired conduction | Yes | No | Predicted to have a loss-of-function effect with reduced sodium current density | 87 |
| SCN5A | Sodium channel α-subunit | Caucasian proband with familial AF | Yes | Yes | Loss-of-function effect with hyperpolarizing shift in channel steady state inactivation | 88 |
| SCN5A | Sodium channel α-subunit | Japanese family with autosomal dominant AF | Yes | Yes | Gain-of-function effect with depolarized shift of voltage dependence of steady-state inactivation | 90 |
| SCN1B | Sodium channel β-subunit | 2 isolated non-familial cases of AF (1 Caucasian, 1 black) | No | Yes | Loss-of-function effect with reduced sodium current and altered channel gating | 89 |
| SCN2B | Sodium channel β-subunit | 2 isolated non-familial cases of AF (Caucasian) | Yes (in 1 of the 2 patients) | Yes | Loss-of-function effect with reduced sodium current and altered channel gating | 89 |
| NUP155 | Nucleoporin | Consanguineous family from Uruguay with early-onset AF | Yes | Yes | Reduction in nuclear membrane permeability | 104 |
| GJA5 | Connexin-40 | 4 isolated non-familial cases (3 somatic mutations, 1 germline mutation) | No | Yes | Impaired intracellular transport and intercellular electrical coupling | 19 |
| NPPA | Mutant atrial natriuretic peptide (mANP) | Caucasian family with autosomal dominant AF | Yes | Yes | Elevated levels of mutant ANP | 21 |
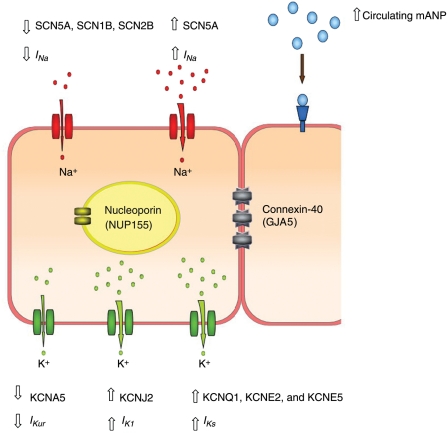
Figure 1.
Pictorial image of adjacent cardiomyocytes illustrating the genes implicated in Mendelian forms of AF and the presumed mechanism of action of the mutation.
There are currently two major hypotheses regarding the electrophysiological mechanisms underlying AF; the multiple wavelet hypothesis and ‘mother rotor’ hypothesis.45 The mechanism of AF as proposed by the ‘mother rotor’ hypothesis involves stable, self-sustaining rotors that generate wavelets of activation which spread throughout the atrial myocardium.46 Alternatively, the multiple wavelet hypothesis proposes that multiple random wavelets of activation with constantly changing re-entrant circuits underlie AF.47 The likelihood is that different mechanisms predominate in different circumstances.48 Alterations in ion channel function are predicted to influence one or both of the proposed mechanisms of AF.
3.1.1. Potassium channel mutations
3.1.1.1. IKs channel mutations
Chen et al.13 provided the first link between ion channelopathies and AF. In a Chinese family with an autosomal dominant pattern of AF inheritance, they reported a missense mutation (S140G) located in the first transmembrane spanning domain of KCNQ1. The KCNQ1 gene encodes a pore-forming α-subunit which associates with ancillary β-subunits, to form a channel responsible for the IKs current. IKs is a delayed-rectifier potassium current which is prominent at higher heart rates and during adrenergic stimulation during the late phase of the action potential. Functionally, the S140G mutant channel was associated with a marked increase in current density suggesting a gain-of-function effect. In a subsequent study, the S140G mutation was demonstrated to cause marked slowing of IKs channel deactivation.49
Previous studies have also reported loss-of-function mutations in KCNQ1 in patients with LQTS type 1.50–54 The (S140G) KCNQ1 gain-of-function mutation identified by Chen et al.13 would therefore be expected to shorten the QT interval. Interestingly, however, in a proportion of the affected family members in the AF kindred, QT interval was prolonged rather than shortened. The molecular basis for this paradoxical observation remains unexplained. These observations highlight the fact that our understanding of cardiac repolarization in the atrium remains incomplete.
Since the original discovery by Chen et al., two further mutations in the KCNQ1 gene have been described. In an unusual case of AF detected in utero, a valine-to-methionine mutation (V141M) adjacent to the aforementioned S140G mutation has been identified.55 More recently, a serine-to-proline mutation (S209P) was reported in a family with an autosomal dominant pattern of inheritance of AF.56 Both KCNQ1 mutations displayed a gain-of-function effect with enhanced IKs current density and altered gating kinetics. The V141M mutation resulted in an expected shortening of the QT interval, whereas the S209P mutation did not alter QT interval.
Mutations in IKs channel β-subunit genes have been described in familial as well as isolated AF cases. As opposed to KCNQ1, which has six transmembrane domains, the KCNE β-subunits have only one transmembrane spanning domain. They are encoded by five genes, KCNE1–KCNE5.57 In a study evaluating 28 unrelated Chinese families with lone AF, Yang et al.14 identified a mutation in the KCNE2 gene which resulted in an arginine-to-cysteine substitution (R27C). More recently, an isolated non-familial case of AF with a missense (L65F) mutation has been identified after a cohort of 158 patients were screened for KCNE5 gene mutations.58 Interaction of both mutant β-subunits (KCNE2 and KCNE5) with the KCNQ1 channel produced a gain-of-function effect with an increased IKs current.
The KCNQ1 α-subunit of the IKs channel can associate with any one of the five accessory β-subunits (KCNE1–5). Previous studies have demonstrated that each of the β-subunits causes a specific alteration in the KCNQ1 current.59,60 On the basis of these observations, it has been proposed that alterations in the patterns of association between KCNQ1 and the β-subunits may allow modulation of the IKs current.61 Interestingly, in normal human cardiac tissue, KCNE4 expression is higher in the atrium as compared with the ventricle;60 however, the precise constituents and regulation of the IKs current in the atrium vs. the ventricle remain unknown.
Presently, information regarding alterations in the IKs current in patients with chronic AF is limited. Transcription of IKs channel subunits has been reported to be altered in patients with AF and may cause significant alterations in atrial electrical activity. However, the results from these studies have been conflicting. Brundel et al.62 reported that mRNA and protein expression of KCNE1 is reduced in AF. In contrast, Lai et al. reported that mRNA expression of KCNE1 is increased in AF. The latter study also reported down-regulation of KCNQ1 transcription.63
From a mechanistic perspective, the gain-of-function mutations in α- and β-subunits of the IKs channel are associated with increased repolarizing potassium currents which in effect would abbreviate the action potential duration as well as the effective refractory period in cardiomyocytes.64 These effects are likely to create a profibrillatory substrate within the atrium.45
In order to further characterize the effect of alterations of IKs on arrhythmia susceptibility, investigators have attempted to use a mouse model. However, due to the fact that IKs is expressed at very low levels in the adult murine heart, it has not been possible to reproduce the effects of IKs mutations in transgenic mouse models.65 Despite these limitations, however, investigators have reported that transgenic mice with ablation of the KCNE1 gene have spontaneous episodes of AF.66
3.1.1.2. IK1 channel mutations
In 2005, Xia et al.16 reported a novel missense mutation in the KCNJ2 gene in a Chinese AF kindred. KCNJ2 encodes the Kir2.1 channel which underlies the inward-rectifier potassium current, IK1.67,68 A valine-to-isoleucine mutation (V93I) was identified which resulted in a gain-of-function effect with increased potassium current amplitudes, both in the inward and outward directions. Interestingly, loss-of-function mutations in Kir2.1 have been reported to cause the Anderson syndrome, a condition characterized by QT interval prolongation, ventricular arrhythmias, multiple bony abnormalities, and intermittent episodes of muscular weakness.69 The gain-of-function mutation in KCNJ2 in the AF kindred was not associated with an alteration in the QT interval. A possible explanation for the missing effect of the V93I mutation at the ventricular level is that the IK1 current is significantly smaller in the atrium as compared with the ventricle.70,71
A number of studies have reported an up-regulation of inward-rectifier current density in AF patients.72–74 In turn, enhanced inward-rectifier currents have been demonstrated to promote AF by accelerating and stabilizing atrial rotors that maintain the arrhythmia.75–77 Further evidence for the role of IK1 in the pathogenesis of AF has come from studies demonstrating chamber-specific differences in inward-rectifier current function (IK1 and IKach). Voigt et al. reported that in patients with paroxysmal AF, inward-rectifier current densities were two-fold larger in left atrial cardiomyocytes when compared with right atrial cardiomyocytes. In contrast, in patients with chronic AF, there were no differences in IK1 between the atria, although they did report elevated basal currents. These observations may suggest that an unequal distribution of inward-rectifier potassium currents in atria supports the transition from paroxysmal to persistent AF.78
3.1.1.3. IKur channel mutations
Olson et al.17 identified a loss-of-function potassium channel gene mutation associated with AF. In a proband with lone AF which was refractory to conventional therapy, they reported a heterozygous nonsense mutation (E375X) in the KCNA5 gene. KCNA5 encodes the Kv1.5 channel which underlies the ultrarapid delayed-rectifier (IKur) current. IKur is an important repolarizing current specific to the atrium.79–81 Functional analysis of the mutant Kv1.5 channel revealed prolongation of the atrial action potential and triggered activity with stress, which would be predicted to promote initiation of AF. More recently, three further loss-of-function KCNA5 gene mutations (T527M A576V and E610K) have been reported in four families after screening a total of 120 families.82
In patients with chronic AF, some studies have reported reduced expression of Kv1.5 in parallel with an attenuation of the ultrarapid delayed-rectifier (IKur) current.83,84 These observations lend further support to the hypothesis that suppression of the IKur current increases susceptibility to AF.
3.1.1.4. Candidate gene screening for potassium channel mutations in AF cohorts
Following the reports of potassium channel gene mutations in rare monogenic kindreds, a number of investigators have performed candidate gene screening to determine the prevalence of such mutations. In 2004, Ellinor et al.85 screened a cohort of 141 unrelated patients with lone AF for KCNQ1 mutations and failed to identify any mutations. In a subsequent study, the same group screened 96 unrelated probands with familial AF for mutations in the KCNJ2 and KCNE1–5 genes and once again found no evidence of causal mutations.86 In a study by Otway et al.,18 four potassium channel genes (KCNQ1, KCNE1, KCNE2, and KCNE3) were screened for mutations in 50 AF families. Only one missense mutation in the KCNQ1 gene was identified. Functional analysis of the mutant gene product did not demonstrate altered channel kinetics, suggesting that the KCNQ1 mutation might not be causative. Taken together, these data suggest that potassium channel mutations are not a major cause of AF in the general population.
3.1.2. Sodium channel mutations
Mutations in the genes encoding both the α- and β-subunits of the voltage-gated sodium channel have been reported in patients with AF.87–91 The pore-forming α-subunit is encoded by the SCN5A gene, whereas four genes designated SCN1B through SCN4B encode the function-modifying β-subunits. In addition to AF, mutations in sodium channel genes are associated with a range of arrhythmias. SCN5A mutations have been reported to cause the Brugada syndrome,92 congenital sick sinus syndrome,93 cardiac conduction disease,94 idiopathic ventricular fibrillation,95 and LQTS type 3 (LQTS3).96 SCN1B and SCN3B mutations are also associated with cardiac conduction system disease and Brugada syndrome.97,98 As a result, patients with AF associated with sodium channel mutations often have complex, overlapping phenotypes.
In 2005, Olson et al.87 reported an SCN5A mutation (D1275N) in a large multigenerational family. The mutation was associated with variable clinical manifestations which included AF, DCM, and abnormal cardiac conduction. On the basis of reports from other studies, the D1275N mutation is expected to cause a loss of cardiac sodium channel function.99 In a more recent study, a cohort of 189 AF patients were screened for SCN5A mutations and a single missense mutation (N1986K) was identified in one AF kindred. Functional analysis of the mutation revealed a loss-of-function effect with a hyperpolarized shift of steady-state inactivation. One family member with the N1986K mutation had associated conduction system disease.88
Loss-of-function mutations in the SCN5A gene are also associated with Brugada syndrome.92 The occurrence of AF in patients with Brugada syndrome appears to be relatively common. However, reports of AF and Brugada syndrome in patients with SCN5A mutations are rare. In a cohort of 38 patients with Brugada syndrome, Makiyama et al.100 reported the occurrence of AF in 10 cases (26.3%). However, they did not identify SCN5A mutations in any of the patients with co-existing AF and Brugada syndrome. Similarly, in 59 Brugada syndrome patients, Bordachar et al.101 reported an incidence of AF in 20%. However, only two of the Brugada syndrome patients with an SCN5A mutation had documented AF. The reasons why loss-of-function mutations in SCN5A cause AF in some cases and ventricular arrhythmic conditions in others are presently unclear.
The role of the function-modifying sodium channel β-subunits in arrhythmic cardiac diseases is less clearly defined. In a recent study of 480 AF patients, Watanabe et al.89 screened the four β-subunit genes (SCN1B–SCN4B) for mutations and reported two mutations in SCN1B (R85H, D153N) and two mutations in SCN2B (R28Q, R28W). Functional analysis of the mutant β1- and β2-subunits demonstrated altered channel gating and a reduction in sodium current indicating a loss-of-function effect. Interestingly, in three of the four mutation carriers, the electrocardiogram demonstrated ST-segment elevation in the right-sided leads. The findings from this study are consistent with previous reports linking decreased sodium current with enhanced AF susceptibility.87,88
Makiyama et al.90 recently reported a gain-of-function SCN5A mutation associated with AF. They identified a novel missense mutation (M1875T) in a Japanese family with autosomal dominant hereditary AF. Analysis of the mutant channel function demonstrated that the voltage dependence of steady-state inactivation was shifted in the depolarizing direction, suggesting a gain-of-function. Gain-of-function SCN5A mutations are also associated with LQTS3.96 However, in contrast to LQTS3 mutations, the M1875T mutation in the AF kindred did not display persistent inward sodium currents. As a result, normal QT interval was observed in the majority of affected individuals.
AF has previously also been described as a concomitant disease in familial LQTS3. Benito et al.24 described a three-generation family with LQTS3 and AF due to a gain-of-function mutation (Y1795C) in SCN5A. Three out of eight family members displayed early-onset paroxysmal AF. Johnson et al.23 reported a mixed phenotype of LQTS and AF in one of 59 patients with genetically proven LQTS3. These findings provide further evidence of the role of gain-of-function SCN5A mutations in AF.
The electrophysiological mechanisms by which sodium channel mutations cause AF are not clearly understood. Increased inward sodium currents induce triggered activity and stabilize high-frequency rotors.102,103 Yet, they also make re-entry less likely. Conversely, reduced sodium current density promotes re-entry by shortening action potential duration and shortening the atrial re-entry wavelength.102 However, the attenuation of sodium current also destabilizes high-frequency rotors.102 Overall, multiple effects in various experimental models make it difficult to predict a priori what the effects of alterations in sodium channel function will be.
Consistent with the data reported for potassium channel gene mutations, mutations in genes coding sodium channel subunits do not appear to be a common cause of AF. Chen et al.31 screened a cohort of 157 lone AF patients and did not identify any SCN5A mutations. Similarly, we identified SCN5A mutations in only one kindred out of a cohort of 189 AF patients and Watanabe et al.89 identified only four patients with SCNB mutations in a cohort of 480 patients.88 Darbar et al.91 sequenced the SCN5A gene in a cohort of 375 AF patients and discovered eight novel variants. However, segregation analysis suggested that only six of the novel SCN5A variants are associated with AF.
3.2. Non-ion channel mutations
3.2.1. Nucleoporin gene (NUP155) mutation
In 2004, Oberti et al.104 identified a large consanguineous family from Uruguay with autosomal recessive inheritance of AF. The pattern of disease was characterized by an early onset of AF at the foetal or infantile stage with severe associated complications including cardiomyopathy, ventricular arrhythmias, and sudden death. The locus was mapped on chromosome 5p13 (arAF1).
In a subsequent study, a homozygous mutation (R391H) in a nucleoporin gene (NUP155) was identified.20 NUP155 encodes a nucleoporin which is an essential molecular component of the nuclear pore complexes (NPCs).105 NPCs mediate exchange of macromolecules between the nucleus and the cytoplasm.106 The mechanistic link between NUP155 mutation and AF remains unclear. It has been proposed that a reduction in nucleocytoplasmic transport due to NUP155 deficiency may alter expression of atrial genes which in turn may influence cellular processes such as maturation of calcium handling proteins and ion channels. These effects may ultimately alter the action potential duration and promote AF. An alternative hypothesis is that altered function of the nuclear envelope due to NUP155 deficiency may reduce myocyte survival by blocking mitosis. Myocyte apoptosis may promote cardiac fibrosis and conduction heterogeneity which may in turn create a substrate for arrhythmia.107 Future studies are required to further define the role of the nucleoporin in AF.
3.2.2. Connexin-40 gene (GJA5) mutations
In a study by Gollob et al.19 involving a small cohort of unrelated patients with lone AF, four novel mutations were identified in the GJA5 gene. GJA5 encodes connexin-40, a gap junction protein in the atrium which plays a critical role in mediating coordinated conduction of the action potential through cell-to-cell electrical coupling.108 Out of 15 AF patients in the study, four patients carried missense GJA5 mutations. Interestingly, only one of the patients had a germ-line sequence variant. The three remaining patients had tissue-specific mutations, suggesting that somatic mutations could also be involved in AF predisposition. Functional analysis of the mutant gene product revealed abnormal intracellular transport in addition to a reduction in electrical coupling between cells. It has been proposed that impaired cell–cell electrical coupling results in conduction heterogeneity, micro-re-entrant circuits, and hence AF.19
A number of studies have investigated connexin-40 expression in patients with chronic AF. The results from such studies have not been consistent. Some investigators have reported increased connexin-40,109,110 others have reported decreased connexin-40,111–114 and yet others have reported no change in the level of connexin.115 Similarly, reports of changes in distribution of connexins in AF have been inconsistent. Some investigators have reported increased lateralization of gap junction distribution,109,112,116 whereas others have reported increased heterogeneity.117
3.2.3. Atrial natriuretic peptide gene (NPPA) mutation
Hodgson-Zingman et al.21 reported on a family with an autosomal dominant pattern of AF which co-segregated with a frameshift mutation in the gene encoding atrial natriuretic peptide (NPPA). The mutation was associated with markedly elevated levels of mutant atrial natriuretic peptide (ANP). ANP is involved in the regulation of sodium and water homeostasis and arterial blood pressure. In response to volume expansion and atrial stretch, ANP release causes natriuresis, diuresis, and vasodilator effects.118 Previous studies have reported that when exposed to pathophysiological levels of ANP, atrial myocytes display altered electrophysiological properties.119–122 The mutant peptide in the AF kindred was demonstrated to shorten atrial action potential duration in an animal model.21 An alternative plausible hypothesis is that excessive ANP may cause structural atrial remodelling due to its pro-apoptotic effect.123 Thus, at this stage, the role of ANP in the pathogenesis of AF remains speculative.
4. Polymorphisms associated with non-familial AF
Case–control association studies have been widely used for genetic analysis of a variety of complex traits including AF in the general population. Association studies involve the comparison of genotype frequencies for candidate genes between a diseased population and a population of healthy controls. In recent years, association studies in AF cohorts have identified a variety of polymorphisms that may influence susceptibility to the arrhythmia. Examples include polymorphisms in the cardiac potassium channel subunit genes,32–35 sodium channel genes,31 genes that regulate ion channel function,36,124 gap junction protein genes,37 genes encoding circulating hormones,125–128 and genes encoding inflammatory mediators.129,130
Interestingly, some of the association studies have identified genetic polymorphisms that are predicted to cause functional alterations in the same ion channels as those implicated in monogenic forms of AF. Examples include polymorphisms in KCNE1 and KCNE5, which encode α- and β-subunits of the IKs channel respectively, and SCN5A, which encodes the α-subunit of the INa channel.31,32,35,131 In addition, one of the reported polymorphisms encodes the β3-subunit of a heterotrimeric G protein (GNB3) which has been linked with an increased inward-rectifier current (IK1).36,132 These results suggest that the same molecular mechanisms may underlie familial and sporadic forms of AF. It should be noted however that the majority of the association studies in AF cohorts have been limited by relatively small sample sizes, inconsistent replication, and a low pre-test probability of the polymorphism actually causing AF. The results from the studies are summarized in Table 2.
Table 2.
Summary of results from association studies in AF cohorts
| Gene | Polymorphism | Cases | Controls | Ethnicity | Comment | P-value | Odds ratio | References |
|---|---|---|---|---|---|---|---|---|
| KCNE1 minK | 38G | 331 | 441 | Caucasian | 0.004 | 1.73 | 32 | |
| KCNE1 minK | 38G | 108 | 108 | Asian | 0.024 | 1.80 | 34 | |
| KCNE5 | 97T | 158 | 96 | Caucasian | 0.007 | 0.52 | 35 | |
| KCNH2 | K897T | 1207 | 2475 | Caucasian | 0.00033 | 1.25 | 33 | |
| GNB3 | C825T | 291 | 292 | Caucasian | 0.02 | 0.46 | 36 | |
| eNOS | 2786C | 331 | 441 | Caucasian | 0.01 | 1.50 | 32 | |
| eNOS | G894T | 51 | 289 | Caucasian | HF patients | 0.001 | 3.2 | 127 |
| SCN5A | H558R | 157 | 314 | Caucasian | 0.002 | 1.6 | 31 | |
| GJA5 | –44AA/+71GG | 173 | 232 | Asian | <0.006 | 1.514 | 37 | |
| AGT | M235T | 250 | 250 | Asian | <0.001 | 2.5 | 39 | |
| AGT | G-6A | 250 | 250 | Asian | 0.005 | 3.3 | 39 | |
| AGT | G-217A | 250 | 250 | Asian | 0.002 | 2.0 | 39 | |
| AGT | T174M | 968 | 8267 | Caucasian | 0.05 | 1.2 | 126 | |
| AGT | 20C/C | 968 | 8267 | Caucasian | 0.01 | 1.5 | 126 | |
| ACE | D/D | 51 | 289 | Caucasian | HF patients | 0.016 | 1.5 | 127 |
| ACE | D/D | 404 | 520 | Caucasian | <0.001 | 1.89 | 128 | |
| MMP2 | C1306T | 196 | 873 | Asian | 1.26 × 10−2 | 8.1 | 129 | |
| IL10 | A-592C | 196 | 873 | Asian | 3.7 × 10−3 | 0.32 | 129 | |
| IL6 | G-174C | 26 | 84 | Caucasian | Post-operative AF (after CABG) | <0.001 | 3.25 | 130 |
| SLN | C-65G | 147 | 92 | Caucasian | 0.011 | 1.98 | 124 |
ACE, angiotensin-converting enzyme; AGT, angiotensinogen; CABG, coronary artery bypass graft surgery; eNOS, endothelial nitric oxide synthase 3; GNB3, guanine nucleotide-binding protein; GJA5, connexin 40; HF, heart failure; IL6, interleukin 6; IL10, interleukin 10; MMP, matrix metalloproteinase; SLN, sarcolipin gene.
5. Summary
In summary, studies involving familial AF kindred have reported several monogenic mutations. The majority of the mutations have been identified in genes encoding ion channels, although some studies have also uncovered mutations in non-ion channel coding genes. Based on available evidence, these rare mutations appear to provide little explanation for the heritability of AF in the general population. However, the identification of these mutations has provided valuable insights into the molecular pathways underlying AF and has also provided a framework for investigating the genetic basis of common forms of the arrhythmia.
The genetic basis of non-familial AF is presently largely unknown. In recent years, GWAS have led to significant advances in our understanding of the genetic basis of complex traits. A recent GWAS for AF has led to the identification of novel variants that appear to confer increased susceptibility to the arrhythmia.40 However, most of the SNPs are located outside the commonly known genes; therefore, the molecular mechanisms underlying their association with AF remains unclear.40
Current attempts to interpret GWAS signals are based on the assumption that common sequence variants are responsible for common traits. However, an interesting alternative hypothesis is that the genetic control of complex traits is due to rare mutations that are either not represented in current GWAS or that cause the observed associations through ‘synthetic’ associations.133,134 This possibility challenges the currently held belief that monogenic mutations are restricted to AF families and rare isolated AF cases. In the future, the use of next-generation sequencing technology to sequence the entire exome or genome may uncover numerous private monogenic mutations that might account for some of the unexplained GWAS signals. In this context, studies in AF families will be of increasing relevance because demonstrating transmission of these private mutations will be essential for proving causality.
Ultimately, the identification of the genes and pathways underlying the familial and more common forms of AF should give us new insights into the development of novel diagnostic tests and targeted therapies for the arrhythmia.
Conflict of interest: none declared.
Funding
This work was supported by a grant from the Netherlands Organization of Scientific Research to M.R. (Rubicon Grant 825.09.020), grants from the National Institutes of Health to S.A.L. (T32 HL007575), D.J.M. (R21HL096009 and R21DA026982), and P.T.E. (R01HL104156, R21DA027021, R01HL092577, and K24HL105780).
References
- 1.Kannel WB, Benjamin EJ. Status of the epidemiology of atrial fibrillation. Med Clin North Am. 2008;92:17–40. doi: 10.1016/j.mcna.2007.09.002. ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott A, Breteler MM, de Bruyne MC, van Harskamp F, Grobbee DE, Hofman A. Atrial fibrillation and dementia in a population-based study. The Rotterdam Study. Stroke. 1997;28:316–321. doi: 10.1161/01.str.28.2.316. [DOI] [PubMed] [Google Scholar]
- 3.Stewart S, Hart CL, Hole DJ, McMurray JJ. A population-based study of the long-term risks associated with atrial fibrillation: 20-year follow-up of the Renfrew/Paisley study. Am J Med. 2002;113:359–364. doi: 10.1016/s0002-9343(02)01236-6. [DOI] [PubMed] [Google Scholar]
- 4.Wang TJ, Larson MG, Levy D, Vasan RS, Leip EP, Wolf PA, et al. Temporal relations of atrial fibrillation and congestive heart failure and their joint influence on mortality: the Framingham Heart Study. Circulation. 2003;107:2920–2925. doi: 10.1161/01.CIR.0000072767.89944.6E. [DOI] [PubMed] [Google Scholar]
- 5.Krahn AD, Manfreda J, Tate RB, Mathewson FA, Cuddy TE. The natural history of atrial fibrillation: incidence, risk factors, and prognosis in the Manitoba Follow-Up Study. Am J Med. 1995;98:476–484. doi: 10.1016/S0002-9343(99)80348-9. [DOI] [PubMed] [Google Scholar]
- 6.Benjamin EJ, Levy D, Vaziri SM, D'Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. JAMA. 1994;271:840–844. [PubMed] [Google Scholar]
- 7.Kannel WB, Abbott RD, Savage DD, McNamara PM. Epidemiologic features of chronic atrial fibrillation: the Framingham study. N Engl J Med. 1982;306:1018–1022. doi: 10.1056/NEJM198204293061703. [DOI] [PubMed] [Google Scholar]
- 8.Kannel WB, Abbott RD, Savage DD, McNamara PM. Coronary heart disease and atrial fibrillation: the Framingham Study. Am Heart J. 1983;106:389–396. doi: 10.1016/0002-8703(83)90208-9. [DOI] [PubMed] [Google Scholar]
- 9.Psaty BM, Manolio TA, Kuller LH, Kronmal RA, Cushman M, Fried LP, et al. Incidence of and risk factors for atrial fibrillation in older adults. Circulation. 1997;96:2455–2461. doi: 10.1161/01.cir.96.7.2455. [DOI] [PubMed] [Google Scholar]
- 10.Wolff L. Familial auricular fibrillation. N Engl J Med. 1943;299:396–397. [Google Scholar]
- 11.Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, et al. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–911. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 12.Ellinor PT, Shin JT, Moore RK, Yoerger DM, MacRae CA. Locus for atrial fibrillation maps to chromosome 6q14–16. Circulation. 2003;107:2880–2883. doi: 10.1161/01.CIR.0000077910.80718.49. [DOI] [PubMed] [Google Scholar]
- 13.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong K, Bjerregaard P, Gussak I, Brugada R. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J Cardiovasc Electrophysiol. 2005;16:394–396. doi: 10.1046/j.1540-8167.2005.40621.x. [DOI] [PubMed] [Google Scholar]
- 16.Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 17.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 18.Otway R, Vandenberg JI, Guo G, Varghese A, Castro ML, Liu J, et al. Stretch-sensitive KCNQ1 mutation A link between genetic and environmental factors in the pathogenesis of atrial fibrillation? J Am Coll Cardiol. 2007;49:578–586. doi: 10.1016/j.jacc.2006.09.044. [DOI] [PubMed] [Google Scholar]
- 19.Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–2688. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, et al. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita H, Kusano-Fukushima K, Nagase S, Fujimoto Y, Hisamatsu K, Fujio H, et al. Atrial fibrillation and atrial vulnerability in patients with Brugada syndrome. J Am Coll Cardiol. 2002;40:1437–1444. doi: 10.1016/s0735-1097(02)02167-8. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JN, Tester DJ, Perry J, Salisbury BA, Reed CR, Ackerman MJ. Prevalence of early-onset atrial fibrillation in congenital long QT syndrome. Heart Rhythm. 2008;5:704–709. doi: 10.1016/j.hrthm.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benito B, Brugada R, Perich RM, Lizotte E, Cinca J, Mont L, et al. A mutation in the sodium channel is responsible for the association of long QT syndrome and familial atrial fibrillation. Heart Rhythm. 2008;5:1434–1440. doi: 10.1016/j.hrthm.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Sebillon P, Bouchier C, Bidot LD, Bonne G, Ahamed K, Charron P, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560–567. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruver EJ, Fatkin D, Dodds GA, Kisslo J, Maron BJ, Seidman JG, et al. Familial hypertrophic cardiomyopathy and atrial fibrillation caused by Arg663His beta-cardiac myosin heavy chain mutation. Am J Cardiol. 1999;83:13H–18H. doi: 10.1016/s0002-9149(99)00251-9. [DOI] [PubMed] [Google Scholar]
- 27.Fox CS, Parise H, D'Agostino RB, Sr, Lloyd-Jones DM, Vasan RS, Wang TJ, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 28.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, et al. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–712. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 29.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 30.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 31.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin Pharmacol Ther. 2007;81:35–41. doi: 10.1038/sj.clpt.6100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatini C, Sticchi E, Genuardi M, Sofi F, Gensini F, Gori AM, et al. Analysis of minK and eNOS genes as candidate loci for predisposition to non-valvular atrial fibrillation. Eur Heart J. 2006;27:1712–1718. doi: 10.1093/eurheartj/ehl087. [DOI] [PubMed] [Google Scholar]
- 33.Sinner MF, Pfeufer A, Akyol M, Beckmann BM, Hinterseer M, Wacker A, et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG) Eur Heart J. 2008;29:907–914. doi: 10.1093/eurheartj/ehm619. [DOI] [PubMed] [Google Scholar]
- 34.Lai LP, Su MJ, Yeh HM, Lin JL, Chiang FT, Hwang JJ, et al. Association of the human minK gene 38G allele with atrial fibrillation: evidence of possible genetic control on the pathogenesis of atrial fibrillation. Am Heart J. 2002;144:485–490. doi: 10.1067/mhj.2002.123573. [DOI] [PubMed] [Google Scholar]
- 35.Ravn LS, Hofman-Bang J, Dixen U, Larsen SO, Jensen G, Haunso S, et al. Relation of 97T polymorphism in KCNE5 to risk of atrial fibrillation. Am J Cardiol. 2005;96:405–407. doi: 10.1016/j.amjcard.2005.03.086. [DOI] [PubMed] [Google Scholar]
- 36.Schreieck J, Dostal S, von Beckerath N, Wacker A, Flory M, Weyerbrock S, et al. C825T polymorphism of the G-protein beta3 subunit gene and atrial fibrillation: association of the TT genotype with a reduced risk for atrial fibrillation. Am Heart J. 2004;148:545–550. doi: 10.1016/j.ahj.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 37.Juang JM, Chern YR, Tsai CT, Chiang FT, Lin JL, Hwang JJ, et al. The association of human connexin 40 genetic polymorphisms with atrial fibrillation. Int J Cardiol. 2007;116:107–112. doi: 10.1016/j.ijcard.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 38.Firouzi M, Ramanna H, Kok B, Jongsma HJ, Koeleman BP, Doevendans PA, et al. Association of human connexin40 gene polymorphisms with atrial vulnerability as a risk factor for idiopathic atrial fibrillation. Circ Res. 2004;95:e29–e33. doi: 10.1161/01.RES.0000141134.64811.0a. [DOI] [PubMed] [Google Scholar]
- 39.Tsai CT, Lai LP, Lin JL, Chiang FT, Hwang JJ, Ritchie MD, et al. Renin–angiotensin system gene polymorphisms and atrial fibrillation. Circulation. 2004;109:1640–1646. doi: 10.1161/01.CIR.0000124487.36586.26. [DOI] [PubMed] [Google Scholar]
- 40.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 41.Bowles KR, Gajarski R, Porter P, Goytia V, Bachinski L, Roberts R, et al. Gene mapping of familial autosomal dominant dilated cardiomyopathy to chromosome 10q21–23. J Clin Invest. 1996;98:1355–1360. doi: 10.1172/JCI118922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowles NE, Bowles KR, Towbin JA. The final common pathway hypothesis and inherited cardiovascular disease. The role of cytoskeletal proteins in dilated cardiomyopathy. Herz. 2000;25:168–175. doi: 10.1007/s000590050003. [DOI] [PubMed] [Google Scholar]
- 43.Sylvius N, Tesson F, Gayet C, Charron P, Benaiche A, Peuchmaurd M, et al. A new locus for autosomal dominant dilated cardiomyopathy identified on chromosome 6q12–q16. Am J Hum Genet. 2001;68:241–246. doi: 10.1086/316929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosch RF, Nattel S. Cellular electrophysiology of atrial fibrillation. Cardiovasc Res. 2002;54:259–269. doi: 10.1016/s0008-6363(01)00529-6. [DOI] [PubMed] [Google Scholar]
- 45.Nattel S. New ideas about atrial fibrillation 50 years on. Nature. 2002;415:219–226. doi: 10.1038/415219a. [DOI] [PubMed] [Google Scholar]
- 46.Jalife J, Berenfeld O, Skanes A, Mandapati R. Mechanisms of atrial fibrillation: mother rotors or multiple daughter wavelets, or both? J Cardiovasc Electrophysiol. 1998;9:S2–S12. [PubMed] [Google Scholar]
- 47.Moe GK. On the multiple wavelet hypothesis of AF. Arch Int Pharmacodyn Ther. 1962;140:183–188. [Google Scholar]
- 48.Nattel S. Atrial electrophysiology and mechanisms of atrial fibrillation. J Cardiovasc Pharmacol Ther. 2003;8(Suppl. 1):S5–S11. doi: 10.1177/107424840300800102. [DOI] [PubMed] [Google Scholar]
- 49.Restier L, Cheng L, Sanguinetti MC. Mechanisms by which atrial fibrillation-associated mutations in the S1 domain of KCNQ1 slow deactivation of IKs channels. J Physiol. 2008;586:4179–4191. doi: 10.1113/jphysiol.2008.157511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang DW, Yazawa K, George AL, Jr, Bennett PB. Characterization of human cardiac Na+ channel mutations in the congenital long QT syndrome. Proc Natl Acad Sci USA. 1996;93:13200–13205. doi: 10.1073/pnas.93.23.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sesti F, Goldstein SA. Single-channel characteristics of wild-type IKs channels and channels formed with two minK mutants that cause long QT syndrome. J Gen Physiol. 1998;112:651–663. doi: 10.1085/jgp.112.6.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 53.Isbrandt D, Leicher T, Waldschutz R, Zhu X, Luhmann U, Michel U, et al. Gene structures and expression profiles of three human KCND (Kv4) potassium channels mediating A-type currents I(TO) and I(SA) Genomics. 2000;64:144–154. doi: 10.1006/geno.2000.6117. [DOI] [PubMed] [Google Scholar]
- 54.Tinel N, Diochot S, Lauritzen I, Barhanin J, Lazdunski M, Borsotto M. M-type KCNQ2–KCNQ3 potassium channels are modulated by the KCNE2 subunit. FEBS Lett. 2000;480:137–141. doi: 10.1016/s0014-5793(00)01918-9. [DOI] [PubMed] [Google Scholar]
- 55.Hong K, Piper DR, Diaz-Valdecantos A, Brugada J, Oliva A, Burashnikov E, et al. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc Res. 2005;68:433–440. doi: 10.1016/j.cardiores.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 56.Das S, Makino S, Melman YF, Shea MA, Goyal SB, Rosenzweig A, et al. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm. 2009;6:1146–1153. doi: 10.1016/j.hrthm.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abbott GW, Goldstein SA. Potassium channel subunits encoded by the KCNE gene family: physiology and pathophysiology of the MinK-related peptides (MiRPs) Mol Interv. 2001;1:95–107. [PubMed] [Google Scholar]
- 58.Ravn LS, Aizawa Y, Pollevick GD, Hofman-Bang J, Cordeiro JM, Dixen U, et al. Gain of function in IKs secondary to a mutation in KCNE5 associated with atrial fibrillation. Heart Rhythm. 2008;5:427–435. doi: 10.1016/j.hrthm.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lundquist AL, Manderfield LJ, Vanoye CG, Rogers CS, Donahue BS, Chang PA, et al. Expression of multiple KCNE genes in human heart may enable variable modulation of I(Ks) J Mol Cell Cardiol. 2005;38:277–287. doi: 10.1016/j.yjmcc.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 60.Bendahhou S, Marionneau C, Haurogne K, Larroque MM, Derand R, Szuts V, et al. In vitro molecular interactions and distribution of KCNE family with KCNQ1 in the human heart. Cardiovasc Res. 2005;67:529–538. doi: 10.1016/j.cardiores.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 61.Lundquist AL, Turner CL, Ballester LY, George AL., Jr Expression and transcriptional control of human KCNE genes. Genomics. 2006;87:119–128. doi: 10.1016/j.ygeno.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Brundel BJ, Van Gelder IC, Henning RH, Tieleman RG, Tuinenburg AE, Wietses M, et al. Ion channel remodeling is related to intraoperative atrial effective refractory periods in patients with paroxysmal and persistent atrial fibrillation. Circulation. 2001;103:684–690. doi: 10.1161/01.cir.103.5.684. [DOI] [PubMed] [Google Scholar]
- 63.Lai LP, Su MJ, Lin JL, Lin FY, Tsai CH, Chen YS, et al. Changes in the mRNA levels of delayed rectifier potassium channels in human atrial fibrillation. Cardiology. 1999;92:248–255. doi: 10.1159/000006982. [DOI] [PubMed] [Google Scholar]
- 64.Moe G. Evidence for reentry as a mechanism of cardiac arrhythmias. Rev Physiol Biochem Pharmacol. 1975;72:55–81. doi: 10.1007/BFb0031546. [DOI] [PubMed] [Google Scholar]
- 65.Nerbonne JM. Molecular basis of functional voltage-gated K+ channel diversity in the mammalian myocardium. J Physiol. 2000;525(Pt 2):285–298. doi: 10.1111/j.1469-7793.2000.t01-1-00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Temple J, Frias P, Rottman J, Yang T, Wu Y, Verheijck EE, et al. Atrial fibrillation in KCNE1-null mice. Circ Res. 2005;97:62–69. doi: 10.1161/01.RES.0000173047.42236.88. [DOI] [PubMed] [Google Scholar]
- 67.Zobel C, Cho HC, Nguyen TT, Pekhletski R, Diaz RJ, Wilson GJ, et al. Molecular dissection of the inward rectifier potassium current (IK1) in rabbit cardiomyocytes: evidence for heteromeric co-assembly of Kir2.1 and Kir2.2. J Physiol. 2003;550:365–372. doi: 10.1113/jphysiol.2002.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopatin AN, Nichols CG. Inward rectifiers in the heart: an update on I(K1) J Mol Cell Cardiol. 2001;33:625–638. doi: 10.1006/jmcc.2001.1344. [DOI] [PubMed] [Google Scholar]
- 69.Andersen ED, Krasilnikoff PA, Overvad H. Intermittent muscular weakness, extrasystoles and multiple developmental abnormalities: a new syndrome? Acta peditr Scand. 1971;60:559–564. doi: 10.1111/j.1651-2227.1971.tb06990.x. [DOI] [PubMed] [Google Scholar]
- 70.Wang Z, Yue L, White M, Pelletier G, Nattel S. Differential distribution of inward rectifier potassium channel transcripts in human atrium vs. ventricle. Circulation. 1998;98:2422–2428. doi: 10.1161/01.cir.98.22.2422. [DOI] [PubMed] [Google Scholar]
- 71.Giles WR, Imaizumi Y. Comparison of potassium currents in rabbit atrial and ventricular cells. J Physiol. 1988;405:123–145. doi: 10.1113/jphysiol.1988.sp017325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang H, Garratt CJ, Zhu J, Holden AV. Role of up-regulation of IK1 in action potential shortening associated with atrial fibrillation in humans. Cardiovasc Res. 2005;66:493–502. doi: 10.1016/j.cardiores.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 73.Bosch RF, Zeng X, Grammer JB, Popovic K, Mewis C, Kuhlkamp V. Ionic mechanisms of electrical remodeling in human atrial fibrillation. Cardiovasc Res. 1999;44:121–131. doi: 10.1016/s0008-6363(99)00178-9. [DOI] [PubMed] [Google Scholar]
- 74.Workman AJ, Kane KA, Rankin AC. The contribution of ionic currents to changes in refractoriness of human atrial myocytes associated with chronic atrial fibrillation. Cardiovasc Res. 2001;52:226–235. doi: 10.1016/s0008-6363(01)00380-7. [DOI] [PubMed] [Google Scholar]
- 75.Pandit SV, Berenfeld O, Anumonwo JM, Zaritski RM, Kneller J, Nattel S, et al. Ionic determinants of functional reentry in a 2-D model of human atrial cells during simulated chronic atrial fibrillation. Biophys J. 2005;88:3806–3821. doi: 10.1529/biophysj.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samie FH, Berenfeld O, Anumonwo J, Mironov SF, Udassi S, Beaumont J, et al. Rectification of the background potassium current: a determinant of rotor dynamics in ventricular fibrillation. Circ Res. 2001;89:1216–1223. doi: 10.1161/hh2401.100818. [DOI] [PubMed] [Google Scholar]
- 77.Sekar RB, Kizana E, Cho HC, Molitoris JM, Hesketh GG, Eaton BP, et al. IK1 heterogeneity affects genesis and stability of spiral waves in cardiac myocyte monolayers. Circ Res. 2009;104:355–364. doi: 10.1161/CIRCRESAHA.108.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Voigt N, Trausch A, Knaut M, Matschke K, Varro A, Van Wagoner DR, et al. Left-to-right atrial inward-rectifier potassium current gradients in patients with paroxysmal versus chronic atrial fibrillation. Circ Arrhythm Electrophysiol. 2010;3:472–480. doi: 10.1161/CIRCEP.110.954636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamkun MM, Knoth KM, Walbridge JA, Kroemer H, Roden DM, Glover DM. Molecular cloning and characterization of two voltage-gated K+ channel cDNAs from human ventricle. FASEB J. 1991;5:331–337. doi: 10.1096/fasebj.5.3.2001794. [DOI] [PubMed] [Google Scholar]
- 80.Wang Z, Fermini B, Nattel S. Delayed rectifier outward current and repolarization in human atrial myocytes. Circ Res. 1993;73:276–285. doi: 10.1161/01.res.73.2.276. [DOI] [PubMed] [Google Scholar]
- 81.Simard C, Drolet B, Yang P, Kim RB, Roden DM. Polymorphism screening in the cardiac K+ channel gene KCNA5. Clin Pharmacol Ther. 2005;77:138–144. doi: 10.1016/j.clpt.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 82.Yang Y, Li J, Lin X, Yang Y, Hong K, Wang L, et al. Novel KCNA5 loss-of-function mutations responsible for atrial fibrillation. J Hum Genet. 2009;54:277–283. doi: 10.1038/jhg.2009.26. [DOI] [PubMed] [Google Scholar]
- 83.Van Wagoner DR, Pond AL, McCarthy PM, Trimmer JS, Nerbonne JM. Outward K+ current densities and Kv1.5 expression are reduced in chronic human atrial fibrillation. Circ Res. 1997;80:772–781. doi: 10.1161/01.res.80.6.772. [DOI] [PubMed] [Google Scholar]
- 84.Dobrev D, Ravens U. Remodeling of cardiomyocyte ion channels in human atrial fibrillation. Basic Res Cardiol. 2003;98:137–148. doi: 10.1007/s00395-003-0409-8. [DOI] [PubMed] [Google Scholar]
- 85.Ellinor PT, Moore RK, Patton KK, Ruskin JN, Pollak MR, Macrae CA. Mutations in the long QT gene, KCNQ1, are an uncommon cause of atrial fibrillation. Heart. 2004;90:1487–1488. doi: 10.1136/hrt.2003.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ellinor PT, Petrov-Kondratov VI, Zakharova E, Nam EG, MacRae CA. Potassium channel gene mutations rarely cause atrial fibrillation. BMC Med Genet. 2006;7:70. doi: 10.1186/1471-2350-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 89.Watanabe H, Darbar D, Kaiser DW, Jiramongkolchai K, Chopra S, Donahue BS, et al. Mutations in sodium channel beta1- and beta2-subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2009;2:268–275. doi: 10.1161/CIRCEP.108.779181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, et al. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–1334. doi: 10.1016/j.jacc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 91.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Akai J, Makita N, Sakurada H, Shirai N, Ueda K, Kitabatake A, et al. A novel SCN5A mutation associated with idiopathic ventricular fibrillation without typical ECG findings of Brugada syndrome. FEBS Lett. 2000;479:29–34. doi: 10.1016/s0014-5793(00)01875-5. [DOI] [PubMed] [Google Scholar]
- 93.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, et al. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 95.Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 96.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 97.Hu D, Barajas-Martinez H, Burashnikov E, Springer M, Wu Y, Varro A, et al. A mutation in the beta 3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ Cardiovasc Genet. 2009;2:270–278. doi: 10.1161/CIRCGENETICS.108.829192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watanabe H, Koopmann TT, Le Scouarnec S, Yang T, Ingram CR, Schott JJ, et al. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Groenewegen WA, Firouzi M, Bezzina CR, Vliex S, van Langen IM, Sandkuijl L, et al. A cardiac sodium channel mutation cosegregates with a rare connexin40 genotype in familial atrial standstill. Circ Res. 2003;92:14–22. doi: 10.1161/01.res.0000050585.07097.d7. [DOI] [PubMed] [Google Scholar]
- 100.Makiyama T, Akao M, Tsuji K, Doi T, Ohno S, Takenaka K, et al. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol. 2005;46:2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 101.Bordachar P, Reuter S, Garrigue S, Cai X, Hocini M, Jais P, et al. Incidence, clinical implications and prognosis of atrial arrhythmias in Brugada syndrome. Eur Heart J. 2004;25:879–884. doi: 10.1016/j.ehj.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 102.Kneller J, Kalifa J, Zou R, Zaitsev AV, Warren M, Berenfeld O, et al. Mechanisms of atrial fibrillation termination by pure sodium channel blockade in an ionically-realistic mathematical model. Circ Res. 2005;96:e35–e47. doi: 10.1161/01.RES.0000160709.49633.2b. [DOI] [PubMed] [Google Scholar]
- 103.Song Y, Shryock JC, Belardinelli L. An increase of late sodium current induces delayed afterdepolarizations and sustained triggered activity in atrial myocytes. Am J Physiol Heart Circ Physiol. 2008;294:H2031–H2039. doi: 10.1152/ajpheart.01357.2007. [DOI] [PubMed] [Google Scholar]
- 104.Oberti C, Wang L, Li L, Dong J, Rao S, Du W, et al. Genome-wide linkage scan identifies a novel genetic locus on chromosome 5p13 for neonatal atrial fibrillation associated with sudden death and variable cardiomyopathy. Circulation. 2004;110:3753–3759. doi: 10.1161/01.CIR.0000150333.87176.C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X, Yang H, Corydon MJ, Pedersen S, Korenberg JR, Chen XN, et al. Localization of a human nucleoporin 155 gene (NUP155) to the 5p13 region and cloning of its cDNA. Genomics. 1999;57:144–151. doi: 10.1006/geno.1999.5741. [DOI] [PubMed] [Google Scholar]
- 106.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 107.Ehrlich JR, Biliczki P, Hohnloser SH, Nattel S. Atrial-selective approaches for the treatment of atrial fibrillation. J Am Coll Cardiol. 2008;51:787–792. doi: 10.1016/j.jacc.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 108.Kanno S, Saffitz JE. The role of myocardial gap junctions in electrical conduction and arrhythmogenesis. Cardiovasc Pathol. 2001;10:169–177. doi: 10.1016/s1054-8807(01)00078-3. [DOI] [PubMed] [Google Scholar]
- 109.Polontchouk L, Haefliger JA, Ebelt B, Schaefer T, Stuhlmann D, Mehlhorn U, et al. Effects of chronic atrial fibrillation on gap junction distribution in human and rat atria. J Am Coll Cardiol. 2001;38:883–891. doi: 10.1016/s0735-1097(01)01443-7. [DOI] [PubMed] [Google Scholar]
- 110.Dupont E, Ko Y, Rothery S, Coppen SR, Baghai M, Haw M, et al. The gap-junctional protein connexin40 is elevated in patients susceptible to postoperative atrial fibrillation. Circulation. 2001;103:842–849. doi: 10.1161/01.cir.103.6.842. [DOI] [PubMed] [Google Scholar]
- 111.Gaborit N, Steenman M, Lamirault G, Le Meur N, Le Bouter S, Lande G, et al. Human atrial ion channel and transporter subunit gene-expression remodeling associated with valvular heart disease and atrial fibrillation. Circulation. 2005;112:471–481. doi: 10.1161/CIRCULATIONAHA.104.506857. [DOI] [PubMed] [Google Scholar]
- 112.Kostin S, Klein G, Szalay Z, Hein S, Bauer EP, Schaper J. Structural correlate of atrial fibrillation in human patients. Cardiovasc Res. 2002;54:361–379. doi: 10.1016/s0008-6363(02)00273-0. [DOI] [PubMed] [Google Scholar]
- 113.Nao T, Ohkusa T, Hisamatsu Y, Inoue N, Matsumoto T, Yamada J, et al. Comparison of expression of connexin in right atrial myocardium in patients with chronic atrial fibrillation versus those in sinus rhythm. Am J Cardiol. 2003;91:678–683. doi: 10.1016/s0002-9149(02)03403-3. [DOI] [PubMed] [Google Scholar]
- 114.Wilhelm M, Kirste W, Kuly S, Amann K, Neuhuber W, Weyand M, et al. Atrial distribution of connexin 40 and 43 in patients with intermittent, persistent, and postoperative atrial fibrillation. Heart Lung Circ. 2006;15:30–37. doi: 10.1016/j.hlc.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 115.Kanagaratnam P, Cherian A, Stanbridge RD, Glenville B, Severs NJ, Peters NS. Relationship between connexins and atrial activation during human atrial fibrillation. J Cardiovasc Electrophysiol. 2004;15:206–216. doi: 10.1046/j.1540-8167.2004.03280.x. [DOI] [PubMed] [Google Scholar]
- 116.Dhein S, Duerrschmidt N, Scholl A, Boldt A, Schulte JS, Pfannmuller B, et al. A new role for extracellular Ca2+ in gap-junction remodeling: studies in humans and rats. Naunyn Schmiedebergs Arch Pharmacol. 2008;377:125–138. doi: 10.1007/s00210-008-0265-z. [DOI] [PubMed] [Google Scholar]
- 117.Takeuchi S, Akita T, Takagishi Y, Watanabe E, Sasano C, Honjo H, et al. Disorganization of gap junction distribution in dilated atria of patients with chronic atrial fibrillation. Circ J. 2006;70:575–582. doi: 10.1253/circj.70.575. [DOI] [PubMed] [Google Scholar]
- 118.Levin ER, Gardner DG, Samson WK. Natriuretic peptides. N Engl J Med. 1998;339:321–328. doi: 10.1056/NEJM199807303390507. [DOI] [PubMed] [Google Scholar]
- 119.Stambler BS, Guo GB. Atrial natriuretic peptide has dose-dependent, autonomically mediated effects on atrial refractoriness and repolarization in anesthetized dogs. J Cardiovasc Electrophysiol. 2005;16:1341–1347. doi: 10.1111/j.1540-8167.2005.00259.x. [DOI] [PubMed] [Google Scholar]
- 120.Crozier I, Richards AM, Foy SG, Ikram H. Electrophysiological effects of atrial natriuretic peptide on the cardiac conduction system in man. Pacing Clin Electrophysiol. 1993;16:738–742. doi: 10.1111/j.1540-8159.1993.tb01653.x. [DOI] [PubMed] [Google Scholar]
- 121.Le Grand B, Deroubaix E, Couetil JP, Coraboeuf E. Effects of atrionatriuretic factor on Ca2+ current and Cai-independent transient outward K+ current in human atrial cells. Pflugers Arch. 1992;421:486–491. doi: 10.1007/BF00370260. [DOI] [PubMed] [Google Scholar]
- 122.Lonardo G, Cerbai E, Casini S, Giunti G, Bonacchi M, Battaglia F, et al. Atrial natriuretic peptide modulates the hyperpolarization-activated current (If) in human atrial myocytes. Cardiovasc Res. 2004;63:528–536. doi: 10.1016/j.cardiores.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 123.Kato T, Muraski J, Chen Y, Tsujita Y, Wall J, Glembotski CC, et al. Atrial natriuretic peptide promotes cardiomyocyte survival by cGMP-dependent nuclear accumulation of zyxin and Akt. J Clin Invest. 2005;115:2716–2730. doi: 10.1172/JCI24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nyberg MT, Stoevring B, Behr ER, Ravn LS, McKenna WJ, Christiansen M. The variation of the sarcolipin gene (SLN) in atrial fibrillation, long QT syndrome and sudden arrhythmic death syndrome. Clin Chim Acta. 2007;375:87–91. doi: 10.1016/j.cca.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 125.Tsai CT, Hwang JJ, Chiang FT, Wang YC, Tseng CD, Tseng YZ, et al. Renin–angiotensin system gene polymorphisms and atrial fibrillation: a regression approach for the detection of gene-gene interactions in a large hospitalized population. Cardiology. 2008;111:1–7. doi: 10.1159/000113419. [DOI] [PubMed] [Google Scholar]
- 126.Ravn LS, Benn M, Nordestgaard BG, Sethi AA, Agerholm-Larsen B, Jensen GB, et al. Angiotensinogen and ACE gene polymorphisms and risk of atrial fibrillation in the general population. Pharmacogenet Genomics. 2008;18:525–533. doi: 10.1097/FPC.0b013e3282fce3bd. [DOI] [PubMed] [Google Scholar]
- 127.Bedi M, McNamara D, London B, Schwartzman D. Genetic susceptibility to atrial fibrillation in patients with congestive heart failure. Heart Rhythm. 2006;3:808–812. doi: 10.1016/j.hrthm.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 128.Fatini C, Sticchi E, Gensini F, Gori AM, Marcucci R, Lenti M, et al. Lone and secondary nonvalvular atrial fibrillation: role of a genetic susceptibility. Int J Cardiol. 2007;120:59–65. doi: 10.1016/j.ijcard.2006.08.079. [DOI] [PubMed] [Google Scholar]
- 129.Kato K, Oguri M, Hibino T, Yajima K, Matsuo H, Segawa T, et al. Genetic factors for lone atrial fibrillation. Int J Mol Med. 2007;19:933–939. [PubMed] [Google Scholar]
- 130.Gaudino M, Andreotti F, Zamparelli R, Di Castelnuovo A, Nasso G, Burzotta F, et al. The −174G/C interleukin-6 polymorphism influences postoperative interleukin-6 levels and postoperative atrial fibrillation. Is atrial fibrillation an inflammatory complication? Circulation. 2003;108(Suppl. 1):II195–II199. doi: 10.1161/01.cir.0000087441.48566.0d. [DOI] [PubMed] [Google Scholar]
- 131.Lai LP, Deng CL, Moss AJ, Kass RS, Liang CS. Polymorphism of the gene encoding a human minimal potassium ion channel (minK) Gene. 1994;151:339–340. doi: 10.1016/0378-1119(94)90685-8. [DOI] [PubMed] [Google Scholar]
- 132.Dobrev D, Wettwer E, Himmel HM, Kortner A, Kuhlisch E, Schuler S, et al. G-Protein beta(3)-subunit 825T allele is associated with enhanced human atrial inward rectifier potassium currents. Circulation. 2000;102:692–697. doi: 10.1161/01.cir.102.6.692. [DOI] [PubMed] [Google Scholar]
- 133.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]