Abstract
Aims
Chloroquine, an anti-malarial quinoline, is structurally similar to quinidine. Both drugs have been shown to block ion channels. We tested the hypothesis that chloroquine's mode of interaction with the vestibule of the cytoplasmic domain of the inward rectifier potassium channel Kir2.1 makes it a more effective IK1 blocker and anti-fibrillatory agent than quinidine.
Methods and results
We used comparative molecular modelling and ligand docking of the three-dimensional structures of quinidine and chloroquine in the intracellular domain of Kir2.1. Simulations predicted that chloroquine effectively blocks potassium flow by binding at the centre of the ion permeation vestibule of Kir2.1. In contrast, quinidine binds the vestibular side, only partially blocking ion movement. We tested the modelling predictions in Kir2.1-expressing human embryonic kidney (HEK)-293 cells. The half-maximal inhibitory concentration for chloroquine block of IK1 was 1.2 µM, while that of quinidine was 57 µM. Finally, we used optical mapping of Langendorff-perfused mouse hearts with cardiac-specific Kir2.1 up-regulation to compare the anti-fibrillatory effects of the drugs. In five of six hearts, 10 μM quinidine slowed the frequency but did not terminate the tachyarrhythmia. In five of five hearts, 10 μM chloroquine terminated the arrhythmia, restoring sinus rhythm.
Conclusion
Quinidine only partially blocks IK1. Chloroquine binds at the centre of the ion permeation vestibule of Kir2.1, which makes it a more effective IK1 blocker and anti-fibrillatory agent than quinidine. Integrating the structural biology of drug-ion channel interactions with cellular electrophysiology and optical mapping is an excellent approach to understand the molecular mechanisms of anti-arrhythmic drug action and for drug discovery.
Keywords: Inward rectifier potassium current, Chloroquine, Quinidine, Fibrillation
1. Introduction
The pharmacologic treatment of cardiac fibrillation is inadequate.1 Arguably, this is in part because of our relatively incomplete understanding of the mechanisms of arrhythmias, and our poor knowledge of the molecular bases of drug effects. Recent advancements in the structural biology of membrane ion channel proteins provide new opportunities to better understand drug-ion channel interactions and hopefully improve anti-arrhythmic therapy.
Quinidine is a class-1 anti-arrhythmic quinoline-type drug that is only partially effective as an anti-fibrillatory agent,2,3 although recently it had a resurgence as being potentially useful in the prevention of arrhythmias associated with the short QT syndrome (SQTS).4–6 Here, we aimed at shedding light on the molecular bases on quinidine's anti-arrhythmic action by comparing its effects with those of the anti-malarial chloroquine, a related quinoline, used briefly in the past to treat arrhythmias in patients.7 More recently, chloroquine was shown to be a potent blocker of the strong inward rectifier potassium channel Kir2.1, which is the major carrier of IK1 in mammalian ventricles.8,9 It was also demonstrated to effectively terminate fibrillatory activity in three different experimental models.9 On the other hand, the gain of function mutations in Kir2.1 has been shown to result in both SQTS type 3 10 and familial atrial fibrillation (AF) in humans.11
The three-dimensional crystal structure of the intracellular domain of Kir2.1 has been solved.12 It has also been shown that amino acids E224, D259, E299, F254, and D255 of the Kir2.1 intracellular domain, which form a highly electronegative ring, are critical for chloroquine's blocking action.8,9 This new information allowed us to directly compare the structural and electrophysiological properties of chloroquine-mediated Kir2.1 block to those of quinidine. Chloroquine and quinidine are closely related in their chemical structure; both have a quinoline ring base but different functional groups. We tested the hypothesis that chloroquine is a more effective blocker of IK1 than quinidine because the geometry of the intracellular ion-permeation pathway in the Kir2.1:chloroquine complex is markedly different from that of Kir2.1:quinidine due to the dissimilarities in the functional groups of the drugs.
We employed a numerical and experimental approach integrating protein structure, single cell, and whole organ electrophysiology. We compared chloroquine's specific mode of interaction with the vestibule of the Kir2.1 cytoplasmic domain12 and the resulting geometry of the vestibule–chloroquine complex with those of quinidine. We substantiated the molecular modelling predictions using patch clamping to compare the effects of chloroquine with those of quinidine on Ba2+ sensitive currents in human embryonic kidney (HEK)-293 cells transiently expressing Kir2.1. Finally, we used isolated Langendorff-perfused hearts from transgenic mice13 with cardiac-specific over-expression of Kir2.114 to compare the anti-fibrillatory actions of the two drugs.
2. Methods
2.1. Molecular modelling
Numerical studies were based on the co-ordinates for chloroquine and quinidine developed in the small molecule topology generator PRODRG15 and the cytoplasmic domain of Kir2.1 (PDB ID: 1U4F). Docking simulations were performed in Autodock 4.0.116 employing the generic algorithm for 10 runs with a population of 150. Prior to these calculations, the ionization of the ligands was adjusted for physiological pH. The same protonated protein model, grid size, and grid placement were used for all docking studies, and the ligands were initially positioned adjacent to the protein model outside of the channel centred at (0,0,0). During calculations, the protein was held rigid and the ligands were allowed to flex. The resulting complexes were ranked according to the binding energy. The complex with the lowest binding energy was further refined by slow-cooled simulated annealing followed by minimization in Crystallography and the nuclear magnetic resonance (NMR) system program.17 During the simulated annealing runs, both protein and ligand were flexible. The complex was heated to 1000 K and then cooled to 300 K in 25 K steps with a surrounding dielectric constant of 80. Solvent accessible surface areas for the complexes were calculated in Access from CCP4 (The CCP4 Suite: Programs for Protein Crystallography) and Solvent accessible surface maps of the complexes were calculated using a 1.4 Å probe and displayed in Pymol with the APBS plug-in (PyMOL Molecular Graphics System 2008; DeLano Scientific, Palo Alto, CA, USA).
2.2. Experimental
2.2.1. Molecular biology and patch clamping
Kir2.1 cDNA was subcloned into pcDNA3.1(+) plasmid (Invitrogen) and expressed in HEK-293 cells (ATCC, Virginia). Macroscopic currents were recorded as detailed earlier8 and Ba2+-sensitive (2 mM) current voltage (IV) relationships were constructed. For the determination of the half-maximal inhibitory concentration (IC50) for the IK1 block by chloroquine (0.3–10 µM) or quinidine (3–1000 µM), the fractional block of current was plotted as a function of drug concentration ([D]) and the data fit with the Hill equation: 1/[1−(IC50)/[D]nh]. Analysis of membrane currents through the action potential (AP) voltage clamp protocol was performed in HEK-293 cells by using the AP recorded from isolated mouse ventricular myocytes as the command signal. Currents elicited in response to the AP voltage clamp were recorded under control conditions, and in the presence of 10 µM chloroquine or quinidine. Kir2.1 currents are represented as Ba2+ sensitive currents elicited by the AP voltage clamp. Alanine mutagenesis of Kir2.1 residues was conducted as detailed earlier8 and the current at −50 mV was recorded in transfected HEK cells in response to voltage steps from −80 mV to −50 mV, and then to −100 mV, in the absence or presence of 100 µM quinidine.
2.2.2. Optical mapping
All animal studies conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and to the University of Michigan Committee on the Use and Care of Animals (protocol 09985). Adult, Kir2.1-overexpressing transgenic mice13 were heparinized and anaesthetized. The excised heart was Langendorff-perfused with warm (36 ± 1°C), and oxygenated Tyrode's solution, and optical mapping was carried out as described earlier18. Recording rate: 600–1000 frames per second, and resolution: 109 µm.
2.2.3. Arrhythmia induction
To induce ventricular tachycardia/fibrillation (VT/VF), we used a bipolar, silver tip stimulation electrode placed at the apex of the heart to deliver short bursts of 3 ms square pulses at a cycle length of 10–20 ms14.
2.2.4. Dominant frequency maps
We used fast Fourier transform analysis on each pixel of the epicardial optical signals to generate dominant frequency (DF) maps19 and constructed maps of spatial DF distribution as previously described.
2.2.5. Statistics
Results are mean ± standard deviation. ANOVA and Fisher's exact and t-tests were used as appropriate. Significance was at P< 0.05.
3. Results
3.1. Chloroquine and quinidine interact differently with the tetrameric Kir2.1 intracellular domain
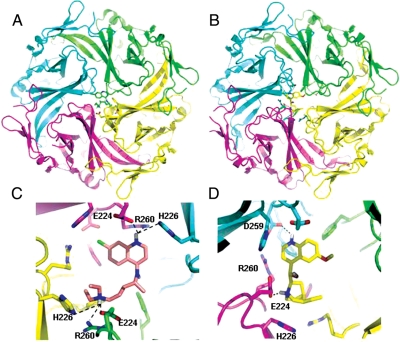
Pegan et al.12 elucidated the three-dimensional crystal structure of Kir2.1. Since there is no crystal or NMR structure of Kir2.1 or any family member complexed with a drug-like molecule, Rodriguez-Menchaca et al.8 and Noujaim et al.9 used modelling to predict the interaction of chloroquine and Kir2.1. Here, we compared the docking of chloroquine and quinidine in the ion permeation of the tetrameric Kir2.1 intracellular domain. Chloroquine and quinidine are structurally related, in that they both have a quinoline ring base. However, their functional groups are different. Consequently, as shown in Figure 1, modelling studies revealed that chloroquine and quinidine have different modes of binding to the Kir2.1 intracellular domain. In Figure 1A, chloroquine represented in green sticks, acts as a plug, fitting in the centre of the tetrameric channel. The drug is suspended in a band of negative charges formed by the acidic amino acids E224, D259, and E299. As shown in Figure 1C, E224 forms charge–charge interactions with chloroquine. D259 is within 4 Å of the drug, and is involved in van der Waals interactions. H226 and R260 form hydrogen bonds with the protonated nitrogens of chloroquine, and F254 from three subunits provide a hydrophobic base for the molecule. E299 is located >5 Å from the chloroquine molecule. Quinidine, yellow sticks in Figure 1B and D, binds in the same zone of negative charge. However, it forms hydrogen bonds and van der Waals interactions with specific residues from two subunits of Kir2.1 (Figure 1D), causing it to be off-centred (in contrast to the centred position of chloroquine in Figure 1A and C) and drawn to one side of the channel (Figure 1B and D). The carboxylate of D259 from one subunit forms a hydrogen bond with the protonated quinoline ring of quinidine, while the carboxylate from E224 of an adjacent subunit forms hydrogen bonds with the caged nitrogen. In addition to the hydrophilic interactions, F254 from two subunits (the unlabelled pink and blue residues underneath quinidine in Figure 1D) form hydrophobic interactions with the quinoline ring and the carbon cage along with the acyl chain of R260 from two subunits. Due to the four-fold symmetry of Kir2.1, the top four docking results were equivalent, simply separated by a 90° rotation about the channel.
Figure 1.
Models of the lowest energy binding complexes. (A) Kir2.1 complexed with chloroquine. Kir2.1; ribbon structure with each chain depicted in a different colour. Chloroquine; green sticks. (B). Kir2.1:quinidine complex. Quinidine: yellow sticks. (C) Detailed view of chloroquine binding with Kir2.1 side chains within 5 Å of chloroquine (sticks.) (D). Close up view. Binding site of quinidine with side chains of Kir2.1 residues within 4 Å of the molecule (sticks). Hydrogen bonds: dashed lines. Kir2.1 amino acids are labelled as D259; F254; E224; R260, and H226.
3.2. Chloroquine and quinidine have different effects on the geometry of the aqueous vestibule of Kir2.1
To unveil the consequences of the different interactions of chloroquine and quinidine with the channel, solvent accessible areas and maps were calculated using CCP4 and Pymol, respectively (Figure 2). Maps were constructed by probing the channels with a probe of 1.4 Å in radius, corresponding to the radius of a water molecule. The probe traces the water accessible regions of the channel. The resulting shell structure is three-dimensional, but for visualization, a slab was shown to expose the shells’ insides. The yellow colours denote the exterior of the ‘solvent accessible shell’ and black is the inside of the shell's cavity or the ion-permeation pathway. The figure shows differences in the cavity's geometries depending on whether chloroquine or quinidine was bound to the ion-permeation pathway. The total surface area decreased by 162 and 217.5 Å2 upon binding of quinidine and chloroquine, respectively. The surface maps defining the channel show that Kir2.1 has a cavity that runs through the centre of the tetramer, but the channel is disrupted upon compound binding. Chloroquine causes a break in the ion pathway (Figure 2 white broken lines), while quinidine partially blocks the channel, but allows room for ions to still pass through albeit at a slower pace (Figure 2 white arrow). It is possible for more than one molecule of quinidine to bind to the electronegative band in the cytoplasmic tail of Kir2.1. However, the restricted space imposes an extremely tight fit, rendering this scenario unlikely.
Figure 2.
Stereoviews of solvent accessible surface maps of the intracellular pore of Kir2.1 in complex with chloroquine and quinidine. Views oriented with respect to a 90° rotation of Figure 1A and B. Kir2.1 cytosolic side is on the left in each figure. Outer surfaces: yellow; cavities: black. Slabbed surfaces reveal the cavity of the inner vestibule of the intracellular domain of Kir2.1. (A) Surface map for Kir2.1. Surface map of Kir2.1 in complex with chloroquine (B) and quinidine (C). Orange arrow: ion-permeation pathway in Kir2.1 alone. Broken white lines: break in the ion-permeation pathway. White arrow: narrowing of the ion-permeation pathway.
3.3. Chloroquine is a more potent blocker of IK1 compared with quinidine
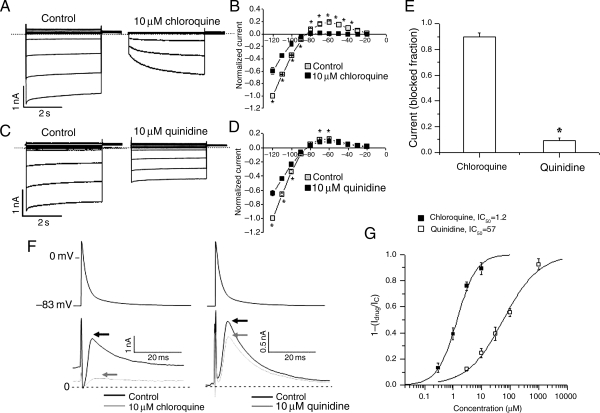
Molecular modelling predicted that chloroquine was a better blocker of IK1 than quinidine. We tested this prediction using patch clamping. Figure 3 shows whole-cell current traces in Kir2.1-expressing HEK-293 cells before and after 10 μM chloroquine (Figure 3A) or 10 μM quinidine (Figure 3C). The IV curves illustrate the ability of chloroquine to block most of the outward component of IK1 (n= 5) (Figure 3B), whereas the effect of 10 μM quinidine was much less pronounced (n= 5) (Figure 3D). Figure 3E shows that the fraction of blocked outward current at −60 mV in Kir2.1-expressing HEK-293 cells by chloroquine was 0.90 ± 0.03 vs. 0.09 ± 0.2 in response to quinidine (n= 5, *P< 0.01). In Figure 3F, we examined the whole-cell IK1 current in Kir2.1-expressing HEK-293 cells in response to a mouse AP clamp protocol. The top traces are voltage-clamp profiles and the bottom traces are the control and chloroquine- (left) and quinidine (right)-sensitive currents. The arrows indicate the maximum outward IK1, which was markedly reduced by 10 μM chloroquine, compared with the more subtle effect of 10 μM quinidine. Figure 3G shows the dose–response curves of the fractional blocked peak outward current in response to the AP voltage clamp with chloroquine and quinidine. Solid lines are Hill equation best fits. IC50: chloroquine = 1.2 ± 0.2 μM, quinidine = 57 ± 3.8 μM. Chloroquine is 48 times more potent as an IK1 blocker than quinidine.
Figure 3.
Effects of chloroquine and quinidine on IK1. Currents in response to 4 s pulses from a holding potential of −80 mV to test potentials from −120 mV to −20 mV, applied at 10 mV increments in HEK-293 cells transfected with Kir2.1 in the absence and presence of 10 μM chloroquine (A) or 10 μM quinidine (C). Scale: 2 s; 1 nA, the dashed lines define the zero current levels. IV curves of currents at the end of pulses are normalized to the current at −120 mV, obtained under control conditions (n = 5 cells, *P< 0.05). Chloroquine (B) reduces both the inward and outward components of IK1. Quinidine (D) reduces the inward component of IK1 but has a modest effect on the outward current. (E) The fraction of blocked outward current at −60 mV by chloroquine was 0.90 ± 0.03 vs. 0.09 ± 0.02 in response to quinidine (n= 5, *P< 0.01). (F) AP voltage clamping in HEK-293 cells transfected with Kir2.1. Top, mouse left ventricular AP. Bottom, IK1 current profile in response to the AP voltage clamp with 10 µM chloroquine or quinidine. Chloroquine reduces the maximum current (indicated by arrows). Quinidine has a modest effect. (G) Dose response curve of the fractional blocked peak outward current in response to the AP voltage clamp with chloroquine and quinidine. Solid lines are Hill equation best fits. IC50: chloroquine = 1.2 ± 0.2, quinidine = 57 ± 3.8 μM.
3.4. Chloroquine but not quinidine terminates IK1-mediated tachyarrhythmias in the mouse heart
Up-regulation of IK1 in the mouse heart via cardiac-specific Kir2.1 over-expression leads to stable and fast re-entrant VT/VF.14 Here, we compared the effects of chloroquine with those of quinidine on VT/VF in this mouse model. On the basis of the molecular simulations, and the patch clamp experiments, we postulated that at the same dose, intracoronary perfusion of 10 μM chloroquine, but not quinidine, should terminate VT/VFs and restore sinus rhythm, even though other currents, such as INa, are affected by both chloroquine and quinidine.20
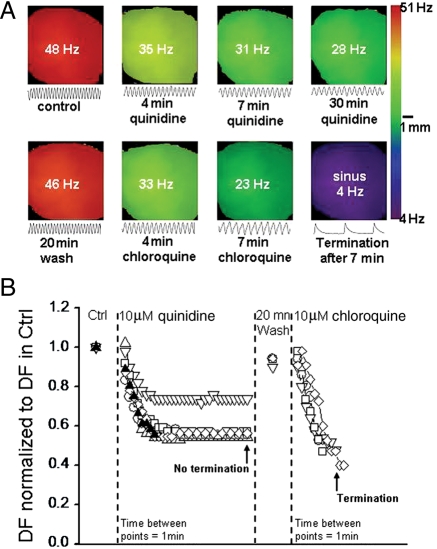
We then proceeded to evaluate the anti-fibrillatory properties of quinidine compared with chloroquine in six mouse hearts. VT/VF was induced by burst pacing the apex of the heart. Figure 4A, top row, is DFmax maps showing the frequency of the tachyarrhythmia during the course of 30 min perfusion of 10 μM quinidine. The tracings below each map are 500 ms single pixel recordings obtained from the same location. At the onset, the frequency was 48 Hz. It decelerated progressively to 28 Hz after 30 min of quinidine. However, sinus rhythm was not restored. In the bottom row of Figure 4A, after a washout of 20 min, the arrhythmia accelerated to 46 Hz. Thereafter, chloroquine perfusion started, and after 7 min, the tachyarrhythmia slowed to 23 Hz, and sinus rhythm resumed. Figure 4B shows a composite data from six experiments. The time elapsed between individual data points after the application of 10 μM quinidine or chloroquine is 1 min. With quinidine, tachyarrhythmias terminated in one heart (filled symbol) at 8 min of quinidine perfusion but not in five other hearts even after 30 min of continuous quinidine perfusion (open symbols). In these hearts, the DFmax decreased by a factor of 0.6 ± 0.1 without stopping, and after a washout period of 20 min, the DFmax recovered back to 0.94 ± 0.05 of control. We then perfused five of these hearts with 10 μM chloroquine. The VT/VF frequency reduced by a factor of 0.55 ± 0.035, and sinus rhythm was restored after an average time of 8 min of chloroquine perfusion. Chloroquine restored sinus rhythm in five of five hearts, while quinidine did so in one of six hearts (χ2 = 0.015, Fisher's test).
Figure 4.
Effects of chloroquine and quinidine on ventricular arrhythmia. (A) Top row: DF maps at the onset of pacing-induced VT (DFmax = 48 Hz), 4 min (35 Hz), 7 min (31 Hz), and 30 min (28 Hz) after starting 10 μM quinidine. VT slowed down but did not terminate. Bottom row: DF maps of VT after 20 min quinidine washout (DFmax = 46 Hz), then at 4 min (33 Hz) and 7 min (23 Hz), followed by sinus rhythm restoration (4 Hz), in the presence of 10 μM chloroquine. Single pixel recordings (500 ms) are shown below each map. (B). Plot of normalized DFmax in six hearts during VT. In five of six hearts (open symbols) 30 min of 10 μM quinidine slowed down but did not terminate VT/VF. In one heart tachyarrhythmia terminated (filled symbol). After 20 min of washout, 10 μM chloroquine applied to five tachyarrhythmic hearts terminated the arrhythmias and sinus rhythm resumed.
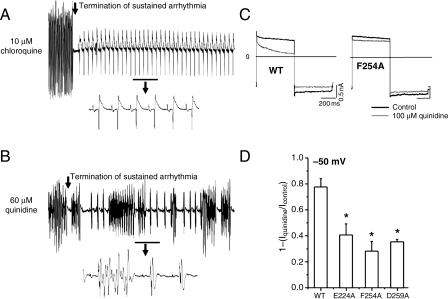
Since 10 μM is below the IC50 for quinidine block of IK1 (Figure 3G), we tested the effects of 60 μM quinidine on induced tachyarrhythmias in three isolated hearts. This high dose of quinidine terminated the sustained tachyarrhythmia; however, in all hearts, the restored sinus beats were repeatedly interrupted by spontaneous, non-sustained bursts of VT/VF. On the other hand, hearts perfused with chloroquine were in normal sinus rhythm after chemical defibrillation. Figure 5A and B shows examples of 10 s electrocardiogram (ECG) runs illustrating that while 10 μM chloroquine restored sinus rhythm, 60 μM quinidine was pro-arrhythmic.
Figure 5.
Arrhythmia termination by 10 μM chloroquine and 60 μM quinidine. (A and B). 10 s ECG runs highlighting the termination of burst pacing-induced tachyarrhythmia by chloroquine and quinidine. The underlined regions are magnified beneath. Choroquine restored sinus rhythm. Quinidine was proarrhythmic. Alanine scanning mutagenesis of amino acids facing the cytoplasmic pore of Kir2.1. (C) Voltage command: 500 ms steps from −80 mV to −50 mV then to −100 mV, in the presence or absence of 100 μM Quinidine. Representative currents elicited in HEK cells expressing WT Kir2.1 (left), Kir2.1 F254A (right). Scales: 200 ms, 0.5 nA. (D) Fractional blocked outward current measured at −50 mV in the presence of E224A, F254A, and D259A. *<0.05, n= 5 cells each.
3.5. Mutation of specific residues suggested by modelling to be involved in quinidine's interaction with Kir2.1 makes the channel less susceptible to block
Figure 5C and D shows that amino acids E224, F254, and D259 proposed by the model to interact with chloroquine are experimentally involved in quinidine's block of the channel. These residues have also been shown earlier to be important for chloroquine's block of Kir2.18. In Figure 5C, we show representative currents recorded in HEK cells transfected with WT Kir2.1 or Kir2.1 carrying the mutation F254A in the presence or absence of 100 µM quinidine. Quinidine blocks the WT channel; however, in the presence of the F254A mutation, the ability of quinidine to block the outward current was reduced. Figure 5D is the quantification of the fraction of blocked current at −50 mV in the presence of E224A, F254A, or D259A mutations, which made the channel resistant to the quinidine block.
4. Discussion
We have taken advantage of knowledge derived from the solved crystal structure of the Kir2.1 intracellular domain to provide a structural basis for the electrophysiological effects of two different quinolines (chloroquine and quinidine) on IK1. Molecular modelling predicts that chloroquine should be a more effective IK1 blocker than quinidine. Although both compounds are envisaged to bind to the negatively charged ring within the vestibule, they have different binding modes, which alter the size of the ion pathway. While chloroquine introduces a break in the ion-permeation pathway, quinidine's interaction with the channel results in a partial block. This was further verified experimentally by patch clamping and optical mapping. The IC50 of chloroquine's block of IK1 was 1.2 μM while that of quinidine was 57 μM. Consequently, in a mouse model of IK1 up-regulation, chloroquine terminated VF/VT in a total of five of five mouse hearts. Quinidine terminated arrhythmias in one of six hearts (χ2 = 0.015).
Integration of information, from the molecule to the organ, derived from the study of the effects of currently available drugs on cardiac electrical properties may help set the stage for the development of new, more effective and safer therapeutic approaches, with the ultimate goal of preventing sudden cardiac death. We have therefore conducted studies at three complementary levels of integration to investigate the anti-fibrillatory effects of chloroquine, an anti-malarial quinoline, which has been suggested to have anti-arrhythmic properties,7 in comparison with quinidine, another quinoline, widely used in the past.2,3
Recently, quinidine was suggested to be beneficial in certain cases of short QT syndrome.4–6 In addition, quinidine was shown to have superior anti-arrhythmic properties in the treatment of VF in animal models of pharmacologically induced short QT compared with sotalol and flecainide because it prolongs refractoriness and reduces dispersion of repolarization.21 Similar to quinidine, chloroquine also prolongs the AP duration and refractoriness in the atria and ventricles by effectively blocking inward rectifier potassium currents, including IK1, the acetylcholine-activated potassium current (IKA,Ch), and the ATP-dependent potassium current (IK,ATP).9,20 Our results using a transgenic mouse model of Kir2.1 overexpression demonstrate that the anti-fibrillatory effects of chloroquine are substantially greater than those of quinidine in IK1-mediated tachyarrhythmia. Those effects correlate strongly with the preferential ability of chloroquine to block IK1 when compared with quinidine. Similarly, by blocking other inward rectifier potassium currents, chloroquine was shown to terminate both VT/VF and AF in three different species, including mouse, rabbit and sheep.9
It seems clear that the anti-fibrillatory effects of chloroquine in the Kir2.1-overexpressing mouse model are due in large measure to preferential IK1 blockade. IK1 is of interest because it plays a significant role in the stability and frequency of re-entrant VT/VF 14,19,22 and in AF. From the clinical standpoint, IK1 is important because mutations in the Kir2.1 protein have been shown to be associated with at least two different inherited arrhythmogenic diseases in humans.10,11 However, one should not underestimate the potential contribution of chloroquine's inhibition of other channels to the mechanism of VF termination. Compared with other inward and outward currents, chloroquine preferentially blocks IK1 (IK1>IKr>INa>ICaL).20 However, while similar comparisons on the relative effectiveness of inhibition on different ionic channels have not been made for quinidine, there is evidence that it also blocks INa, ICaL, Ito, IKr, IKs, IKur, and IK1.23
IK1 is important in the control of the rate and frequency of functional re-entry.14,19 Experiments and numerical simulations demonstrated that IK1 mediates electrotonic interactions between the rotor core and its immediate surroundings.14,19 When IK1 is increased, the rate of the terminal phase of repolarization of the AP is accelerated, and the resting membrane potential is slightly hyperpolarized.13,14,24The space constant is also decreased14 resulting in a smaller core size. The small core, combined with the short AP duration, reduces the likelihood of wave front-wave tail interactions, allowing a fast and relatively stable rotor.14 The opposite occurs when IK1 is reduced, such as by chloroquine9 or BaCl2,14,22 both of which lead to slowing and eventual termination of rotor activity.
Rodriguez-Menchaca et al.8 have shown that amino acids E224, D259, E299, F254, and D255 of the Kir2.1 intracellular domain are critical for chloroquine blockade. Noujaim et al.9 also showed that chloroquine interacts with electronegative residues in the intracellular domains of Kir2.1, Kir3.1, and Kir6.2, leading to IK1, IKACh, and IKATP blockade, and termination of arrhythmias mediated by these three inward rectifier currents in the mouse, rabbit, and sheep hearts. This study elucidates the structural basis for the different IK1 blocking efficacies of chloroquine and quinidine. Similarly to the case of chloroquine, E224, D259, F254, H266, R260, and D255 are also critical for a partial block of Kir2.1 by quinidine. The atoms of E299 are not solvent accessible, therefore the role of E299 is most likely to maintain the structural integrity of the protein and to offset the positive charges of R228 and H226. Such residues help to form an electronegative band that interacts with the protonated quinoline ring and caged nitrogen of quinidine. Hence, both quinidine and chloroquine block Kir2.1 by interacting with the band of negative charge located in the inner vestibule of the intracellular pore.12 However, chloroquine is a more potent IK1 blocker, and is thus more effective than quinidine in terminating IK1-mediated re-entrant VT/VF.
Drug-ion channel interaction studies that span multiple levels of integration that include structure/function relationships, up to the whole heart may be a helpful strategy in furthering our understanding of current anti-arrhythmics, and the development of novel, potentially useful anti-fibrillatory pharmacophores.
4.1. Limitations
Molecular simulations cannot account for all the drug-channel interactions that can take place or the possible effects of modulatory agents, such as PIP2, on Kir channels.25 Crystallization and determination of the 3D structure of the drugs in complex with the Kir channels would be essential to understand all aspects of drug-channel interactions.
The mouse is a useful animal model. However, several differences exist between the assemblage of ion channels underlying the AP of the mouse compared with other species.26,27 Care should be taken in translating to the human, basic electrophysiological findings in the mouse. Chloroquine and quinidine are not specific IK1 blockers. Their effects on other currents can contribute to the results we observe in the heart.
While reducing IK1 may be antifibrillatory, there is also evidence that IK1 loss-of-function, such as in Anderson–Tawil syndrome (ATS), may be proarrhythmic.28 However, whilst ATS patients may develop ventricular tachyarrhythmias, including torsades de pointes, sudden cardiac death is a very rare event.28–30
Does the above suggest that the ventricles of patients with ATS are incapable of undergoing stable, long-standing, functional re-entry? And does our study suggest that compared with chloroquine, quinidine's relatively lower efficacy as an anti-fibrillatory agent is due to its relatively modest effect on IK1? Further studies in animal models of ATS, together with pharmacologic analyses of the type presented here should help answer this question. Such knowledge, together with the new information derived from our study, may lead to novel integrative and multidisciplinary approaches in search of new anti-fibrillatory pharmacophores that increase the inward going rectification of IK1, preferably in a cardiac-specific manner.
Conflict of interest: none declared.
Funding
This work was supported by National Institutes of Health (P01-HL039707, P01-HL087226 to J.J. and K99-HL105574 to S.F.N.) and Leducq Foundation grants to J.J.; American Heart Association (Scientist Development Grant to S.V.P. and Postdoctoral Fellowship to S.F.N.); The Secretarίa de Educación Pública-Consejo Nacional de Ciencia y Tecnologίa México (CB-2008-01-105941 to J.A.S.C.]; The University of Michigan Center for Structural Biology to J.A.S.
References
- 1.Benjamin EJ, Chen PS, Bild DE, Mascette AM, Albert CM, Alonso A, et al. Prevention of atrial fibrillation: Report from a National Heart, Lung, and Blood Institute Workshop. Circulation. 2009;119:606–618. doi: 10.1161/CIRCULATIONAHA.108.825380. doi:10.1161/CIRCULATIONAHA.108.825380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coplen SE, Antman EM, Berlin JA, Hewitt P, Chalmers TC. Efficacy and safety of quinidine therapy for maintenance of sinus rhythm after cardioversion. A meta-analysis of randomized control trials. Circulation. 1990;82:1106–1116. doi: 10.1161/01.cir.82.4.1106. [DOI] [PubMed] [Google Scholar]
- 3.Flaker GC, Blackshear JL, McBride R, Kronmal RA, Halperin JL, Hart RG. Antiarrhythmic drug therapy and cardiac mortality in atrial fibrillation. The Stroke Prevention in Atrial Fibrillation Investigators. J Am Coll Cardiol. 1992;20:527–532. doi: 10.1016/0735-1097(92)90003-6. doi:10.1016/0735-1097(92)90003-6. [DOI] [PubMed] [Google Scholar]
- 4.Wolpert C, Schimpf R, Giustetto C, Antzelevitch C, Cordeiro J, Dumaine R, et al. Further insights into the effect of quinidine in short QT syndrome caused by a mutation in HERG. J Cardiovasc Electrophysiol. 2005;16:54–58. doi: 10.1046/j.1540-8167.2005.04470.x. doi:10.1046/j.1540-8167.2005.04470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giustetto C. Quinidine to treat short QT syndrome: a real alternative to ICD? In: Raviele A, editor. Cardiac Arrhythmias. Italy: Springer Verlag; 2005. pp. 333–335. [Google Scholar]
- 6.Kaufman ES. Quinidine in short QT syndrome: an old drug for a new disease. J Cardiovasc Electrophysiol. 2007;18:665–666. doi: 10.1111/j.1540-8167.2007.00815.x. doi:10.1111/j.1540-8167.2007.00815.x. [DOI] [PubMed] [Google Scholar]
- 7.Burrell ZL, Jr, Martinez AC. Chloroquine and hydroxychloroquine in the treatment of cardiac arrhythmias. N Engl J Med. 1958;258:798–800. doi: 10.1056/NEJM195804172581608. doi:10.1056/NEJM195804172581608. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Menchaca AA, Navarro-Polanco RA, Ferrer-Villada T, Rupp J, Sachse FB, Tristani-Firouzi M, et al. The molecular basis of chloroquine block of the inward rectifier Kir2.1 channel. Proc Natl Acad Sci USA. 2008;105:1364–1368. doi: 10.1073/pnas.0708153105. doi:10.1073/pnas.0708153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noujaim SF, Stuckey JA, Ponce-Balbuena D, Ferrer-Villada T, Lopez-Izquierdo A, Pandit S, et al. Specific residues of the cytoplasmic domains of cardiac inward rectifier potassium channels are effective antifibrillatory targets. FASEB J. 2010;24:4302–4312. doi: 10.1096/fj.10-163246. : [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priori SG, Pandit SV, Rivolta I, Berenfeld O, Ronchetti E, Dhamoon A, et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ Res. 2005;96:800–807. doi: 10.1161/01.RES.0000162101.76263.8c. doi:10.1161/01.RES.0000162101.76263.8c. [DOI] [PubMed] [Google Scholar]
- 11.Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054. doi:10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 12.Pegan S, Arrabit C, Zhou W, Kwiatkowski W, Collins A, Slesinger PA, et al. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8:279–287. doi: 10.1038/nn1411. doi:10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 13.Li J, McLerie M, Lopatin AN. Transgenic upregulation of IK1 in the mouse heart leads to multiple abnormalities of cardiac excitability. Am J Physiol Heart Circ Physiol. 2004;287:H2790–H2802. doi: 10.1152/ajpheart.00114.2004. doi:10.1152/ajpheart.00114.2004. [DOI] [PubMed] [Google Scholar]
- 14.Noujaim SF, Pandit SV, Berenfeld O, Vikstrom K, Cerrone M, Mironov S, et al. Up-regulation of the inward rectifier K+ current (IK1) in the mouse heart accelerates and stabilizes rotors. J Physiol. 2007;578:315–326. doi: 10.1113/jphysiol.2006.121475. doi:10.1113/jphysiol.2006.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. doi:10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 16.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [Google Scholar]
- 17.Brunger AT, Admas PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Vaidya D, Morley GE, Samie FH, Jalife J. Reentry and fibrillation in the mouse heart. A challenge to the critical mass hypothesis. Circ Res. 1999;85:174–181. doi: 10.1161/01.res.85.2.174. [DOI] [PubMed] [Google Scholar]
- 19.Samie FH, Berenfeld O, Anumonwo J, Mironov SF, Udassi S, Beaumont J, et al. Rectification of the background potassium current: a determinant of rotor dynamics in ventricular fibrillation. Circ Res. 2001;89:1216–1223. doi: 10.1161/hh2401.100818. doi:10.1161/hh2401.100818. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Chapula JA, Salinas-Stefanon E, Torres-Jacome J, Benavides-Haro DE, Navarro-Polanco RA. Blockade of currents by the antimalarial drug chloroquine in feline ventricular myocytes. J Pharmacol Exp Ther. 2001;297:437–445. [PubMed] [Google Scholar]
- 21.Milberg P, Tegelkamp R, Osada N, Schimpf R, Wolpert C, Breithardt G, et al. Reduction of dispersion of repolarization and prolongation of postrepolarization refractoriness explain the antiarrhythmic effects of quinidine in a model of short QT syndrome. J Cardiovasc Electrophysiol. 2007;18:658–664. doi: 10.1111/j.1540-8167.2007.00813.x. doi:10.1111/j.1540-8167.2007.00813.x. [DOI] [PubMed] [Google Scholar]
- 22.Warren M, Guha PK, Berenfeld O, Zaitsev A, Anumonwo JM, Dhamoon AS, et al. Blockade of the inward rectifying potassium current terminates ventricular fibrillation in the guinea pig heart. J Cardiovasc Electrophysiol. 2003;14:621–631. doi: 10.1046/j.1540-8167.2003.03006.x. doi:10.1046/j.1540-8167.2003.03006.x. [DOI] [PubMed] [Google Scholar]
- 23.Clark RB, Sanchez-Chapula J, Salinas-Stefanon E, Duff HJ, Giles WR. Quinidine-induced open channel block of K+ current in rat ventricle. Br J Pharmacol. 1995;115:335–343. doi: 10.1111/j.1476-5381.1995.tb15882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miake J, Marban E, Nuss HB. Functional role of inward rectifier current in heart probed by Kir2.1 overexpression and dominant-negative suppression. J Clin Invest. 2003;111:1529–1536. doi: 10.1172/JCI17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logothetis DE, Jin T, Lupyan D, Rosenhouse-Dantsker A. Phosphoinositide-mediated gating of inwardly rectifying K+ channels. Pflugers Arch. 2007;455:83–95. doi: 10.1007/s00424-007-0276-5. doi:10.1007/s00424-007-0276-5. [DOI] [PubMed] [Google Scholar]
- 26.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. doi:10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 27.Nerbonne JM. Molecular basis of functional voltage-gated K+ channel diversity in the mammalian myocardium. J Physiol. 2000;525:285–298. doi: 10.1111/j.1469-7793.2000.t01-1-00285.x. doi:10.1111/j.1469-7793.2000.t01-1-00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tristani-Firouzi M, Jensen JL, Donaldson MR, Sansone V, Meola G, Hahn A, et al. Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome) J Clin Invest. 2002;110:381–388. doi: 10.1172/JCI15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Benson DW, Tristani-Firouzi M, Ptacek LJ, Tawil R, Schwartz PJ, et al. Electrocardiographic features in Andersen-Tawil syndrome patients with KCNJ2 mutations: characteristic T-U-wave patterns predict the KCNJ2 genotype. Circulation. 2005;111:2720–2726. doi: 10.1161/CIRCULATIONAHA.104.472498. doi:10.1161/CIRCULATIONAHA.104.472498. [DOI] [PubMed] [Google Scholar]
- 30.Fodstad H, Swan H, Auberson M, Gautschi I, Loffing J, Schild L, et al. Loss-of-function mutations of the K(+) channel gene KCNJ2 constitute a rare cause of long QT syndrome. J Mol Cell Cardiol. 2004;37:593–602. doi: 10.1016/j.yjmcc.2004.05.011. doi:10.1016/j.yjmcc.2004.05.011. [DOI] [PubMed] [Google Scholar]