Abstract
Chronic kidney disease (CKD) has a heritable component and is an important global public health problem because of its high prevalence and morbidity.1 We conducted genome-wide association studies (GWAS) to identify susceptibility loci for glomerular filtration rate estimated by serum creatinine (eGFRcrea), cystatin C (eGFRcys), and CKD (eGFRcrea<60 ml/min/1.73m2) in European-ancestry participants of four populations-based cohorts (ARIC, CHS, FHS, RS; n=19,877, 2,388 CKD cases), and tested for external replication in 21,466 participants (1,932 CKD cases). Significant associations (p<5*10−8) were identified for SNPs with [1] CKD at the UMOD locus; [2] eGFRcrea at the UMOD, SHROOM3, and GATM/SPATA5L1 loci; [3] eGFRcys at the CST and STC1 loci. UMOD encodes the most common protein in human urine, Tamm-Horsfall protein,2 and rare mutations in UMOD cause Mendelian forms of kidney disease.3 Our findings provide new insights into CKD pathogenesis and underscore the importance of common genetic variants influencing renal function and disease.
Keywords: chronic kidney disease, renal function, epidemiology, genetics, genome-wide association study, single nucleotide polymorphism
CKD affects 10–3% of US adults.4 Estimates from Europe are similar,5 and incidence and prevalence are increasing worldwide. Its most severe form, end-stage renal disease, requires dialysis and currently affects over 500,000 US adults.6 In addition to conferring risk for end-stage renal disease, CKD increases the risk of cardiovascular disease7 and all-cause mortality.8
Multiple studies such as familial aggregation studies have provided evidence for a genetic component to kidney disease. Heritability estimates of eGFRcrea are reported between 0.41 and 0.75 in individuals with the major CKD risk factors hypertension or diabetes,9, 10 and as 0.33 in a population-based sample.11 Heritability estimates of GFRcys are similar. While rare genetic variants causing different forms of monogenetic kidney disease have been identified, common CKD susceptibility variants have been difficult to detect reproducibly by linkage or candidate gene studies.12
To discover such variants, we conducted meta-analyses of study-specific GWAS for indices of renal function, eGFRcrea and eGFRcys, and for CKD from four population-based, unselected cohorts of the CHARGE Consortium13: Atherosclerosis Risk in Communities Study, Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), and Rotterdam Study (RS). As a direct measurement of kidney function is not feasible in population-based studies, we applied commonly used estimating equations to determine eGFRcrea14 and eGFRcys.15 Population-based measures of GFR are imperfect,16 and using two different biomarkers to estimate GFR can therefore help to uncover true signals. CKD was defined as eGFRcrea <60 ml/min/1.73m2 according to National Guidelines,17 as detailed in the methods. Genotypes for >2.5 million SNPs were imputed within each study using reference genotype data from the HapMap CEU population. Study-specific details on genotyping and imputation are provided in Supplementary Table 1. SNPs showing evidence of suggestive (p<4*10−7) or significant (p<5*10−8) genome-wide association were tested for in silico replication in independent study samples, the Age Gene/Environment Susceptibility-Reykjavik Study (AGES) and the Women’s Genome Health Study (WGHS). Detailed information on the study samples are provided in the Supplementary Methods.
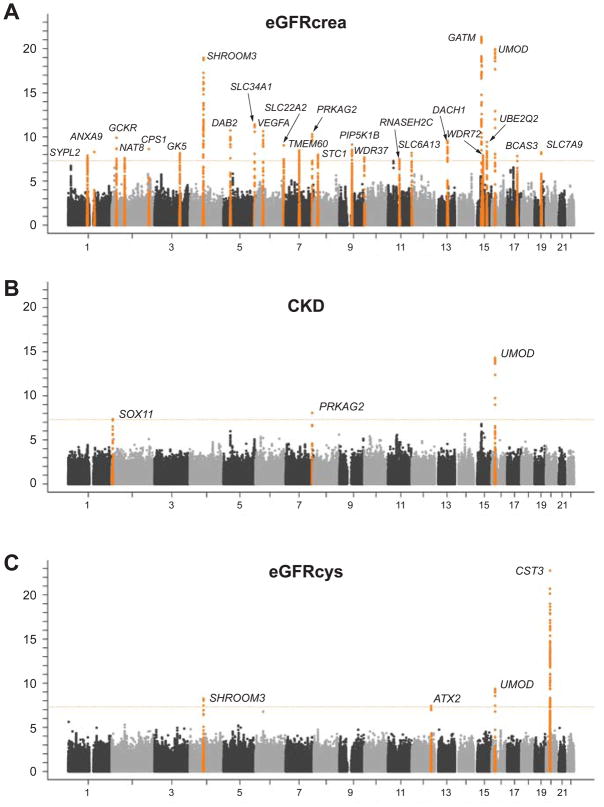
Characteristics of the four discovery and two validation study samples are shown in Table 1; 19,877 participants with 2,388 CKD cases and 21,466 participants with 1,932 CKD cases contributed information, respectively. CKD prevalence was higher in cohorts with older participants, ranging from 6.3% (WGHS) to 24.3% (AGES). Characteristics among CKD cases are provided in Supplementary Table 2. Figure 1 summarizes meta-analysis results for CKD, eGFRcrea, and eGFRcys across the discovery samples. The observed versus expected p-value distributions (quantile-quantile plots) are shown in Supplementary Figure 1: study-specific genomic inflation factors did not indicate substantial inflation of the test statistics for any of the traits.
Table 1.
Characteristics of the study samples
| ARIC | CHS | FHS | RS | AGES | WGHS | |
|---|---|---|---|---|---|---|
| Cohort information | discovery | replication | ||||
| Study Desiagn | prospective multi-center | Population-based prospective multi-center | Community-based family multi-generation | Population-based prospective | Population-based prospective | Trial |
| Analyzed sample size, CKD/eGFRcrea/eGFRcys | 8069 / 6525 / 6430 | 3278 / 3278 / 2844 | 4140 / 3934 / 2992 | 4390 / 4390 / NA | 3219 / 3219 / NA | 18247 / 18247 / NA |
| Sample characteristics* | mean (SD) / % (n) | mean (SD) / % (n) | mean (SD) / % (n) | mean (SD) / % (n) | mean (SD) / % (n) | mean (SD) / % (n) |
| Age, years | 63.1 (5.6) | 72.4 (5.4) | 62.4 (11.6) | 70.0 (9.0) | 76.4 (5.5) | 54.7 (7.1) |
| Male | 46.7 (3047) | 39.2 (1286) | 45.5 (1890) | 38.6 (1694) | 42 (1352) | 0 |
| Hypertension prevalence | 43.0 (2793) | 52.4 (1719) | 57.4 (2377) | 34.1 (1497) | 81 (2596) | 24.4 (4460) |
| Diabetes mellitus | 13.7 (893) | 11.8 (385) | 9.8 (406) | 10.7 (470) | 11.5 (368) | 2.6 (474) |
| eGFR cystatin, ml/min/1.73m2 | 84.1 (19.7) | 79.9 (18.3) | 77.9 (16.9) | NA | NA | NA |
| eGFR creatinine, ml/min/1.73m2 | 80.6 (17.2) | 80.0 (22.6) | 85.2 (23.5) | 77.1 (17.2) | 73.0 (20) | 90.4 (22.8) |
| CKD, eGFR<60 ml/min/1.73m2 | 9.1 (731) | 18.7 (612) | 10.7 (445) | 13.7 (600) | 24.3 (781) | 6.3 (1151) |
Information on demographics, hypertension, and diabetes are ascertained at the visit eGFRcrea was measured. Sample sizes for CKD and eGFRcrea in ARIC and FHS differ as these studies use a cumulative definition of CKD as detailed in the method section.
Abbreviations: ARIC: Atherosclerosis Risk in Communities Study, CHS: Cardiovascular Health Study, FHS: Framingham Heart Study, RS: Rotterdam Study, AGES: Age Gene/Environment Susceptibility-Reykjavik Study, WGHS: Women’s Genome Health Study, SD: standard deviation, CKD: chronic kidney disease, eGFR: estimated glomerular filtration rate.
Figure 1. Meta-analysis -log10(P-value) vs. genomic position plots for CKD (A), eGFRcrea (B), and eGFRcys (C) in the discovery samples.
Genomic loci with evidence of suggestive association (p<4*10−7) are plotted in orange and with significant association (p<5*10−8) in red, with the exception of the SNP at the JAG1 locus on chromosome 20 (panel B, grey) which did not replicate.
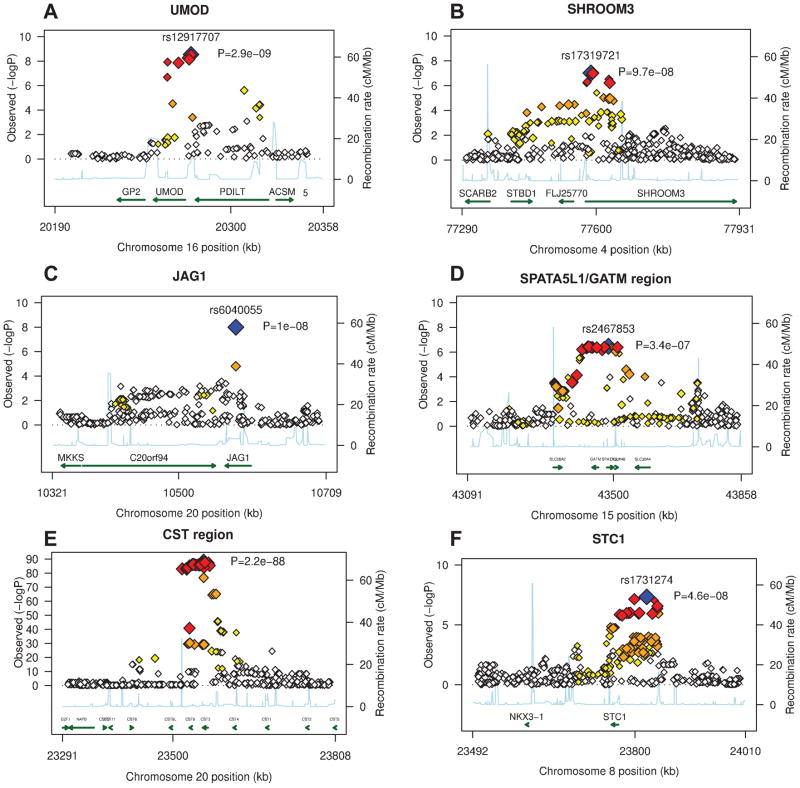
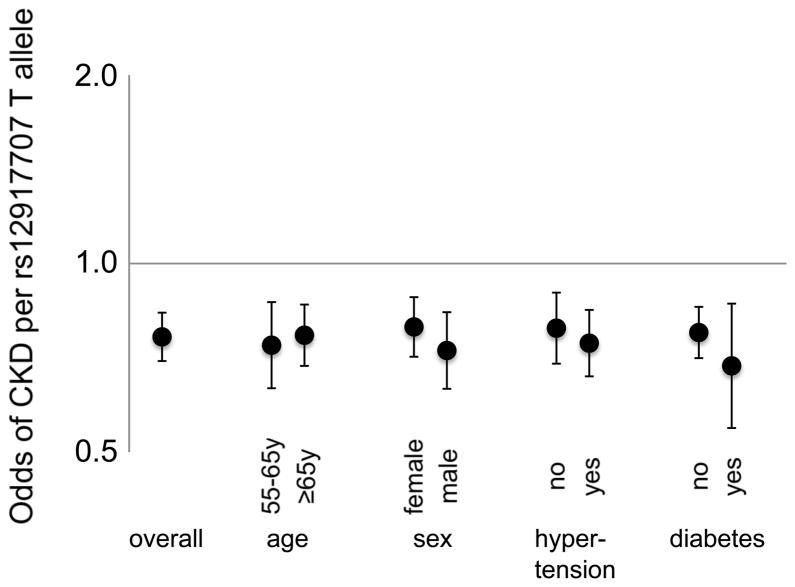
Table 2 lists the most significant SNP at each genomic locus associated with CKD, eGFRcrea, and eGFRcys, and replication results. Study-specific results are presented in Supplementary Table 3. For CKD, we identified SNP rs12917707 in a highly evolutionary conserved region 3.6 kb upstream from the uromodulin (UMOD) gene on chromosome 16 (Fig. 2A, p=5*10−16 across discovery and replication samples, Table 2). Seven SNPs in or upstream of UMOD in high LD (r2>0.8) with rs12917707 were also associated with CKD at a genome-wide significant level. The minor T allele at rs12917707 was associated with 20% reduced risk of CKD (meta-analysis OR=0.80, p-value=2*10−12, Table 2). The association of rs12917707 with CKD, which was not significant in the FHS Study, showed some heterogeneity across studies (p-heterogeneity=0.02). Findings were consistent in models adjusting for major CKD risk factors including systolic blood pressure, hypertension medication intake and diabetes mellitus, as well as stratified for age, sex, hypertension, and diabetes status (Figure 3). Prospective information from the ARIC Study demonstrated that the T allele of rs12917707 was associated with a lower relative risk of incident CKD (HR 0.81, 95%CI 0.72–.92, p=0.001) over 14.7 years of follow-up (n=952 cases, see Methods).
Table 2.
Results from meta-analyses of top GWAS signals at each locus (p<4*10−7) for CKD, eGFRcrea, and eGFRcys in discovery samples and after replication
| Locus | GWAS meta-analysis | GWAS discovery and replication meta-analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | position | in (near) gene | minor / major allele | OE var ratio* | MAF | beta / OR | se / 95% CI | p-value | beta / OR | se / 95% CI | p-value |
| CKD (n=19,877) | ||||||||||||
| rs12917707 | 16 | 20275191 | (UMOD) | T/G | 0.96 | 0.18 | 0.76 | 0.70–.83 | 2.8E-09 | 0.80 | 0.75–.85 | 2.3E-12 |
| eGFRcrea (n=18,127) | ||||||||||||
| rs17319721 | 4 | 77587871 | SHROOM3 | A/G | 0.96 | 0.44 | −0.014 | 0.003 | 9.7E-08 | −0.012 | 0.002 | 1.2E-12 |
| rs2467853 | 15 | 43486085 | SPATA5L1/GATM | G/T | 0.97 | 0.38 | −0.013 | 0.003 | 3.4E-07 | −0.013 | 0.002 | 6.2E-14 |
| rs12917707 | 16 | 20275191 | (UMOD) | T/G | 0.96 | 0.18 | 0.022 | 0.003 | 3.0E-11 | 0.018 | 0.002 | 5.2E-16 |
| rs6040055 | 20 | 10581313 | JAG1 | T/C | 0.73 | 0.39 | −0.017 | 0.003 | 1.0E-08 | −0.005 | 0.002 | 5.9E-03 |
| eGFRcys (n=12,266) | ||||||||||||
| rs1731274 | 8 | 23822264 | (STC1) | G/A | 0.96 | 0.43 | −0.017 | 0.003 | 4.6E-08 | NA | NA | NA |
| rs12917707 | 16 | 20275191 | (UMOD) | T/G | 0.96 | 0.18 | 0.021 | 0.004 | 2.0E-07 | NA | NA | NA |
| rs13038305 | 20 | 23558262 | (CST3/CST9) | T/C | 0.95 | 0.21 | 0.076 | 0.004 | 2.2E-88 | NA | NA | NA |
Meta-analysis p-values are adjusted for the trait- and study-specific genomic-control parameters (see Suppl Fig 1). For eGFR, betas indicate the change in eGFR per minor allele for the natural logarithmic transformation of eGFR. NA: eGFRcys was not available in the replication samples, but rs1731274 was associated with eGFRcrea after replication at p=2*10−7, beta −0.009, se 0.002.
Abbreviations: *OE var ratio: sample-size weighted mean of the observed by expected variance ratio for each SNP across the discovery samples; MAF: minor allele frequency; OR: odds ratio; CI: confidence interval; se: standard error.
Figure 2. Genetic architecture of the genome-wide significant susceptibility loci for renal disease in the discovery samples: (A): UMOD gene region, (B): SHROOM3 gene region, (C): GATM/SPATA5L1 gene region, (D) CST genes region, (E) STC1 gene region.
−log10 P-values are plotted versus genomic position (Build 36). The most significant SNP in each region is plotted in blue. LD based on the HapMap CEU sample is color-coded: red (r2 to top SNP 0.8–.0), orange (0.5–.8), yellow (0.2–.5), and white (<0.2). Gene annotations are based on Build 36 and arrows present direction of transcription. P-values are obtained from the discovery traits: CKD (UMOD), eGFRcrea (SHROOM3, GATM), eGFRcys (CST, STC1).
Figure 3. Meta-analysis of the odds of CKD per each additional copy of the minor T allele at UMOD rs12917707 across strata of major kidney disease risk factors.
Error bars correspond to 95% confidence intervals. Meta-analysis values obtained from the discovery samples.
Rare UMOD mutations cause autosomal-dominant forms of kidney disease, medullary cystic kidney disease type 2 (MCKD2), familial juvenile hyperuricemic nephropathy (FJHN), and glomerulocystic kidney disease (GCKD) (OMIM #603860; #162000; #609886).3, 18, 19 As the syndromes caused by rare UMOD mutations are often accompanied by hyperuricemia and gout, we explored the association of rs12917707 with these traits; no significant associations were observed. While this does not exclude the presence of rare UMOD variants among our study participants, our study identifies another example of a genomic risk locus containing susceptibility variants across the spectrum of risk allele frequencies.
UMOD knock-out mice are reported to have 63% lower creatinine clearance than wildtype mice.20 UMOD encodes for the most abundant protein in the urine of healthy individuals, Tamm-Horsfall protein. The physiological functions of Tamm-Horsfall protein are not well understood but may include protection against inflammation and infection.2 A possible role of the UMOD gene in renal development was also recently reported.21 UMOD is transcribed exclusively in renal tubular cells of the thick ascending limb of the loop of Henle. Our findings therefore suggest a common mechanism for CKD pathogenesis localized to the nephron’s loop of Henle, which has previously received little attention. The major risk factors for kidney disease, hypertension and diabetes, are thought to affect the glomerulus primarily, and glomerular damage is typically characterized by albuminuria. Our findings, however, indicate that the association is consistent across strata of hypertension and diabetes, and we observed no association with albuminuria. Thus, our findings provide new insights into CKD pathogenesis and highlight the need to understand the production and functions of Tamm-Horsfall protein within the kidney. The very broad CKD definition we chose, including a variety of causes of CKD such as hypertension and diabetes, indicates that the search for susceptibility variants for complex diseases may not only benefit from phenotypic refinement, but also from evaluating a broad phenotype definition in order to identify common disease mechanisms.
Four loci were identified in association with eGFRcrea: the strongest association was for SNP rs12917707 at the UMOD locus (p-overall=5*10−16, Table 2). The top SNP at the second significant eGFRcrea locus was the intronic SNP rs17319721 located in a highly evolutionary conserved region in shroom family member 3 (SHROOM3) on chromosome 4 (Fig. 2B, p-overall=1*10−12, Table 2). The SHROOM3 gene product is expressed in human kidney and reported to play a role in epithelial cell shape regulation.22 The association at the third eGFRcrea locus, the intronic SNP rs6040055 in jagged 1 (JAG1) on chromosome 20 (Supplementary Figure 2, p=1*10−8, Table 2), did not replicate (p-overall=0.006). The finding may therefore be a false positive, although a biological role of JAG1 in kidney disease is supported by rare JAG1 mutations causing Alagille syndrome (OMIM #118450).23 Lastly, the intronic SNP rs2467853 in spermatogenesis associated 5-like 1 (SPATA5L1) at the GATM/SPATA5L1 locus on chromosome 15 was significantly associated with eGFRcrea (Fig. 2C, p-overall=6*10−14, Table 2). GATM encodes glycine amidinotransferase, an enzyme involved in creatine biosynthesis. SNPs at this locus are therefore likely related to serum levels of creatinine without influencing susceptibility to kidney disease (Table 3). Although rs2467853 is located in SPATA5L1, strong LD extends into the region of the GATM gene.
Table 3.
Association of significant SNPs (p<5*10−8) across indices of renal function and CKD in up to 19,877 participants of the ARIC, CHS, FHS, and RS studies
| Gene | UMOD | SHROOM3 | SPATA5L1 / GATM | STC1 | CST3/CST9 | |
|---|---|---|---|---|---|---|
| rs12917707 | rs17319721 | rs2467853 | rs1731274 | rs13038305 | ||
| Trait | ||||||
| CKD | OR | 0.76 | 1.07 | 1.13 | 1.06 | 0.94 |
| 95% CI | (0.70–0.83) | (1.00–1.15) | (1.06–1.21) | (0.99–1.13) | (0.87–1.03) | |
| p-value | 2.8E-09 | 0.04 | 3.9E-04 | 0.09 | 0.18 | |
| eGFRcrea | beta | 0.022 | −0.014 | −0.013 | −0.009 | 0.004 |
| se | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | |
| p-value | 3.0E-11 | 9.7E-08 | 3.4E-07 | 1.8E-04 | 0.22 | |
| eGFRcys | beta | 0.021 | −0.013 | −0.0001 | −0.017 | 0.076 |
| se | 0.004 | 0.003 | 0.003 | 0.003 | 0.004 | |
| p-value | 2.0E-07 | 4.2E-05 | 0.95 | 4.6E-08 | 2.2E-88 |
Values for the discovery trait for each SNP are shaded in light right gray; values correspond to table 2. Abbreviations: OR: odds ratio, CI: confidence interval, se: standard error.
We identified three loci in association with eGFRcys: the strongest association was for the intergenic SNP rs13038305 between cystatin C (CST3) and cystatin 9 (CST9) (Fig. 2D, p=2.2*10−88, Table 2). SNPs within the cystatin (CST) superfamily gene cluster on chromosome 20 have been previously reported as associated with serum cystatin C levels.24 The genes in the CST super-family encode cystatin proteins. SNPs in these genes likely influence serum levels of cystatin C and therefore estimated eGFRcys, but not true GFR or CKD susceptibility (Table 3). Secondly, we identified the intergenic SNP rs1731274, located 54 kb from the stanniocalcin 1 (STC1) gene on chromosome 8 (Fig. 2E, p=4.6*10−8, Table 2). STC1 encodes stanniocalcin 1, a hormone regulating calcium homeostasis in fish. In mammals, it is highly expressed in the renal nephron and may influence local calcium and phosphate homeostasis via a paracrine mechanism.25 A recent study in STC1 transgenic mice reported STC1 as a renal protective protein with a potent anti-inflammatory role.26 As the replication samples did not have cystatin C measurements available, we explored the association of rs1731274 with eGFRcrea across the discovery and replication samples (p=2*10−7, Table 2). Finally, rs12917707 at the UMOD locus was associated with eGFRcys at p=2*10−7.
Table 3 presents the association of all genome-wide significant SNPs across the three renal traits. SNPs in UMOD, SHROOM3, and STC1 showed direction-consistent association across traits. For example, rs12917707 at UMOD was associated with both higher eGFRcrea and eGFRcys representing better kidney function, and with lower odds of CKD, conferring disease-protection. SNPs at the GATM/SPATA5L1 and CST regions were only associated with the respective discovery trait, the association of rs2467853 at the GATM/SPATA5L1 locus with CKD likely results from the eGFRcrea-based definition of CKD. All SNPs associated with CKD, eGFRcrea, and eGFRcys at p<4*10−7 are listed in Supplementary Tables 4, 5, and 6, respectively.
Together, loci for eGFRcrea explain 0.7% of the eGFRcrea variance [0.43% without the GATM locus], and loci for eGFRcys explain 3.2% of the eGFRcys variance [0.24% without the CST locus], suggesting that additional yet undiscovered genetic variants impact variability in renal function. In accordance with small absolute differences observed in other GWAS for continuous human traits, the multivariable adjusted eGFR difference across genotypes for any one locus was small. Since risk alleles may act in an additive fashion, we created a risk score for each individual as the sum of risk alleles at UMOD, SHROOM3, and STC1. These analyses were performed in the ARIC Study, the largest individual study contributing data and with available prospective information. The mean eGFRcrea was 10 ml/min/1.73m2 lower in individuals with all 6 risk alleles across the 3 loci compared to those with 0 risk alleles (p=2*10−8 per each unit score increase). CKD prevalence ranged from 0% in those without any risk alleles to 12.1% in individuals carrying all six risk alleles.
The association of SNPs in the UMOD gene with indices of renal function and CKD implicate a common pathophysiologic mechanism localized to the nephron’s loop of Henle. As opposed to the renal glomerulus, this region has previously received little attention. Thus, studies to understand the production and functions of Tamm-Horsfall protein are warranted to eventually lead to novel prevention and intervention options to reduce CKD risk.
In summary, we identified and validated common variants at several novel loci conferring susceptibility for kidney dysfunction and CKD in large, unselected population-based studies.
Methods
Study Samples
Four large, population-based cohorts of the CHARGE consortium had GWAS data available and formed the discovery sample: ARIC, CHS, FHS, and RS. Detailed information about these cohorts, including the design papers, is provided in the Supplementary Methods. Briefly, the studies were initiated to study cardiovascular disease and its risk factors and diseases related to aging. The population-based ARIC cohort recruited 15,792 middle-aged participants from 1987–1989 in four US communities. The population-based CHS cohort recruited 5,888 participants 65 years from 1989–1990 and 1992–1993 in four US communities. The FHS is a community-based study with a family component, including the Original (n=5,209, recruited 1948) and Offspring (n=5,214, recruited 1971) component. The community-based RS recruited 7,983 participants aged 55 years or older from 1990–1993. Two independent study samples were used to replicate results. In AGES, 5,764 survivors of the Reykjavik Study were examined from 2002–2006 and contributed to information. The WGHS is a sample drawn in 2006 from the original Women’s Health Study. Each participant provided written informed consent, and Institutional Review Boards of the participating institutions approved the study protocols. African American participants from ARIC and CHS did not contribute information to the present study.
Genotyping and Imputation
Detailed information about genotyping and imputation methods is provided in Supplementary Table 1, and details about data cleaning are provided in the Supplementary Methods. Briefly, all studies directly genotyped between 300,000 and 900,000 SNPs using whole-genome genotyping arrays by either Affymetrix (6.0 [ARIC], 500K and 50K gene-centric [FHS]) or Illumina (Human CNV370 [AGES, CHS], 550K [RS], HumanHap300 Duo-Plus or a combination of HumanHap300 and iSelect [WGHS]). All genotyping was performed according to the manufacturer’s instructions between 2006–2008. Using the Phase II CEU HapMap individuals as a reference panel, genotypes were imputed to a common set of ~2.5 million high-quality HapMap SNPs. Software used for imputation were BimBam v0.9927 (CHS) and MACH v1.0.15/16 (all others, http://www.sph.umich.edu/csg/abecasis/MACH/); FHS accounted for relatedness of participants. Imputed genotypes were expressed as an allelic dosage, a fractional value between 0–2. The WGHS did not impute genotypes.
Outcomes: eGFRcrea, eGFRcys, CKD
Serum creatinine was measured using a modified kinetic Jaffe reaction in all studies but AGES, where an enzymatic method was used. eGFRcrea was calculated using the Modification of Diet in Renal Disease (MDRD) Study equation14: eGFRcrea (ml/min/1.73m2) = 186.3*serum creatinine (mg/dl)−1.154 * age−0.203 * 0.742 (if female). To be comparable across studies, creatinine values in all studies were calibrated using regression to age, sex, and race adjusted mean values from a nationally representative US survey as described previously.28 Cystatin C was measured by a particle-enhanced immunonephelometric assay (N Latex Cystatin C, Dade Behring) at ARIC visit 4, CHS baseline exam, and FHS offspring exam 7 with a nephelometer (BNII, Dade-Behring). eGFRcys was then calculated using the formula eGFRcys = 76.7*(serum cystatin C)−1.19.15 CKD was defined as eGFRcrea <60 ml/min/1.73m2 according to National Kidney Foundation guidelines.17
CKD in CHS, RS, WGHS, and AGES was defined based on a single measurement of serum creatinine at the baseline visit. FHS and ARIC used a cumulative definition of CKD based on serum creatinine measurements at several study visits as detailed in the Supplementary Methods. Incident CKD in ARIC was defined as eGFRcrea <60 ml/in/1.73m2 at study visits 2 or 4 in individuals with eGFRcrea ≥60 ml/in/1.73m2 at study visit 1, or a kidney-disease specific ICD code on a hospital discharge record or death certificate from study inception in 1987 through January 1, 2005.29
Information on age and sex was collected at each study visit, and race was self-reported. Potential population stratification was assessed as detailed in the Supplementary Methods
Statistical analysis
GWAS was conducted within each cohort for eGFRcrea, eGFRcys, and CKD, followed by meta-analysis of the study-specific associations for each trait. SNPs showing genome-wide significant association with any of the three traits in meta-analyses were then explored for their association with the other two traits.
The phenotype for the eGFR analyses in all studies was created by calculating a natural logarithmic transformation of eGFR obtained from the respective equations for eGFRcrea and eGFRcys. All studies but CHS then created sex-specific age- and study-site (ARIC) or cohort (FHS) adjusted residuals. CHS adjusted for age, sex, and study site in multivariable regression models. Incident CKD in ARIC was analyzed using multivariable-adjusted Cox proportional hazards regression. Software packages used by the individual studies to conduct linear and logistic regression are listed in Supplementary Table 1. FHS accounted for the relatedness of individuals in the analyses as detailed in the Supplementary Methods. Pedigree correlations were adjusted for using the robust variance option. All studies used an additive genetic model.
Meta-analysis was conducted using inverse-variance weighting as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html). Prior to meta-analysis, the genomic control parameter was calculated within each study for each trait to assess potential inflation of the test statistics. If the parameter was larger than 1, an adjustment was performed by scaling the test statistics to the inflation factor. Only SNPs with minor allele frequency (MAF) ≥2% were analyzed based on the number of CKD cases, corresponding to approximately 50 carriers of the minor allele with CKD. Statistical heterogeneity was evaluated using Cochrane’s chi-square test (Q-test).
The most significant SNP at genomic loci with evidence of suggestive association (p<4*10−7) for any of the traits were tested for replication in the independent samples. This threshold corresponds to1/2.5 million tests conducted and corresponds to one or less expected false positive findings.30 A threshold of 5*10−8 was used to indicate genome-wide significance, corresponding to a Bonferroni correction for the estimated 1 million common independent SNPs across the genome (0.05/1 million).31 The SNAP program with the HapMap CEU sample as a reference was used to identify the best proxy in the WGHS dataset, to evaluate LD to nearby coding SNPs, and to evaluate LD between imputed SNPs and proxy SNPs that were directly genotyped (http://www.broad.mit.edu/mpg/snap/ldsearch.php).
Supplementary Material
Acknowledgments
We are indebted to the staff and participants of the AGES Reykjavik Study, the ARIC Study, the CHS Study, the FHS Study, the Rotterdam Study, and the WGHS Study for their important contributions.
.AGES: The Age, Gene/Environment Susceptibility Reykjavik Study has been funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). We thank Thor Aspelund and Gudny Eiriksdottir for their contribution to collecting, analyzing and preparing the AGES Reykjavik Study data.
ARIC: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. AK was supported by a German Research Foundation Fellowship. WHLK was supported by K01DK067207.
CHS: The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01-HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652, and R01 AG027002 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01RR00069 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
FHS: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
RS: The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO) (175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was further supported by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Dr Michael Moorhouse, Marijn Verkerk and Sander Bervoets for their help in creating the Rotterdam database and Maxim Struchalin for his contributions to the imputations of the Rotterdam data.
WGHS: The WGHS was supported by the National Heart, Lung, and Blood Institute (HL 043851) and the National Cancer Institute (CA 047988). Collaborative scientific and genotyping support was provided by Amgen, Inc.
Footnotes
Author Contributions:
AK, NLG, AD, SJH, QY, IHdB, TL, DS, DL, VG, JC, MGS, and CSF contributed to the design of this analysis; AK, NLG, AD, SJH, QY, EB, IR, IHdB, TL, DS, DL, AVS, VG, WHLK, JCW, JC, MGS, and CSF contributed to the interpretation of the results; AK, NLG, AD, SJH, JC, MGS, and CSF drafted the manuscript; all others reviewed and commented on the manuscript; AK, NLG, AD, SJH, RK, ML, QY, DEA, GBE, IR, RBS, TL, FR, CMvD, AVS, DIC, GP, and JC contributed to statistical methods and analysis; DS, EJB, DL, VG, JC, and CSF contributed to recruitment and follow up of subjects; EB, YIC, TH, FR, AGU contributed to genotyping; and ML, QY, YSA, FR, AGU, AVS, and GP contributed to bioinformatics.
References
- 1.Levey AS, et al. Chronic kidney disease as a global public health problem: approaches and initiatives - a position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007;72:247–259. doi: 10.1038/sj.ki.5002343. [DOI] [PubMed] [Google Scholar]
- 2.Serafini-Cessi F, Malagolini N, Cavallone D. Tamm-Horsfall glycoprotein: biology and clinical relevance. Am J Kidney Dis. 2003;42:658–676. doi: 10.1016/s0272-6386(03)00829-1. [DOI] [PubMed] [Google Scholar]
- 3.Hart TC, et al. Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet. 2002;39:882–892. doi: 10.1136/jmg.39.12.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coresh J, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 5.Zhang QL, Rothenbacher D. Prevalence of chronic kidney disease in population- based studies: systematic review. BMC Public Health. 2008;8:117. doi: 10.1186/1471-2458-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Institutes of Health. US Renal Data System, USRDS 2008 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: [Google Scholar]
- 7.Sarnak MJ, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation. 2003;108:2154–2169. doi: 10.1161/01.CIR.0000095676.90936.80. [DOI] [PubMed] [Google Scholar]
- 8.Weiner DE, et al. Chronic kidney disease as a risk factor for cardiovascular disease and all-cause mortality: a pooled analysis of community-based studies. J Am Soc Nephrol. 2004;15:1307–1315. doi: 10.1097/01.asn.0000123691.46138.e2. [DOI] [PubMed] [Google Scholar]
- 9.Langefeld CD, et al. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am J Kidney Dis. 2004;43:796–800. doi: 10.1053/j.ajkd.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 10.Bochud M, et al. Heritability of renal function in hypertensive families of African descent in the Seychelles (Indian Ocean) Kidney Int. 2005;67:61–69. doi: 10.1111/j.1523-1755.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 11.Fox CS, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol. 2004;15:2457–2461. doi: 10.1097/01.ASN.0000135972.13396.6F. [DOI] [PubMed] [Google Scholar]
- 12.Freedman BI, Satko SG. Genes and renal disease. Curr Opin Nephrol Hypertens. 2000;9:273–277. doi: 10.1097/00041552-200005000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from five cohorts. Circulation: Cardiovascular Genetics. 2009 doi: 10.1161/CIRCGENETICS.108.829747. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levey AS, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 15.Stevens LA, et al. Estimating GFR using serum cystatin C alone and in combination with serum creatinine: a pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis. 2008;51:395–406. doi: 10.1053/j.ajkd.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function--measured and estimated glomerular filtration rate. N Engl J Med. 2006;354:2473–2483. doi: 10.1056/NEJMra054415. [DOI] [PubMed] [Google Scholar]
- 17.National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39:S1–266. [PubMed] [Google Scholar]
- 18.Vylet’al P, et al. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70:1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 19.Rampoldi L, et al. Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet. 2003;12:3369–3384. doi: 10.1093/hmg/ddg353. [DOI] [PubMed] [Google Scholar]
- 20.Bachmann S, et al. Renal effects of Tamm-Horsfall protein (uromodulin) deficiency in mice. Am J Physiol Renal Physiol. 2005;288:F559–67. doi: 10.1152/ajprenal.00143.2004. [DOI] [PubMed] [Google Scholar]
- 21.Benetti E, et al. Immature renal structures associated with a novel UMOD sequence variant. Am J Kidney Dis. 2009;53:327–331. doi: 10.1053/j.ajkd.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura T, Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 2008;135:1493–1502. doi: 10.1242/dev.019646. [DOI] [PubMed] [Google Scholar]
- 23.Anad F, et al. Alagille syndrome and deletion of 20p. J Med Genet. 1990;27:729–737. doi: 10.1136/jmg.27.12.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang SJ, Yang Q, Meigs JB, Pearce EN, Fox CS. A genome-wide association for kidney function and endocrine-related traits in the NHLBI’s Framingham Heart Study. BMC Med Genet. 2007;8 (Suppl 1):S10. doi: 10.1186/1471-2350-8-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshiko Y, Aubin JE. Stanniocalcin 1 as a pleiotropic factor in mammals. Peptides. 2004;25:1663–1669. doi: 10.1016/j.peptides.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Huang L, et al. Anti-Inflammatory and Renal Protective Actions of Stanniocalcin-1 in a Model of Anti-Glomerular Basement Membrane Glomerulonephritis. Am J Pathol. 2009 doi: 10.2353/ajpath.2009.080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coresh J, et al. Calibration and random variation of the serum creatinine assay as critical elements of using equations to estimate glomerular filtration rate. Am J Kidney Dis. 2002;39:920–929. doi: 10.1053/ajkd.2002.32765. [DOI] [PubMed] [Google Scholar]
- 29.Hsu CC, et al. Apolipoprotein E and progression of chronic kidney disease. JAMA. 2005;293:2892–2899. doi: 10.1001/jama.293.23.2892. [DOI] [PubMed] [Google Scholar]
- 30.Gordon A, Glazko G, Qui X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. The Annals of Applied Statistics. 2007;1:179–190. [Google Scholar]
- 31.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.