Abstract
Cerium oxide nanoparticles (CNPs) have been recently studied for their potent superoxide scavenging properties in both cell and animal model systems. Data from these model systems have shown that exposure of cells to CNPs results in the protection against reactive oxygen species (ROS). Despite these exciting findings, very little is known regarding the uptake or subcellular distribution of these nanomaterials inside cells. In this study we utilized fluorophore (carboxyfluorescein) conjugated cerium oxide NPs (CCNPs) to study the mechanism of uptake and to elucidate the subcellular localization of CNPs using a keratinocyte model system. We observed rapid uptake (within 3 h) of CCNPs that was governed by energy-dependent, clathrin-mediated and caveolae-mediated endocytic pathways. We found CCNPs co-localized with mitochondria, lysosomes and endoplasmic reticulum as well as being abundant in the cytoplasm and the nucleus. Given the radical scavenging properties of cerium oxide and the widespread cellular disposition we observed, CNPs likely act as cellular antioxidants in multiple compartments of the cell imparting protection against a variety of oxidant injuries.
Introduction
Recent years have seen an increasing interest in biomedical and biological applications of rare earth oxide nanoparticles especially CNPs.1 Unlike most metal and metal oxide nano-materials, CNPs act as antioxidants at ambient conditions while displaying low cytotoxicity. CNPs have been shown to protect primary cells against radiation exposure, to provide protection of multiple cell types to oxidative stress, to decrease inflammation induced by chemokines and to provide neuroprotection to spinal cord neurons in the presence of peroxide.1–6 Recently, CNPs have also been shown to scavenge superoxide and exhibit oxidase like activity.1,7–11 The ability to alternate between Ce3+ and Ce4+ states at the surface of CNPs, depending upon the environment and their synthesis process, makes them suitable to provide radical scavenging functions.10 Despite these exciting results in recent publications, the uptake and metabolism of these nanomaterials remains a mystery. Several reports have reported the conjugation of NPs with fluorescent probes, such as fluorescent dyes and proteins, for in vivo imaging.12 A recent study focused on the toxicity of metal oxides and found that FITC labeled CNPs localized in lysosomes in phagocytic cells.13 CNPs exhibit very weak fluorescence, thus they need to be tagged with a fluorescent probe to efficiently track their uptake and subcellular localization.
The plasma membrane of cells defines the external boundaries of cells which segregate the chemically discrete intracellular cytoplasm from the extracellular environment. This border mediates the uptake of nutrients and cell–cell communication. The uptake of macromolecules is mediated by the action of integral membrane proteins.14 Membrane bound vesicles derived from the invagination and pinching-off in the form of pieces of the plasma membrane (endocytosis). Endocytosis can be broadly categorized into ‘phagocytosis’ or ‘pinocytosis’. Pinocytosis (cell drinking) can be further categorized into four distinct mechanisms: (1) clathrin-mediated, (2) caveolae-mediated, (3) clathrin and caveolae independent and (4) macropinocytosis. Once internalized, molecules are further sorted and trafficked to different organelles inside the cell depending surface charge, protein–protein interactions. Nanoparticles in the range of few nanometres have been shown to be taken up by a variety of cell types. Recently Kostarelos et al. observed that the uptake of carbon nanotubes in mammalian cells is independent of cell type and functional groups.15 Endocytosis is a prevalent process of the uptake of nanomaterials into mammalian cells, such as gold, iron oxide, quantum dots,16–18 silica and carbon nanotubes.19–27
In this work we prepared CNPs conjugated with CFL as described previously.28 CNPs exhibit weak fluorescence and CFL as free molecules is impermeable to cell membrane, which gave us significant rationale to conjugate CFL with CCNPs. CFL serves a protective layer to improve particle stability, reducing particle agglomeration, thus, increasing the efficiency of particle internalization by cells. The in vitro biocompatibility and intracellular uptake of CCNPs were investigated in HaCat cells. By covalently modifying the surface of CNPs with CFL, the prepared nanoparticles enable in situ monitoring of uptake processes and intracellular fate under confocal microscope. The time dependent intracellular uptake was also studied using photoluminescence spectroscopy. Our results demonstrate that CFL modified CNPs are not toxic at doses needed to observe their uptake and distribution and uptake processes are mainly controlled by temperature of surrounding medium, cellular ATP levels and clathrin and caveolae mediated endocytosis pathways. Sub cellular distribution study revealed the interaction of CCNPs with mitochondria, localization with lysosomes further supports our observation that CCNPs uptake is by endocytosis. However, localization of CCNPs with ER indicates the interaction of nanoparticles with cellular protein synthesis machinery also. Nuclear uptake of CCNPs was also observed suggesting that CFL modified CNPs could be useful for delivery applications, especially cellular component sensing and nuclear targeting. This study contributes to in vitro biocompatibility and probable action of CNPs as antioxidant agent for intracellular protection against free radicals.
Results and discussion
Evidence of fluorophore conjugation and physico chemical characterization of the CCNPs
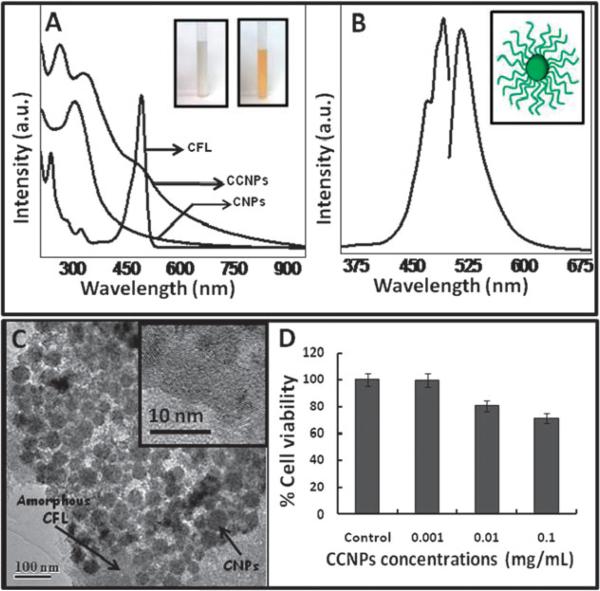
Successful conjugation of CFL (see Experimental) on CNPs surface was confirmed by UV-visible (Fig. 1A) and photoluminescence (Fig. 1B) spectra. CNPs absorb light in the range from ~250 nm to ~350 nm, depending on their oxidation states. UV-visible spectra of CNPs, prior to conjugation with CFL, absorbed maximally at ~305 nm, which is ascribed to the Ce4+ oxidation state in CNPs. UV-visible spectra of CCNPs, when compared to CNPs, showed a clear band at ~265 nm and a broad absorption edge ~490 nm confirming the presence of CFL coating on the nanoparticle surface. The color of CNPs suspension before (white, CNPs) and after (yellow, CCNPs) modification with fluorophores can be seen in Fig. 1A (inset). Further, the fluorescent nature of CNPs after functionalization was verified by PL (photoluminescence) spectra (Fig. 1B). The excitation spectra show maximum emission at 492 nm and the emission spectra exhibit an intense peak at 523 nm. These emission spectra are red-shifted by 2 nm as compared to the properties of free carboxyfluorescein in alkaline medium.29 The probable arrangement of CFL on CNPs surface is presented in Fig. 1B (inset). The chemistry of attachment of CFL was also investigated using XPS (Fig. S1, ESI†). The oxygen (1s) and carbon (1s) spectra were deconvoluted to find the various peaks corresponding to different chemical species present in the sample. The deconvolution of O1s spectra showed peaks corresponding to Ce3+–O at 532.2 eV and Ce4+–O at 530.5 eV which suggests the mixed valence state of cerium in CNPs with CFL. Also the deconvoluted O1s revealed peaks at 531.1 eV corresponding to C=O which is expected only on successful functionalization of CNPs with CFL, wherein the carboxyl group of CFL bonds at the amine site available by treating CNPs with epichlorohydrin.28 The deconvolution of C1s revealed peaks corresponding to different position of carbon in CFL conjugated CNPs as shown in Fig. S1 (ESI†) (C1s) further confirms the predicted functionalization. The deconvolution of Ce 3d spectrum (Fig. S1c, ESI†) shows the presence of Ce in 3+ and 4+ states. The multiple d-splitting is shown by the deconvoluted peaks where the spin–orbit doublet Ce 3d3/2 is denoted by uo, u, u′, u″, u′″ peaks and Ce3d5/2 is denoted by vo, v, v′, v″, v′″ peaks. The integrated area under the curve of each deconvoluted peak was used to calculate the ratio of Ce3+ to Ce4+ and was found to be 0.22.30 To further characterize the conjugation of CFL to CNPs was confirmed by FTIR spectroscopy. FTIR spectrum (Fig. S2, ESI†) obtained from CCNPs clearly shows the presence of broad amide band at ca. 1575 cm−1 which could be assigned to amide bond formation31 between amine terminated CNPs and free carboxylic acid functional groups terminated CFL. FTIR spectrum from CFL clearly shows a broad transmittance peak at ca. 1700 cm−1 which confirms the presence of free carboxylic acid functional group. This peak is not present in FTIR spectrum of CCNPs after the conjugation with CNPs which again confirms the formation of amide bond.
Fig. 1.
UV-visible spectra, photoluminescence spectra, TEM and toxicity of CCNPs. (A) Absorbance spectra of CFL and CNPs recorded before (CNPs) and after (CCNPs) modification with CFL. Inset shows the respective color of NP suspension in aqueous medium. (B) An excitation maxima (at ~490 nm) and emission maxima (at ~520 nm), characteristic for CFL, were observed from the functionalized NPs. A schematic representation showing probable arrangement of CFL on CNPs have shown (inset, B). A thin organic coating (CFL) over roughly spherically agglomerated nanoparticles (CNPs) was observed using TEM (C) and high magnification image of CCNPs (inset, C) depicting the individual particle size of CNPs (3–5 nm) is unchanged by the chemical treatment. The amorphous border covering the nanoparticles confirms the presence of CCNPs over agglomerated and individual particles. HaCat cells were incubated with different concentrations of CCNPs suspended in PBS for 3 h and cell viability was determined using MTT assay (D).
The preparation of CCNPs was well dispersed without any significant aggregation. High resolution TEM (Fig. 1C) micrographs clearly reveal well separated and roughly uniform clusters of CCNPs with an average size of ~50 nm. A high magnification image (Fig. 1C, inset) from one such cluster shows that these clusters are made up of several individual crystals of CNPs 3–5 nm in diameter. The appearance of clusters could be attributed to the air drying effect of nanoparticles over TEM grid. The zeta potential of well dispersed CCNPs in phosphate buffer was found to be negative (−18.2 mV).
CCNPs are not toxic to the cells at levels needed to visualize uptake
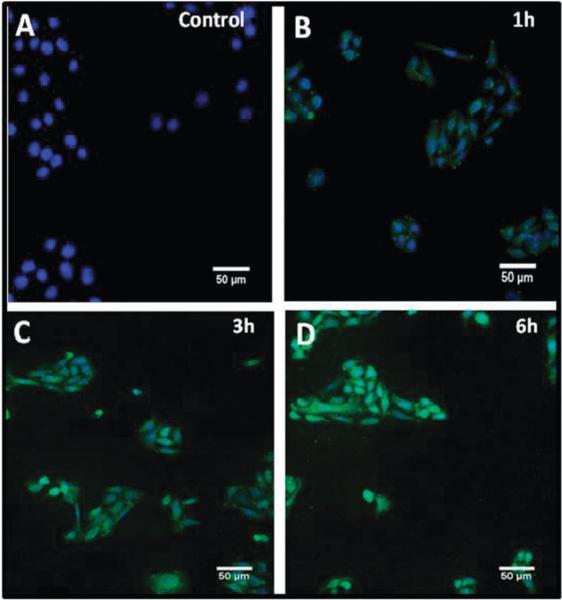
It has been well established that CNPs, as compared to other metal oxide nanoparticles such as zinc oxide and titania (exposed to UV-light), are non-toxic to mammalian cells.13 Nonetheless, we determined the biocompatibility of CCNPs using HaCat cells by MTT assay (Fig. 1D). CCNPs did not affect cellular metabolic activity at concentrations up to 0.1 mg mL−1. With this knowledge we chose a significantly lower concentration of CCNPs (0.01 mg mL−1) since this was the minimum concentration at which we could detect CCNPs inside the cells by microscopy (data not shown). At a dose of 0.1 mg mL−1 cell viability was reduced to 50% (data not shown). We followed the kinetics of uptake of CCNPs using confocal microscopy (Fig. 2). Cells incubated (3 h) with CFL alone did not yield increases in intracellular fluorescence above the native auto fluorescence of cells (data not shown). A control experiment where cells were not exposed to CCNPs (Fig. 2A) did not exhibit any fluorescence. Incubation with CCNPs for 1 h resulted in accumulation of CCNPs (Fig. 2B). We found that incubation for 3 h was sufficient for maximum uptake (Fig. 2C) as no significant increase in uptake was observed beyond this time point. We confirmed the presence of CCNPs in the cytosol of HaCat cells incubated under the same experimental conditions (1–12 h) and these results are shown in ESI† (Fig. S3). The uptake of CCNPs was carried out both in suspended cells (after trypsinization) and adherent cells. As expected, suspended cells showed more rapid uptake of CCNPs than adherent cells, likely due to the increased surface area of the cell exposed to the CCNPs (Fig. S3, ESI†). Nevertheless the kinetics of uptake was similar. Some efflux of nanoparticles was apparent at later time points, based on cytosolic fluorescent patterns (Fig. S3, ESI†).
Fig. 2.
Efficient uptake of CCNPs within 1–3 hours of exposure. Cells were incubated with CCNPs for 1 h (B), 3 h (C) and 6 h (D) in PBS. Nuclei are stained using DAPI. A control experiment without CCNPs exposure to cells (A) shows no increases in fluorescence.
Penetration and distribution of CCNPs in the nucleus
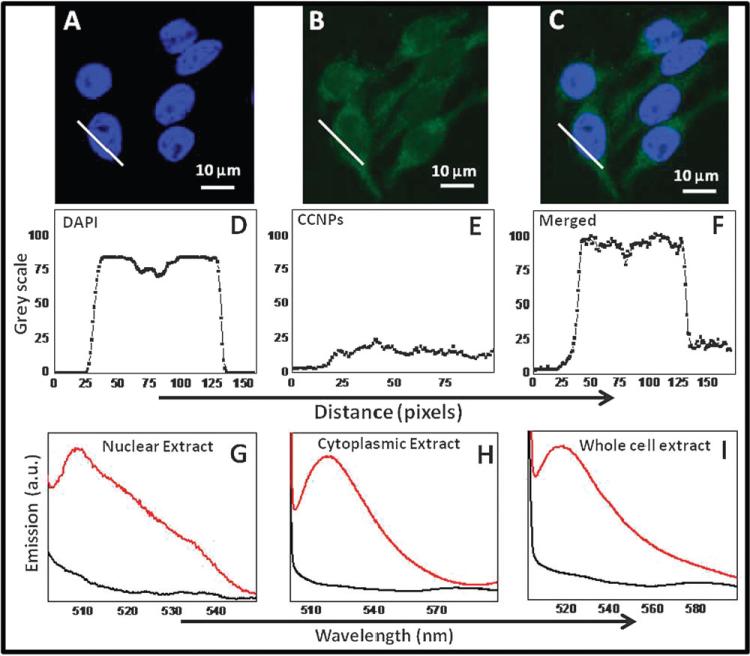
Our initial observations suggested, based on co-localization with DAPI staining, that CCNPs were present in cytoplasm and inside the nucleus as well by confocal images (Fig. 3A and B). PL spectra from untreated (Fig. 3I, black curve) CCNPs and treated (Fig. 3I, red curve) whole cell extract were also recorded and found that only treated cells showed emission spectra centering at 520 nm. Further, we separated the nuclear extracts and confirmed using photoluminescent spectra that CCNPs were present in both the cytosolic fraction (Fig. 3H, red curve) and the nuclear fraction (Fig. 3G, red curve). The corresponding controls (untreated cellular/nuclear extract) did not exhibit (black curve) any emission. Line plots drawn across the cytoplasm and nucleus (Fig. 3E and D) and merged (Fig. 3F) show clear increase of signal in these respective regions, corresponding to CCNPs. Diameter of a mammalian nuclear pore complex is approximately 120 nm whereas the actual diameter of pore through which molecules (inert molecules) are allowed to enter is 9 nm. This diameter can also dilate to 26 nm during internalization.32 TEM images show that a cluster of CCNPs is about 50 nm (Fig. 1C) which is a complex of several ~3–5 nm (Fig. 1C, inset) individual nanoceria particles. Thus it is possible that individual particles could pass through nuclear pores if not present as an aggregate inside the cytosol.
Fig. 3.
Widespread distribution of CCNPs in the cytoplasm and nucleus. Confocal images (A, B and C) showed the distribution of CCNPs in the nucleus and cytoplasm. Corresponding line plot (D, E and F) across the cytoplasm and nucleus suggests strongly the presence of CCNPs in the nucleus. PL spectra recorded from nuclear extract (G), cytosolic extract (H) and crude cell extract (I) confirmed the presence of CCNPs in these compartments.
Energy dependent uptake of CCNPs
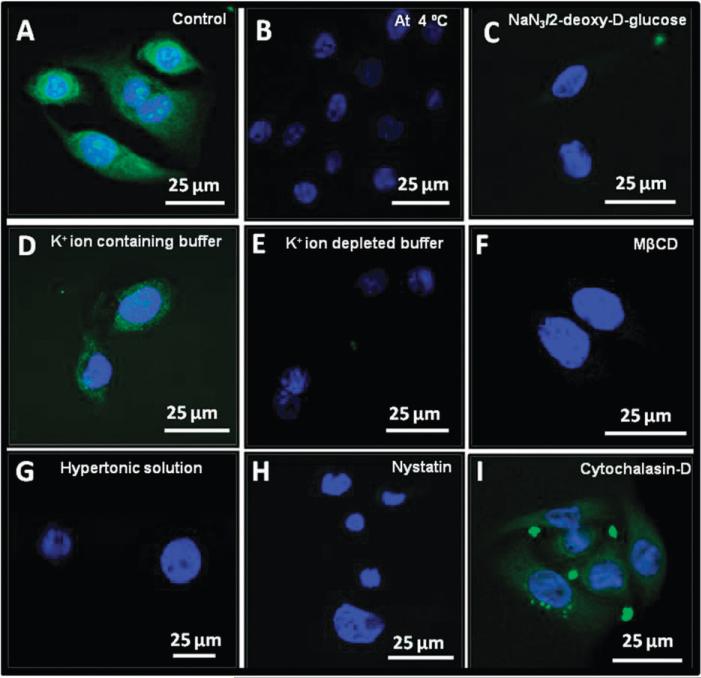
First we determined the requirement of energy for the uptake of CCNPs. As shown in Fig. 4B, internalization of CCNPs was completely abolished when cells were incubated at 4 °C. Low temperature incubation of cells not only slows the metabolism of the cells, it also imparts rigidification of the lipid membrane.33 Therefore, we chose to deplete ATP level, a multifunctional nucleotide and main energy source within cells, using 2-deoxy-d-glucose and 0.02% sodium azide, known for ATP disruption. In an alternative experiment at 37 °C, we pre-incubated the cells with NaN3/2-deoxy-d-glucose, and found negligible internalization of CCNPs (Fig. 4C). The corresponding control experiment (Fig. 4A), where cells were exposed to CCNPs and incubated for 3 h at 37 °C, showed clear uptake. These results suggest that the uptake of CCNPs requires energy and an active metabolic state.
Fig. 4.
Uptake of CCNPs is mediated by energy-dependent, clathrin and caveolae-dependent pathways. Confocal images of HaCat cells incubated with CCNPs at 37 °C (A), 4 °C (B) and cells pre-incubated with NaN3/2-deoxy-d-glucose (C). Cells pre-treated with K+ ion containing buffer (D) or K+ ion depleting buffer (E) or hypertonic sucrose solution (G). Pretreatment with MβCD (F), or nystatin (H) as described in detailed methods section (ESI†). Cells pretreated with cytochalasin-D (I) and subsequently exposed to CCNPs.
Pinning the endocytosis mechanism—dependency on clathrin and lipid rafts
Endocytosis is a process of internalization of macromolecules performed by plasma membrane through the formation of membrane bound vesicles derived by the invagination and pinching-off of the pieces of the plasma membrane. This process is usually separated into two major categories: phagocytosis (uptake of larger particles) and pinocytosis (uptake of fluid and solutes). Phagocytosis is typically being carried out by specialized mammalian cells such as macrophages, monocytes or neutrophils. Pinocytosis can be further subdivided into four parts: macropinocytosis, clathrin-mediated endocytosis, caveolae-mediated endocytosis and clathrin and caveolae-independent endocytosis.
Clathrin dependent endocytosis involves the formation of coated pits on the plasma membrane, which are formed by the assembly of cytosolic coat proteins. These clathrin coated pits invaginate and pinch-off to form endocytic vesicles.14 In order to investigate whether clathrin was involved in uptake of CCNPs, we treated cells to disrupt the formation of cellular clathrin-coated pits and vesicles by pre-incubating cells with a hypertonic solution of sucrose or a K+ ion depleted buffer prior to exposure of CCNPs. Exposure to a hypertonic sucrose solution (Fig. 4G) clearly leads to complete inhibition of the uptake of CCNPs. Similarly, pre-incubation with K+ ion depleted buffer also inhibits CCNPs uptake as no fluorescence signals were observed (Fig. 4E). The corresponding controls (without any prior treatment with either sucrose or K+ ion depleting buffer) displayed a rapid and efficient uptake of CCNPs (Fig. 4D). These results strongly suggest clathrin-dependent endocytosis is required for efficient internalization of CCNPs.
Caveolae are flask-shaped invaginations of the plasma membrane, consists of cholesterol binding protein caveolin with a bilayer enriched in cholesterol and glycolipids. Caveolae represent a kind of cholesterol-rich membrane micro domain, generally referred as lipid-rafts. To determine whether lipid-rafts play a role in CCNPs uptake, we depleted membrane cholesterol by pretreating cells with methyl-beta-cyclodextrin (MβCD) or nystatin for 30 minutes. Depletion of membrane cholesterol led to a complete inhibition of internalization of CCNPs (Fig. 4F and H). These results indicate the uptake of CCNPs is caveolae dependent.
The actin cytoskeleton is essential for many important cellular processes including muscle contraction, cell motility, cell division cytokinesis, vesicle and organelle movement and cell shape. To determine the role of the actin cytoskeleton in CCNP uptake we pre-treated cells with cytochalasin-D (Fig. 4I) or LATB (data not shown) to facilitate depolymerization of actin filaments. Neither treatment significantly altered uptake of CCNPs, suggesting the uptake of CCNPs is independent of actin filaments. It should be noted that actin has only a partial role in clathrin coated vesicle formation.34 It should be noted that in each of these experiments some clusters of CCNPs appear to be bound to the extracellular matrix (EM) around the cells (punctate spots found outside the cells). It is unclear if these aggregates are deposits that occur due to non-specific binding of CCNPs to EM, or due to cellular extrusion of particles after uptake.
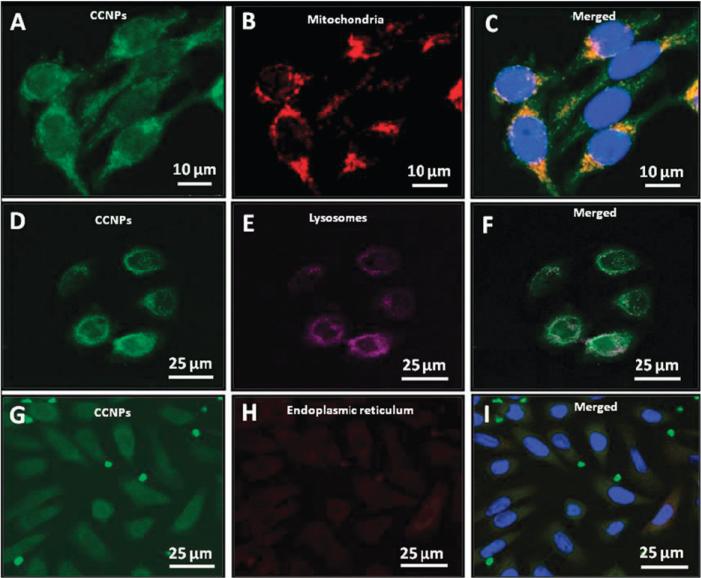
Widespread subcellular localization of CCNPs
To determine the subcellular distribution of CCNPs, we performed labeling experiments with dyes selective for organelles, lysosomes (Fig. 5E), endoplasmic reticulum (Fig. 5H), and mitochondria (Fig. 5B). It is well established that one of the dominant sources of ROS is mitochondria. Leakage of electrons from the respiratory chain to form ROS can result in significant oxidative stress.35 This ultimately contributes to several degeneration diseases and might be the root cause of aging. As shown in Fig. 5, CCNPs co-localize with mitochondria based on the merged image (Fig. 5C). The interaction of CNPs with mitochondria indicates that these unique antioxidants should be studied for the treatment of mitochondria-related diseases. Recently, Hirst et al.36 have observed CNPs around the mitochondria and subsequently found they quenched ROS and the inflammatory response, and these results are in parallel agreement with our findings with CCNPs.
Fig. 5.
Sub cellular distribution of CCNPs. Subcellular distribution of CCNPs (A, D and G) was investigated by co-labeling cells with CCNPs and Mito-tracker dye (B), Lyso-tracker dye (E) or ER-tracker dye (H).
Macromolecules internalized either by phagocytosis or endocytosis usually localized at least transiently in lysosomes. Therefore, we used the Lyso-tracker dye to investigate whether CCNPs co-localize with lysosomes (Fig. 5E). Fig. 5F clearly reveals that a few CCNPs were found to co-localize with lysosomes whereas the majority of the lysosomes do not co-localize with CCNPs. This is likely due to changes in surface charge from cationic (due to acidic pH in lysosomes) leading to release of the CCNPs from the endo-lysosome to the cytosol.28 It has been shown that nanoparticles which remain negatively charged at low pH (in lysosomes) were retained mostly in the endosomal compartment.37
The endoplasmic reticulum plays a major role in protein synthesis and folding, providing an oxidizing environment for disulfide bond formation before transit to the Golgi compartment. Protein disulfide isomerase and ER oxidoreductase 1 are two major enzymes that participate in disulfide bond formation, which ultimately leads to the formation of ROS as a product of electron transport from thiol groups in proteins. It has also been shown that during disulfide bond formation in the ER, oxidation of cysteine residues may significantly contribute to oxidative stress.38,39 We used ER-tracker dye (Fig. 5H) to probe whether CCNPs would co-localize in this compartment. From the merged image in Fig. 5I, it appears that CCNPs localize at the ER periphery which suggest the interaction of CCNPs with ER. Recently it has been reported that antioxidant treatment reduces unfolded protein response, oxidative stress and apoptosis40 which may contribute to the pathogenesis of many diseases. CNPs have the potential to be used as a novel therapeutic nanomaterial against ER-related oxidative injury if they can reach this compartment of the cell.
Experimental
Materials
Carboxyfluorescein was purchased from Fluka chemicals and methyl-β-cyclodextrin (MβCD), 2-deoxy-d-glucose, nystatin, cytochalasin-D, amiloride, latrunculin B (LATB) and sucrose were from Sigma-Aldrich. Hoechst 33342 (DAPI), Mito-tracker, Lyso-tracker and ER-tracker were purchased from Molecular Probes. Defined keratinocyte medium (DKM), trypsin/EDTA, and phosphate buffer saline (PBS) were also obtained from Invitrogen. All reagents were of analytical grade or higher. Milli-Q purified water (18.2 MΩ) was used to prepare all the solutions.
Functionalization of CNPs with carboxyfluorescein
Functionalization was performed as described earlier14 with little modification and can be summarized as: 250 mg of cerium oxide nanopowder was suspended in 10 mL of 0.1 M NaOH solution for 5 min, then, 5 mL of epichlorohydrin was added, followed by the addition of 0.5 mL of 2 M NaOH. The suspension was stirred at room temperature for 6–8 h, centrifuged (15 000 rpm), and the supernatant was decanted. Nanoparticles were then washed with water followed by centrifugation as above. This was repeated until the pH was approximately 7.0. The nanopowder was subsequently suspended in 20 mL of water, 25 mL of 30% ammonium hydroxide solution was added, and this reaction mixture was stirred for 12–15 h. Nanoparticles were captured by centrifugation, and washed with water. 200 mg of the carboxyfluorescein was dissolved in 15 mL of dimethylformamide (DMF) and 5 mL of dichloromethane. Three hundred and seventy-five microlitres (3.5 mM) of N-methyl morpholine (NMP) was added followed by the addition of 442.5 mg (1 mmol) of the benzotriazol-1-yloxy-tris-(dimethylamino)phosphonium hexafluorophosphate (BOP) reagent. The reaction mixture was stirred for 10 min at room temperature. The above mentioned modified nano-particles were then added. This mixture was stirred for approximately 24 h at room temperature. The reaction was stopped with 1 mL of water, and the mixture was centrifuged. It was then washed with DMF, water, and acetone three times each to remove the unattached carboxyfluorescein and CCNPs were isolated by centrifugation. A concentration of 0.01 mg mL−1 (~100 ng mL−1) and 0.001 mg mL−1 (~10 ng mL−1) of CCNPs (wet weight) were used for cellular uptake and co-localization study respectively.
Characterization of CCNPs
The UV-Vis spectra were recorded using Lambda 650 UV/Vis spectrophotometer (Perkin Elmer) before and after treatment of CNPs with CFL to verify the functionalization. The fluorescence properties were investigated by taking the photoluminescence (PL) spectra using Hitachi-7000 spectrophotometer at room temperature at photomultiplier voltage of 700 V and scan speed of 2400 nm min−1 using excitation wavelength of 490 nm and following the emission at 520 nm. The surface chemistry of the CNPs with CFL was studied using X-ray photoelectron spectroscopy (XPS) using PHI-5400 ESCA spectrometer at a base pressure of approximately 10−9 Torr and monochromatic X-ray source of MgKα (1253.6 eV) and a power of 200 W. The binding energy (B.E.) scale of the instrument was calibrated using a standard gold sample with the binding energy at 84.0 ± 0.1 eV for Au (4f7/2). The samples for XPS experiments were mounted on a carbon tape and any charging shift was removed using a B.E. scale of adventitious carbon with the baseline at 284.6 eV for the C1s.41 The XPS spectra were deconvoluted into Gaussian/Lorentzian peak shape using Peak Fit (version 4.0) software. HRTEM images were collected using Tecnai F30 electron microscope operated at 300 KV with a point to point resolution of 0.2 nm. The samples were prepared by drop coating a sample of CCNPs solution on the holy carbon coated copper grids followed by overnight drying in vacuum. Zeta-potential measurements were carried out using dynamic light scattering measurements from Zeta Sizer Nano (Malvern Instruments) which uses a laser of wavelength 633 nm.
Cultivation of keratinocytes
A normal human keratinocyte cell line (HaCat) was obtained from Dr Norbert Fusenig of the division of differentiation and carcinogenesis, German Cancer Research Center. For cultivation in defined medium, cells were transitioned from DKM (defined keratinocyte medium) (Invitrogen, Carlsbad, CA) supplemented with 10% FBS to FBS deficient DKM, by 50% stepwise decrease in FBS concentration (5% FBS in DKM first passage, 2.5% FBS in DKM second passage and so forth). This gradual decrease in FBS conditioned allowed HaCat cells for optimal growth in DKM without serum, as direct transition to FBS deficient DKM resulted into cell death. HaCat cells were cultured in DKM supplemented with 1 mL growth supplement (Invitrogen) and (10 μg mL−1 streptomycin and 10 IU mL−1 penicillin (Mediatech, Herndon, VA). The cultures were incubated at 37 °C in a humidified 5% CO2 atmosphere to maintain the proper pH.
Cell viability assays
In order to determine the toxicity of CCNPs, MTT assay was performed. In a typical experiment, 2500 cells were seeded per well in a 96-well plate. Cells were allowed to adhere to well plate surface for 24 h, and different concentrations (1 mg mL−1, 0.1 mg mL−1, 0.01 mg mL−1, and 0.001 mg mL−1) of CCNPs were separately dispersed in DKM or PBS and added to the cells for 24 h and 3 h respectively. A tetrazolium dye (MTT) was added (1.2 mM) and cells were subsequently incubated at 37 °C in 5% CO2 for 4 h. To solubilize the dye, 100 μL cell lysis solution (10% SDS, 5 mM HCl) was added to each well and the plate was incubated overnight at 37 °C. Absorbance of the soluble dye was recorded at 570 nm using a Spectra Max 190 spectrophotometer (Molecular Devices, Sunnyvale, CA). Cell viability was determined by dividing the absorbance of treated samples to untreated controls and reported as a percentage with error being the standard deviation from triplicate wells.
Uptake and subcellular localization studies
HaCat cells were cultured in media described above and after reaching ~90% confluency, cells were harvested and plated on glass cover slips (~2000 cells per cover slip per well) in a 6-well culture dish. After incubation for 24 h, media was replaced with PBS supplemented with desired inhibitors followed by the incubation with CCNPs (dispersed in PBS) with cells for 3 h. Inhibitors were used essentially as previously described by Chen et al.42 with slight modifications. (a) Low temperature incubation: for low temperature studies, incubation of cells with CCNPs in PBS buffer was carried out at 4 °C for 3 h, followed by washing with PBS at 4 °C prior to fixation and staining. For ATP-depletion, cells were pre-incubated in PBS buffer supplemented with 50 mM 2-deoxy-d-glucose and 0.02% sodium azide for 30 min at 37 °C.27 (b) Hypertonic incubation: cells were pre-incubated for 30 min at 37 °C in PBS buffer supplemented with 0.45 M sucrose before exposure to CCNPs in PBS buffer.27 (c) K+ ion depletion: K+ depletion was achieved by pre-incubation of the cells in the K+-free HEPES (HEPES − K+) for 1 h and subsequently rinsing in the hypotonic buffer (HEPES − K+ diluted 1 : 1 with Milli-Q water) for 5 min.43 Cells were then quickly washed with HEPES − K+ buffer three times and incubated in the same for 10 min at 37 °C. After K+ depletion treatment, cells were incubated with CCNPs in HEPES − K+ as mentioned above. Control experiments were carried out with the K+-containing buffer (HEPES + K+). (d) MβCD and nystatin treatment: cells were pretreated with 10 mM MβCD or 25 μg mL−1 nystatin in PBS buffer for 35 min at 37 °C to deplete membrane cholesterol44 before incubation with CCNPs. (e) Macropinocytosis inhibition: Cells were pre-incubated for 35 min with 5 μM cyto-D45 and 2 μM LATB,46 respectively, followed by incubation with CCNPs. The drugs were maintained during the incubation with CCNPs. Cells were washed twice with PBS followed by fixation and staining (see below).
For determining the location of CCNPs after uptake, cells were again plated on cover slips as described above followed by incubation with nanoparticles (in PBS) for 3 h. After incubation cells were treated with Mito-tracker, Lyso-tracker and ER-tracker dyes (as per the guidelines provided by Invitrogen) separately and incubated for 30 min, 60 min and 30 min, respectively, followed by washing with excess PBS. Following these treatments, cells were fixed and stained (see below).
Isolation of cytoplasmic and nuclear extracts
Adherent cells (HaCat) were grown in 25 cm2 flask till ~90% confluency (~1.5 × 106 cells) followed by incubation with CCNPs for different time intervals (1, 3, 6, 9 and 12 h) in CO2 incubator at 37 °C. The corresponding control flasks were also treated in same way except for exposure to CCNPs. Next, cells were trypsinized and finally washed thrice with PBS and dispersed in PBS. To follow uptake kinetics in suspended cells, HaCat cells were grown till ~90% confluency followed by trypsinization. A fixed number (1.5 × 106) of cells per treatment were exposed to CCNPs for different time intervals (1, 3, 6, 9 and 12 h) in a 5% CO2 environment at 37 °C. After incubation, cells were washed thrice with PBS and finally dispersed in PBS. Further, for whole cell extract, cells were sonicated for 10 s and PL spectra were recorded directly from the lysate. For cytoplasmic and nuclear extract isolation, we used the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo) and followed the protocol provided.
Confocal microscopy
Cells were fixed with chilled (−20 °C) methanol for 8 min followed by incubation with DAPI for 8 min. Cells were washed and mounted in anti-fade mounting media (Calbiochem) and slides were prepared. These slides were stored at 4 °C until confocal imaging by Leica TCS SP5 laser scanning confocal microscope with 40 × oil objective lens.
Conclusion
The uptake of cerium oxide based nanoparticles is energy-dependent and mediated via clathrin-dependent endocytosis from lipid rafts. CCNPs are widely distributed in multiple compartments of the cell, thus indicating that their recent efficacy as a novel antioxidant may be in part due to this wide distribution. These results, combined with increasing evidence that these nanomaterials scavenge multiple reactive oxygen and potentially nitrogen species, further suggest that nanoceria is a promising novel nanomaterial for biomedical applications.
Supplementary Material
Acknowledgements
These studies were supported by and NIH grant (1R01AG031529-01) to WS and SS and by an NSF grant (NIRT 0708172 CBET) to SS and WS.
Footnotes
Electronic supplementary information (ESI) available: Detailed experimental procedures and supplementary figures are available for this work. See DOI: 10.1039/c0mb00014k
References
- 1.Karakoti AS, Monteiro-Riviere NA, Aggarwal R, Davis JP, Narayan RJ, Self WT, McGinnis J, Seal S. JOM. 2008;60:33–37. doi: 10.1007/s11837-008-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das M, Patil S, Bhargava N, Kang J-F, Riedel LM, Seal S, Hickman JJ. Biomaterials. 2007;28:1918–1925. doi: 10.1016/j.biomaterials.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heckert EG, Karakoti AS, Seal S, Self WT. Biomaterials. 2008;29:2705–2709. doi: 10.1016/j.biomaterials.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korsvik C, Patil S, Seal S, Self WT. Chem. Commun. 2007:1056–1058. doi: 10.1039/b615134e. [DOI] [PubMed] [Google Scholar]
- 5.Niu J, Azfer A, Rogers LM, Wang X, Kolattukudy PE. Cardiovasc. Res. 2007;73:549–559. doi: 10.1016/j.cardiores.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarnuzzer RW, Colon J, Patil S, Seal S. Nano Lett. 2005;5:2573–2577. doi: 10.1021/nl052024f. [DOI] [PubMed] [Google Scholar]
- 7.Asati A, Santra S, Kaittanis C, Nath S, Perez JM. Angew. Chem., Int. Ed. 2009;48:2308–2312. doi: 10.1002/anie.200805279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heckert EG, Karakoti AS, Seal S, Self WT. Biomaterials. 2008;29:2705–2709. doi: 10.1016/j.biomaterials.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakoti AS, Kuchibhatla S, Babu KS, Seal S. J. Phys. Chem. C. 2007;111:17232–17240. [Google Scholar]
- 10.Karakoti AS, Singh S, Kumar A, Malinska M, Kuchibhatla SVNT, Wozniak K, Self WT, Seal S. J. Am. Chem. Soc. 2009;131:14144–14145. doi: 10.1021/ja9051087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korsvik C, Patil S, Seal S, Self WT. Chem. Commun. 2007:1056–1058. doi: 10.1039/b615134e. [DOI] [PubMed] [Google Scholar]
- 12.Xu X-HN, Chen J, Jeffers RB, Kyriacou S. Nano Lett. 2002;2:175. [Google Scholar]
- 13.Xia T, Kovochich M, Liong M, Mädler L, Gilbert B, Shi H, Yeh JI, Zink JI, Nel AE. ACS Nano. 2008;2:2121. doi: 10.1021/nn800511k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner SD, Schmid SL. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 15.Kostarelos K, Lacerda L, Pastorin G, Wu W, Wieckowski S, Luangsivilay J, Godefroy S, Pantarotto D, Briand J-P, Muller S, Prato M, Bianco A. Nat. Nanotechnol. 2007;2:108–113. doi: 10.1038/nnano.2006.209. [DOI] [PubMed] [Google Scholar]
- 16.Hanaki K, Momo A, Oku T, Komoto A, Maenosono S, Yamaguchi Y, Yamamoto K. Biochem. Biophys. Res. Commun. 2003;302:496–501. doi: 10.1016/s0006-291x(03)00211-0. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat. Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 18.Parak WJ, Boudreau R, Gros ML, Gerion D, Zanchet D, Micheel CM, Williams SC, Alivisatos AP, Larabell C. Adv. Mater. 2002;14:882–885. [Google Scholar]
- 19.Chithrani BD, Ghazani AA, Chan WCW. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 20.Sulin Z, Ju L, George L, Gang B, Subra S. Adv. Mater. 2009;21:419–424. [Google Scholar]
- 21.Choy J-H, Jung J-S, Oh J-M, Park M, Jeong J, Kang Y-K, Han O-J. Biomaterials. 2004;25:3059–3064. doi: 10.1016/j.biomaterials.2003.09.083. [DOI] [PubMed] [Google Scholar]
- 22.Oh J-M, Park M, Kim S-T, Jung J-Y, Kang Y-G, Choy J-H. J. Phys. Chem. Solids. 2006;67:1024–1027. [Google Scholar]
- 23.Berry CC. J. Mater. Chem. 2005;15:543–547. [Google Scholar]
- 24.Xing X, He X, Peng J, Wang K, Tan W. J. Nanosci. Nanotechnol. 2005;5:1688–1693. doi: 10.1166/jnn.2005.199. [DOI] [PubMed] [Google Scholar]
- 25.Kam NWS, Dai H. J. Am. Chem. Soc. 2005;127:6021–6026. doi: 10.1021/ja050062v. [DOI] [PubMed] [Google Scholar]
- 26.Choy J-H, Kwak S-Y, Park J-S, Jeong Y-J. J. Mater. Chem. 2001;11:1671–1674. [Google Scholar]
- 27.Nadine Wong Shi K, Zhuang L, Hongjie D. Angew. Chem., Int. Ed. 2006;45:577–581. [Google Scholar]
- 28.Patil S, Reshetnikov S, Haldar MK, Seal S, Mallik S. J. Phys. Chem. C. 2007;111:8437–8442. [Google Scholar]
- 29.Silcoff E. R. a. T. S. New J. Chem. 1999;23:1187–1192. [Google Scholar]
- 30.Kumar A, Suresh B, Karakoti AS, Schulte A, Seal S. Langmuir. 2009;25:10998–11007. doi: 10.1021/la901298q. [DOI] [PubMed] [Google Scholar]
- 31.Bansal V, Ahmad A, Sastry M. J. Am. Chem. Soc. 2006;128:14059–14066. doi: 10.1021/ja062113+. [DOI] [PubMed] [Google Scholar]
- 32.Peters R. Xenopus Protocols. Springer; 2006. pp. 235–258. [Google Scholar]
- 33.Letoha T, Gaál S, Somlai C, Venkei Z, Glavinas H, Kusz E, Duda E, Czajlik A, Peták F, Penke B. J. Pept. Sci. 2005;11:805–811. doi: 10.1002/psc.678. [DOI] [PubMed] [Google Scholar]
- 34.Fujimoto LM, Roth R, Heuser JE, Schmid SL. Traffic. 2000;1:161–171. doi: 10.1034/j.1600-0854.2000.010208.x. [DOI] [PubMed] [Google Scholar]
- 35.Balaban RS, Nemoto S, Finkel T. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Hirst SM, Karakoti AS, Tyler RD, Sriranganathan N, Seal S, Reilly CM. Small. 2009;5:2848–2856. doi: 10.1002/smll.200901048. [DOI] [PubMed] [Google Scholar]
- 37.Panyam J, Zhou W, Prabha S, Sahoo SK, Labhasetwar V. FASEB J. 2002;16:1217–1226. doi: 10.1096/fj.02-0088com. [DOI] [PubMed] [Google Scholar]
- 38.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 39.Tu BP, Weissman JS. J. Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18525–18530. doi: 10.1073/pnas.0809677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barr TL, Seal S. J. Vac. Sci. Technol., A. 1995;A13:1239–1246. [Google Scholar]
- 42.Chen X, Kis A, Zettl A, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8218–8222. doi: 10.1073/pnas.0700567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puri V, Watanabe R, Singh R, Dominguez M, Brown JC, Wheatley CL, Marks DL, Pagano RE. J. Cell Biol. 2001;154:535–548. doi: 10.1083/jcb.200102084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma X, Wang Q, Jiang Y, Xiao Z, Fang X, Chen Y. Biochem. Biophys. Res. Commun. 2007;356:67–71. doi: 10.1016/j.bbrc.2007.02.080. [DOI] [PubMed] [Google Scholar]
- 45.Rinne J, Albarran B, Jylhava J, Ihalainen T, Kankaanpaa P, Hytonen V, Stayton P, Kulomaa M, Vihinen-Ranta M. BMC Biotechnol. 2007;7:1–14. doi: 10.1186/1472-6750-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spector I, Nava RS, Dina B, Yoel K. Cell Motil. Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.