Abstract
Dysregulation of ligand-independent receptor tyrosine kinase (RTK) dimerization, which is the first step in RTK activation, leads to pathologies. A mechanistic understanding of the dimerization process is lacking, and this lack of basic knowledge is one bottleneck in developing effective RTK-targeted therapies. For instance, the roles and the relative contributions of the different RTK domains to RTK dimerization are unknown. Here we use quantitative imaging Förster resonance energy transfer (QI-FRET) to determine the contribution of the extracellular (EC) domain of fibroblast growth factor receptor 3 (FGFR3) to FGFR3 dimerization. We provide the first direct experimental evidence that the contribution of FGFR3 EC domain to dimerization is repulsive in the absence of ligand, and on the order of 1 kcal/mole. The magnitude of this repulsive contribution is similar to the dimer over-stabilization that can occur due to pathogenic single amino acid mutations, and therefore significant for biological function.
INTRODUCTION
RTKs are the second largest family of membrane receptors, after the G protein-coupled receptor family. They are single-pass transmembrane (TM) proteins, with an N-terminal extracellular (EC) domain usually composed of several motifs and involved in ligand binding, a single TM domain followed by a juxtamembrane region, and an intracellular tyrosine kinase domain 1-3. RTK activation is a two-step process involving (1) lateral dimerization in the membrane plane and (2) ligand binding, which is believed to stabilize the RTK dimers and likely alter their structure and activity. RTK monomers are inactive, while RTK dimers are active because the interactions between the two catalytic domains in the dimer stimulate catalytic activity 4-6. Thus, RTK dimerization regulates RTK activity.
Ligand-induced dimerization and activation has been long recognized as critical in signaling by RTKs. In many cases, however, ligands are not necessary for RTK dimerization and activation, and stable dimers have been shown to exist in cellular membranes in the absence of ligand 7;8. Furthermore, dysregulation of ligand-independent dimerization, due to RTK overexpression or due to RTK mutations, had been linked to human pathologies 9-13. For example, mutations in FGFR3, an RTK which is critical for skeletal development 14-16, are believed to cause skeletal dysplasias and cancer by enhancing ligand-independent dimerization 16;17. Yet, a mechanistic understanding of ligand-independent FGFR3 dimerization is lacking.
The extracellular (EC) domain of FGFR3, which consists of three Immunoglobulin-like (Ig-like) domains (D1-D3), has been crystallized in the presence of the ligand fgf1 18. The structure has revealed that D2 and D3 mediate ligand binding, while D1 inhibits it. The behavior of FGFR3 EC domain in the process of ligand-independent dimerization, however, is unknown.
While dimerization studies have been carried out for isolated RTK transmembrane (TM) domains, and FGFR3 TM domain in particular has been shown to possess a propensity for sequence-specific dimerization 19-22, the dimerization energetics of RTK EC domains has not been characterized thus far due to experimental limitations. Tanner and Kyte have argued that the role of ErbB1 extracellular domain is inhibitory in the absence of ligand 23. However, their experiments have been scrutinized, and their conclusions questioned 24. The effect of the reduced dimensionality on interactions in membranes is believed to be large 25, but is unknown. Thus, measurements of interactions between isolated soluble extracellular domains in 3D aqueous solutions26;27, and measurements of interactions between isolated TM domains in 2D membranes28-31 cannot be compared.
Recently, we introduced a methodology that yields dimerization free energies of proteins in mammalian membranes 32. This method allows us to measure membrane protein interactions in two dimensions. The measurements utilize the quantitative imaging FRET (QI-FRET) method which yields the FRET efficiency E, as well as the concentration of donors and acceptors, CD and CA (the three parameters needed to calculate association constants) 33. Experiments are carried out in single plasma membrane-derived vesicles, which bud off from cells upon treatments that disrupt the cytoskeleton 34;35. Their membranes exhibit a homogeneous distribution of the fluorescently-tagged proteins 36, which allows us to compare the fluorescence intensity of the vesicles with that of purified fluorescent protein solutions of known concentration, and implement the QI-FRET method 32.
Here we use QI-FRET to determine the contribution of FGFR3 EC domain to FGFR3 dimerization in plasma membrane vesicles derived from CHO cells. We measure this contribution by comparing the dimerization of two FGFR3 constructs, with and without the three Ig-like motifs of FGFR3 EC domain, in plasma membrane-derived vesicles. We provide the first direct experimental demonstration that FGFR3 EC domain inhibits ligand-independent dimerization.
RESULTS
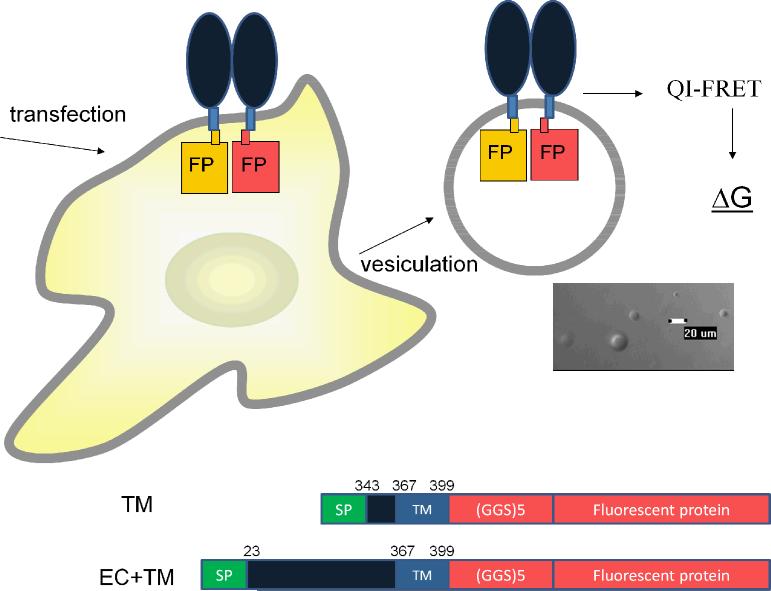
The two gene constructs used in this study are shown in Figure 1, and the design of the plasmids is described in Supplementary Material. The two constructs used were (i) a EC+TM construct, consisting of the signal peptide, the EC domain and TM domain of FGFR3, a 15 amino acid flexible linker (GGS)5 37, and the fluorescent protein, and (ii) a TM construct lacking the three immunoglobulin-like motifs, consisting of the signal peptide, a short 24 amino acid C-terminal segment from the EC domain, the TM domain of FGFR3, a 15 amino acid flexible linker (GGS)5, and the fluorescent protein. The attachment of the fluorescent proteins via the 15 amino acid flexible linkers has been used in a previous study 32, and thus the distance between the fluorescent proteins in the dimers is known, ~ 48.5 Å.
Figure 1.
Overview of measurements of dimerization free energy in plasma membrane-derived vesicles. Two different constructs were used (i) A EC+TM construct consisting of the signal peptide, the EC domain, the TM domain, a 15 amino acid-long (GGS)5 flexible linker, and the fluorescent proteins and (ii) a TM construct lacking the three Ig-like motifs of FGFR3 extracellular domain, consisting of the signal peptide, a 24 amino acid segment of the EC domain, the TM domain, a 15 amino acid (GGS)5 flexible linker, and the fluorescent proteins. We determine the contribution of the EC domain to FGFR3 dimerization by measuring the difference in dimerization free energy for these two constructs.
The homodimerization of these two constructs was assessed in the following two sets of experiments. In the first set of experiments, EC+TM_EYFP and EC+TM_mCherry were co-transfected into CHO cells. In the second set of experiments, TM_EYFP and TM_mCherry were co-transfected into CHO cells. Twenty-four hours after transfection, vesiculation was carried out as described in Li et al 36. Vesicles loaded with EC+TM_EYFP/EC+TM_mCherry and TM_EYFP/TM_mCherry were collected and imaged in a Nikon confocal microscope as described 32. Briefly, the intensity of the donor emission was collected from 500 to 530 nm in the donor scan by exciting with a 488 nm laser. The sensitized emission was obtained from the FRET scan, which used the 488 nm laser to excite EYFP, and recorded the emission of mCherry in the interval 565 – 615 nm. The acceptor scan utilized the 543 nm laser to excite mCherry and collected photons with wavelengths >650 nm to yield the emission intensity of the acceptor mCherry.
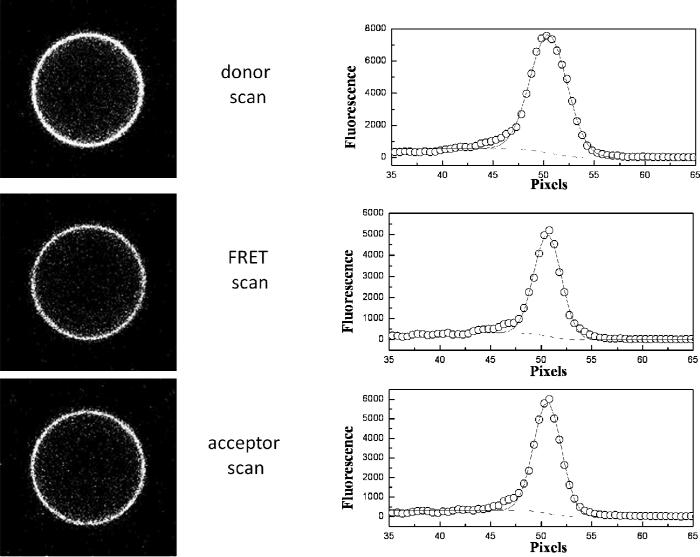
Typical images are seen in Figure 2. The fluorescence is uniform, indicative of homogeneous protein distribution, and is concentrated on the membrane, as expected for membrane proteins. The homogeneous protein distribution in the vesicles is expected, since the cytoskeleton, which plays a role in sustaining lateral membrane heterogeneity, is not present in the vesicles. As discussed previously, the uniform fluorescence allows the calibration of the donor and acceptor concentrations in the vesicles, and is critical for the successful implementation of the QI-FRET method 32;36.
Figure 2.
One vesicle loaded with FGFR3 TM_EYFP and TM_mCherry. Intensities per unit membrane area were obtained by integrating the Gaussian intensity profiles across the membrane after correcting for the background.
The images were processed with a Matlab program written in the lab, which automatically recognized the vesicles and integrated the membrane intensity 32. The signal that originates from the membrane was fitted to a Gaussian function (solid lines in Figure 2), while the background fluorescence inside the vesicle was modeled by an error function (dashed lines in Figure 2). The integration of the Gaussian yielded the three intensities, , per unit membrane area 32. The concentrations of donor-labeled and acceptor-labeled RTK constructs were determined from these intensities as described in detail in Chen et al 32. Briefly, the concentration of acceptor-labeled proteins per unit membrane area (normal to the focal plane) was calculated for each vesicle as:
| (1) |
where iA is a calibration constant obtained by imaging purified mCherry solution of known concentration using the acceptor scan 36. On the other hand, the concentration of donor-labeled proteins per unit membrane area () was determined as 32:
| (2) |
Next, the FRET efficiency (E) was calculated for each vesicle according to 36:
| (3) |
In these equations, βD and βA are the bleedthrough coefficients, determined by imaging vesicles expressing only donor or only acceptor 36. The Gauge factor, GF, which correlates the sensitized acceptor emission with donor quenching, was determined by analyzing vesicles expressing a construct of the donor fused to the acceptor 36.
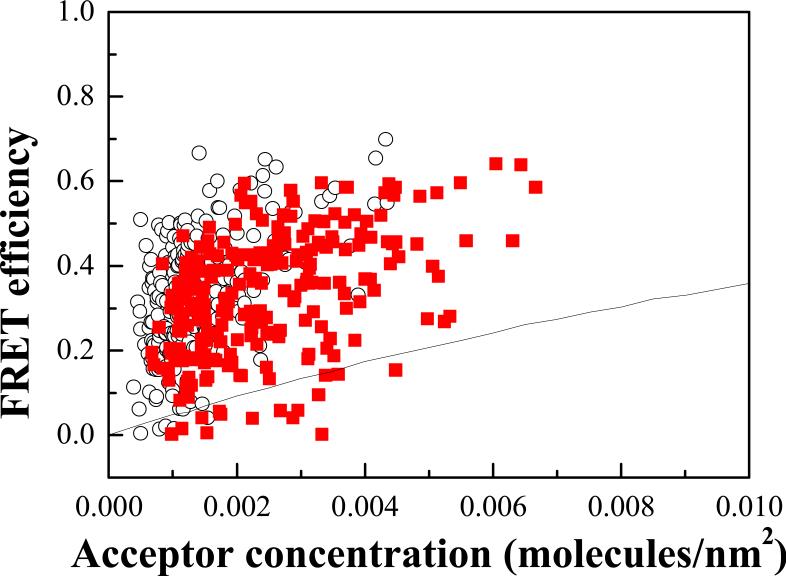
The FRET efficiency E is plotted as a function of the acceptor concentration in the vesicles in Figure 3 for both the EC+TM and TM constructs. Each data point represents a single vesicle. The acceptor concentration in Figure 3 varies over more than an order of magnitude, yielding a functional dependence of the measured FRET efficiency as a function of the concentration. The FRET efficiency increases with the acceptor concentration, as expected for dimerization 20;38;39. The data scatter can be explained by the random noise associated with imaging 32, and is reducible by acquiring a large number of data points (the approach taken in single-molecule experiments). Despite data scatter, we see that the FRET efficiency measured for the TM construct is higher than the EC+TM efficiency, demonstrating that the EC domain inhibits dimerization.
Figure 3.
FRET data and proximity contribution for FGFR3 EC+TM and TM in CHO plasma membrane-derived vesicles. Each data point represents a single vesicle, for which E, CA and CD are determined using the QI-FRET method. Filled squares: FRET efficiencies measured for the FGFR3 EC+TM construct. Hollow circles: FRET efficiencies for the FGFR3 TM construct. QI-FRET data analysis has demonstrated that the scatter is largely due to random noise in image acquisition, and is reducible by collecting a large number of data points 32.
The FRET efficiencies E given by equation (3) contain two contributions: ED, the FRET efficiency due to sequence-specific dimerization, and Eproximity, the FRET efficiency due to random co-localization of donor and acceptor 28;38. The proximity contribution, arising from random co-localization of donors and acceptors has been discussed in detail previously 19;40;41, and is plotted in Figure 3 with the solid line. Next, it is subtracted from the measured FRET to obtain FRET due to sequence-specific dimerization, ED:
| (4) |
The dimeric fraction f in each vesicle is determined from the corrected FRET efficiency ED as 42:
| (5) |
where xA is the acceptor fraction, a known parameter since the donor and acceptor concentrations are determined with the QI-FRET method. The parameter Ẽ is the FRET efficiency in a dimer with a donor and an acceptor 36. Ẽ depends on the distance between the donor and the acceptor in the dimer, i.e. on the structure of the dimer. In both constructs, the fluorescent proteins are attached to the TM domains via a 15 aa flexible linker 43, the same attachment used in previous studies of GpA dimerization 32. Ẽ for this attachment was determined as Ẽ = 0.63 ± 0.04, corresponding to 48.5 Å separation distance between the two fluorophores in the dimer. With xA and Ẽ known, the dimeric fractions in each vesicle can be calculated from equation (5).
Next, the dimeric fraction 2[D]/[T] was calculated as a function of the total concentration [T] for various values of KD, based on the following dimerization model :
| (6) |
where the dimerization constant is:
| (7) |
and the total concentration is given by:
| (8) |
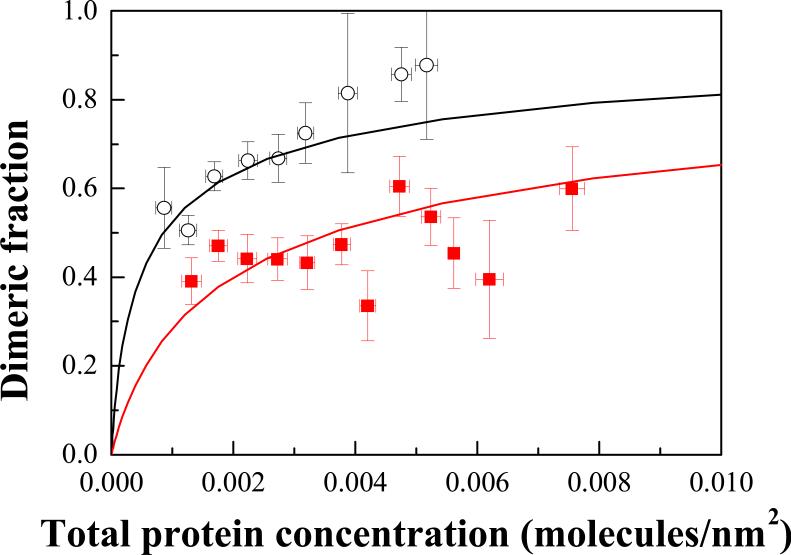
We fitted the calculated prediction to the experimentally measured single-vesicle dimeric fractions, while optimizing for the dimerization constant KD. The optimal values for the two constructs determined in the fit were KD(EC+TM) = 227 ± 45 nm2 and KD(TM) = 1170 ± 260 nm2 (see curves in Figure 4). These optimal KD values were independent of the initial guesses used in the fit.
Figure 4.
Filled squares: Averaged dimeric fractions measured for the FGFR3 EC+TM construct. Hollow circles: averaged dimeric fractions measured for the FGFR3 TM construct. The data were fitted to the dimerization model given by equations (6)-(8), yielding the dimerization constants KD. The contribution of FGFR3 extracellular domain to dimerization is inhibitory, ΔΔG = 0.9 ± 0.2 kcal/mole.
Since the probability of having exactly the same total RTK concentration in several vesicles is very low, the average dimeric fractions and the respective standard errors were calculated by averaging data with similar concentrations, within bins of size 0.0005 molecules/nm2 that had 3 to 20 data points. The average values of the calculated dimeric fractions are shown in Figure 4, as open circles for the TM construct and filled squares for the EC+TM construct. Note that the curves in Figure 4 were obtained by fitting all the single-vesicle data points, and are shown together with the experimental averages to facilitate comparison between the fits and the data. We see that the averaged data and the fits are in agreement. We also see that the dimeric fraction is higher for the TM construct, indicating that the propensity for dimerization is reduced in the presence of the EC domain.
As discussed previously 32, the units of the dimerization constants are area (units of dissociation constants are 1/area), because the lateral interactions between the RTK constructs are interactions in two dimensions. Defining the standard state as KD = 1 nm2, we obtained a free energy of dimerization ΔG(TM) = -RTlnKD(TM) = -4.2 ± 0.2 kcal/mole and ΔG(EC+TM) = -3.3 ± 0.1 kcal/mole. Thus, the contribution of FGFR3 extracellular domain to dimerization is positive, ΔΔG = 0.9 ± 0.2 kcal/mole, and therefore inhibitory.
DISCUSSION
The role of the TM domains in the RTK dimerization process has been controversial. For many years, they were believed to be passive anchors. These views were based on experiments in cells which showed that alterations in TM domains had negligible effects on signaling 44;45. In other cases, however, changes in the sequences of the TM domains affected signaling 46;47. Furthermore, pathogenic mutations in TM domains were discovered and proposed to promote ligand-independent dimerization 48;49. Studies of the isolated TM domains in lipid bilayers or bacterial membranes invariably showed that these domains can dimerize by themselves 19;29;31;50-52, and they were proposed to be a major driving force for RTK dimerization in the absence of ligand 22;53. However, a formal proof for such a role has been missing due to experimental challenges in measuring dimerization energetics of RTK constructs in mammalian membranes.
In the absence of ligand, RTK extracellular domains are believed to explore two different conformations, a dimerization-competent conformation with the dimerization interface exposed, and a dimerization-incompetent conformation with the dimerization interface hidden 54-56. The crystal structures of RTKs in the absence of ligand are locked in the inactive conformation 57-59. In cellular membranes, the time spent in these two conformations determines the propensity for dimerization of the extracellular domain. Based on the crystal structures and biochemical evidence, Ferguson and colleagues have argued that an RTK EC domain (other than ErbB2) spends most of the time in the dimerization-incompetent conformation54. A question remains, however, whether the dimerization-incompetent EC conformation is repulsive, attractive, or neutral in the dimerization process.
Here we present the first direct experimental evidence that the contribution of FGFR3 EC domain to dimerization is repulsive in the absence of ligand. We accomplish this by measuring directly the dimerization energetics of two FGFR3 constructs, with and without the three Ig-like motifs of the EC domain, using a new methodology.
The dimerization of the isolated FGFR3 TM domain (residues 367 to 399) has been characterized previously in lipid bilayers 19;38. Its free energy of dimerization is about -3 kcal/mole, and can now be compared to the value reported here for the TM construct, ~ -4 kcal/mole. We see that the TM construct used here dimerizes more strongly than the shorter TM domain studied in lipid bilayers. There may be different reasons for the observed difference. First, the native membrane environment is complex and crowded with membrane proteins, unlike the model lipid bilayer, and crowding is known to have a strong effect on macromolecular interactions 60. Second, the short EC sequence (residues 343 to 366) that is a part of the TM construct also may be contributing to FGFR3 dimerization.
Studies of the effect of pathogenic mutations on FGFR3 TM domain dimerization have provided new insights into the mechanism behind human pathologies 22;61. One mutation that has been studied using this approach is the Ala391→Glu mutation, associated with Crouzon syndrome with acanthosis nigricans, an autosomal dominant disorder characterized by premature ossification of the skull (craniosynostosis), as well as hyperpigmentation and hyperkeratosis of the skin 62. The change in the free energy of dimerization of the isolated FGFR3 TM domain in lipid bilayers due to this mutation is -1.3 kcal/mole 19. Thus, the Ala391→Glu mutation over-stabilizes FGFR3 TM domain dimers19. Since dimerization and activation are tightly coupled, it can be expected that dimer over-stabilization leads to FGFR3 overactivation and the observed pathology.
Interestingly, the magnitude of the EC contribution measured here, about 1 kcal/mole, is similar in magnitude and opposite in sign to the effect of the Ala391→Glu mutations on dimerization (i.e. it is repulsive instead of stabilizing). It therefore appears that changes in dimerization propensities on the order of 1 kcal/mole can have a significant effect on RTK signaling. Therefore, the repulsive contribution of the EC domain in the absence of ligand is likely critical for maintaining signaling at normal levels.
MATERIALS AND METHODS
Plasmid constructs
pRSET_mCherry was obtained from the laboratory of Roger Tsien (University of California, San Diego), and pEYFP was obtained from the laboratory of Michael Betenbaugh (Johns Hopkins University, Baltimore). PcDNA3.1_FGFR3 was a kind gift from Dr. Daniel Donoghue from the University of California, San Diego. All plasmids used for mammalian expression were engineered using the pcDNA3.1(+) vector (Invitrogen). All primers were purchased from Invitrogen. For a detailed description of plasmid design, see Supplemental Material.
The FGFR3 TM construct consisted of the FGFR3 signal peptide (MGAPACALALCVAVAIVAGA), a 24 amino acid-long C-terminal sequence of the EC domain, the TM domain (DEAGSVYAGILSYGVGFFLFILVVAAVTLCRLR), a flexible 15 amino acid-long linker (GGS)5 and fluorescent proteins at the C terminus. The gene was constructed between the multicloning sites HindIII and XbaI of pcDNA3.1(+), as described in Supplemental Material.
FGFR3 EC+TM construct contained the signal peptide, the extracellular and transmembrane domains of FGFR3, a linker and fluorescent proteins at the C terminus. The linker was same as the linker used in the FGFR3 TM construct.
CHO cell growth, transfection and vesiculation
Chinese hamster ovary (CHO) cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1 mM non-essential amino acids, 1.8 g/L D-glucose, and 1.5 g/L sodium bicarbonate. 2×104 cells/well were seeded in a 6-well plate one day before transfection.
Transfection was carried out using Fugene® HD transfection reagent (Roche Applied Science), following the manufacturer's protocol. Vesiculation was carried out using a previously described protocol 63. Cells were rinsed three times with PBS (pH 7.4) containing 0.75 mM calcium and 0.5 mM magnesium (CM-PBS), and incubated with 1 ml of vesiculation buffer at 37 °C. To quench the formaldehyde, a glycine solution in PBS was added to the vesiculation buffer to a final concentration of 0.125 M. The vesiculation buffer consisted of CM-PBS with 25 mM formaldehyde and 0.5 mM 1,4-dithiothreitol (DTT).
Image acquisition
Vesicles were imaged using a Nikon Eclipse confocal laser scanning microscope with a 60X water immersion objective. All the images were collected and stored at a 512×512 resolution. Three different scans were performed for each vesicle: (1) excitation at 488 nm, with a 500-530 nm emission filter (donor scan); (2) excitation at 488 nm, with a 565-615 nm emission filter (FRET scan); and (3) excitation at 543 nm, with a 650 nm long-pass filter (acceptor scan). Gains of 8.0 were used for all the three scans. To minimize the bleaching of fluorescent proteins, ND8 filters were used during excitation with the 488 nm laser, and pixel dwell time was set to the minimum (1.68 μs).
Image analysis
The fluorescence intensities of vesicles were analyzed with a Matlab program developed in the lab, which automatically recognizes the vesicles and calculates the intensities in the three channels.
Supplementary Material
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Reference List
- 1.Fantl WJ, Johnson DE, Williams LT. Signaling by Receptor Tyrosine Kinases. Annu.Rev.Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 2.L'Horte CGM, Knowles MA. Cell responses to FGFR3 signaling: growth, differentiation and apoptosis. Experim.Cell Res. 2005;304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends in Cell Biology. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 6.Zhang XW, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J.Mol.Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 8.Tao RH, Maruyama IN. All EGF(ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J.Cell Sci. 2008;121:3207–3217. doi: 10.1242/jcs.033399. [DOI] [PubMed] [Google Scholar]
- 9.Harari D, Yarden Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene. 2000;19:6102–6114. doi: 10.1038/sj.onc.1203973. [DOI] [PubMed] [Google Scholar]
- 10.Browne BC, O'Brien N, Duffy MJ, Crown J, O'Donovan N. HER-2 Signaling and Inhibition in Breast Cancer. Current Cancer Drug Targets. 2009;9:419–438. doi: 10.2174/156800909788166484. [DOI] [PubMed] [Google Scholar]
- 11.Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 Receptor and Breast Cancer: Ten Years of Targeted Anti-HER-2 Therapy and Personalized Medicine. Oncologist. 2009;14:320–368. doi: 10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 12.Sakai K, Yokote H, Murakami-Murofushi K, Tamura T, Saijo N, Nishio K. Pertuzumab, a novel HER dimerization inhibitor, inhibits the growth of human lung cancer cells mediated by the HER3 signaling pathway. Cancer Science. 2007;98:1498–1503. doi: 10.1111/j.1349-7006.2007.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 14.Colvin JS, Bohne BA, Harding GW, Mcewen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat.Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 15.Vajo Z, Francomano CA, Wilkin DJ. The molecular and genetic basis of fibroblast growth factor receptor 3 disorders: The achondroplasia family of skeletal dysplasias, Muenke craniosynostosis, and Crouzon syndrome with acanthosis nigricans. Endocrine Reviews. 2000;21:23–39. doi: 10.1210/edrv.21.1.0387. [DOI] [PubMed] [Google Scholar]
- 16.Horton WA, Hall JG, Hecht JT. Achondroplasia. Lancet. 2007;370:162–172. doi: 10.1016/S0140-6736(07)61090-3. [DOI] [PubMed] [Google Scholar]
- 17.Webster MK, Donoghue DJ. FGFR activation in skeletal disorders: Too much of a good thing. Trends Genet. 1997;13:178–182. doi: 10.1016/s0168-9525(97)01131-1. [DOI] [PubMed] [Google Scholar]
- 18.Olsen SK, Ibrahimi OA, Raucci A, Zhang FM, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, Mohammadi M. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li E, You M, Hristova K. FGFR3 dimer stabilization due to a single amino acid pathogenic mutation. J.Mol.Biol. 2006;356:600–612. doi: 10.1016/j.jmb.2005.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You M, Spangler J, Li E, Han X, Ghosh P, Hristova K. Effect of pathogenic cysteine mutations on FGFR3 transmembrane domain dimerization in detergents and lipid bilayers. Biochemistry. 2007;46:11039–11046. doi: 10.1021/bi700986n. [DOI] [PubMed] [Google Scholar]
- 21.He L, Hristova K. Pathogenic activation of receptor tyrosine kinases in mammalian membranes. J.Mol.Biol. 2008;384:1130–1142. doi: 10.1016/j.jmb.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner KG, Kyte J. Dimerization of the extracellular domain of the receptor for epidermal growth factor containing the membrane-spanning segment in response to treatment with epidermal growth factor. J.Biol.Chem. 1999;274:35985–35990. doi: 10.1074/jbc.274.50.35985. [DOI] [PubMed] [Google Scholar]
- 24.Stanley AM, Fleming KG. The transmembrane domains of ErbB receptors do not dimerize strongly in micelles. J.Mol.Biol. 2005;347:759–772. doi: 10.1016/j.jmb.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 25.Grasberger B, Minton AP, DeLisi C, Metzger H. Interaction Between Proteins Localized in Membranes. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemmon MA, Bu ZM, Ladbury JE, Zhou M, Pinchasi D, Lax I, Engelman DM, Schlessinger J. Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J. 1997;16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang XW, Kuriyan J. Mechanism for Activation of the EGF Receptor Catalytic Domain by the Juxtamembrane Segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You M, Li E, Wimley WC, Hristova K. FRET in liposomes: measurements of TM helix dimerization in the native bilayer environment. Analytical Biochemistry. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artemenko EO, Egorova NS, Arseniev AS, Feofanov AV. Transmembrane domain of EphA1 receptor forms dimers in membrane-like environment. Biochim.Biophys.Acta. 2008;1778:2361–2367. doi: 10.1016/j.bbamem.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Gerber D, Sal-Man N, Shai Y. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J.Biol.Chem. 2004;279:21177–21182. doi: 10.1074/jbc.M400847200. [DOI] [PubMed] [Google Scholar]
- 31.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J.Biol.Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Novicky L, Merzlyakov M, Hristov T, Hristova K. Measuring the Energetics of Membrane Protein Dimerization in Mammalian Membranes. J.Am.Chem.Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merzlyakov M, Chen L, Hristova K. Studies of receptor tyrosine kinase transmembrane domain interactions: The EmEx-FRET method. J.Membr.Biol. 2007;215:93–103. doi: 10.1007/s00232-007-9009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holowka D, Baird B. Structural Studies on the Membrane-Bound Immunoglobulin E-Receptor Complex .1. Characterization of Large Plasma-Membrane Vesicles from Rat Basophilic Leukemia-Cells and Insertion of Amphipathic Fluorescent-Probes. Biochemistry. 1983;22:3466–3474. doi: 10.1021/bi00283a025. [DOI] [PubMed] [Google Scholar]
- 35.Holowka D, Baird B. Structural Studies on the Membrane-Bound Immunoglobulin E-Receptor Complex .2. Mapping of Distances Between Sites on Ige and the Membrane-Surface. Biochemistry. 1983;22:3475–3484. doi: 10.1021/bi00283a025. [DOI] [PubMed] [Google Scholar]
- 36.Li E, Placone J, Merzlyakov M, Hristova K. Quantitative measurements of protein interactions in a crowded cellular environment. Anal.Chem. 2008;80:5976–5985. doi: 10.1021/ac800616u. [DOI] [PubMed] [Google Scholar]
- 37.Evers TH, van Dongen EMWM, Faesen AC, Meijer EW, Merkx M. Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry. 2006;45:13183–13192. doi: 10.1021/bi061288t. [DOI] [PubMed] [Google Scholar]
- 38.Li E, You M, Hristova K. SDS-PAGE and FRET suggest weak interactions between FGFR3 TM domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 39.You M, Li E, Hristova K. The achondroplasia mutation does not alter the dimerization energetics of FGFR3 transmembrane domain. Biochemistry. 2006;45:5551–5556. doi: 10.1021/bi060113g. [DOI] [PubMed] [Google Scholar]
- 40.Posokhov YO, Merzlyakov M, Hristova K, Ladokhin AS. A simple “proximity” correction for Forster resonance energy transfer efficiency determination in membranes using lifetime measurements. Analytical Biochemistry. 2008;380:134–136. doi: 10.1016/j.ab.2008.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolber PK, Hudson BS. An analytic solution to the Förster energy transfer problem in two dimensions. Biophys.J. 1979;28:197–210. doi: 10.1016/S0006-3495(79)85171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merzlyakov M, Hristova K. Forster Resonance Energy Transfer Measurements of Transmembrane Helix Dimerization Energetics. Methods in Enzymology: Fluorescence Spectroscopy. 2008;450:107–127. doi: 10.1016/S0076-6879(08)03406-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Crouy-Chanel A, El Yaagoubi A, Kohiyama M, Richarme G. Reversal by GroES of the GroEL preference from hydrophobic amino acids toward hydrophilic amino acids. J.Biol.Chem. 1995;270:10571–10575. doi: 10.1074/jbc.270.18.10571. [DOI] [PubMed] [Google Scholar]
- 44.Kashles O, Szapary D, Bellot F, Ullrich A, Schlessinger J, Schmidt A. Ligand-Induced Stimulation of Epidermal Growth-Factor Receptor Mutants with Altered Transmembrane Regions. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:9567–9571. doi: 10.1073/pnas.85.24.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carpenter CD, Ingraham HA, Cochet C, Walton GM, Lazar CS, Sowadski JM, Rosenfeld MG, Gill GN. Structural-Analysis of the Transmembrane Domain of the Epidermal Growth-Factor Receptor. J.Biol.Chem. 1991;266:5750–5755. [PubMed] [Google Scholar]
- 46.Petti LM, Irusta PM, DiMaio D. Oncogenic activation of the PDGF beta receptor by the transmembrane domain of p185(neu). Oncogene. 1998;16:843–851. doi: 10.1038/sj.onc.1201590. [DOI] [PubMed] [Google Scholar]
- 47.Cheatham B, Shoelson SE, Yamada K, Goncalves E, Kahn CR. Substitution of the Erbb-2 Oncoprotein Transmembrane Domain Activates the Insulin-Receptor and Modulates the Action of Insulin and Insulin-Receptor Substrate-1. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7336–7340. doi: 10.1073/pnas.90.15.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner DB, Liu J, Cohen JA, Williams WV, Greene MI. A Point Mutation in the Neu Oncogene Mimics Ligand Induction of Receptor Aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 49.Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys.J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escher C, Cymer F, Schneider D. Two GxxxG-Like Motifs Facilitate Promiscuous Interactions of the Human ErbB Transmembrane Domains. J.Mol.Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Finger C, Escher C, Schneider D. The Single Transmembrane Domains of Human Receptor Tyrosine Kinases Encode Self-Interactions. Science Signaling. 2009:2. doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- 53.Bell CA, Tynan JA, Hart KC, Meyer AN, Robertson SC, Donoghue DJ. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol.Biol.Cell. 2000;11:3589–3599. doi: 10.1091/mbc.11.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Molecular Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 55.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 56.Bouyain S, Longo PA, Li SQ, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garrett TPJ, Mckern NM, Lou MZ, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 58.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 59.Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TPJ, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Molecular Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 60.Minton AP. Implications of macromolecular crowding for protein assembly. Cur.Opinion Struc.Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 61.Li E, Hristova K. Receptor Tyrosine Kinase transmembrane domains: function, dimer structure, and dimerization energetics. Cell Adhesion and Migration. 2010;4:249–254. doi: 10.4161/cam.4.2.10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyers GA, Orlow SJ, Munro IR, Przylepa KA, Jabs EW. Fibroblast-Growth-Factor-Receptor-3 (Fgfr3) Transmembrane Mutation in Crouzon-Syndrome with Acanthosis Nigricans. Nat.Genet. 1995;11:462–464. doi: 10.1038/ng1295-462. [DOI] [PubMed] [Google Scholar]
- 63.Scott RE, Perkins RG, Zschunke MA, Hoerl BJ, Maercklein PB. Plasma-Membrane Vesiculation in 3T3-Cells and Sv3T3-Cells .1. Morphological and Biochemical Characterization. J.Cell Sci. 1979;35:229–243. doi: 10.1242/jcs.35.1.229. [DOI] [PubMed] [Google Scholar]
- 64.Supported by NSF MCB-0718841 and NIH GM068619.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.