Abstract
Apolipoprotein mediated formation of nanodisks was studied in detail using apolipophorin III (apoLp-III), thereby providing insight in apolipoprotein-lipid binding interactions. The spontaneous solubilization of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) vesicles occured only in a very narrow temperature range at the gel-liquid-crystalline phase transition temperature, exhibiting a net exothermic interaction based on isothermal titration calorimetry analysis. The resulting nanodisks were protected from proteolysis by trypsin, endoproteinase Glu-C, chymotrypsin and elastase. DMPC solubilization and the simultaneous formation of nanodisks was promoted by increasing the vesicle diameter, protein to lipid ratio and concentration. Inclusion of cholesterol in DMPC dramatically enhanced the rate of nanodisk formation, presumably by stabilization of lattice defects which form the main insertion sites for apolipoprotein α-helices. The presence of fully saturated acyl chains with a length of 13 or 14 carbons in phosphatidylcholine allowed the spontaneous vesicle solubilization upon apolipoprotein addition. Nanodisks with C13:0-phosphatidylcholine were significantly smaller with a diameter of 11.7 ± 3.1 nm compared to 18.5 ± 5.6 nm for DMPC nanodisks determined by transmission electron microscopy. Nanodisk formation was not observed when the phosphatidylcholine vesicles contained acyl chains of 15 or 16 carbons. However, using very high concentrations of lipid and protein (> 10 mg/ml), 1,2,-dipalmitoyl-sn-glycero-3-phosphocholine nanodisks could be produced spontaneously although the efficiency remained low.
Keywords: apolipoprotein, apolipophorin III, DMPC, nanodisk, phosphatidylcholine
1. Introduction
Apolipoproteins bear the intrinsic ability to solubilize phospholipid vesicles forming small discoidal complexes, more recently termed nanodisks [1]. The complexes can be seen as the most basic lipoprotein particle containing one circular bilayer of phospholipid surrounded by the apolipoprotein, which resembles the macromolecular structure of nascent high density lipoprotein (HDL) found in vivo [2]. Nanodisks are excellent particles for studies towards understanding the structure of apolipoproteins in a lipid-bound state as they are composed of one type of apolipoprotein and one phospholipid species [3–5]. They can be used as platforms for insertion of other membrane proteins or peptides, which can then be analyzed in a well-defined lipid environment [6]. The ability of apolipoproteins to solubilize lipid vesicles has often been employed to gain insight into the lipid binding properties of wild-type and mutant apolipoproteins. It has been well established that vesicles of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) can be solubilized spontaneously at the phosphatidylcholine (PC) transition of the ordered gel to the liquid crystalline phase by exchangeable apolipoproteins such as apolipoprotein (apo)A-I, apoE and insect apolipophorin III (apoLp-III) [7–9]. ApoLp-III has been used as a model apolipoprotein for structural and functional analysis [10]. This well studied apolipoprotein is relatively small (18 kDa), composed of a bundle of amphipathic α-helices, and is monomeric in solution [11]. X-ray crystal and NMR solution structures are available for the apolipoprotein from two different species [12–14]. The structure of apoLp-III is similar to that of human apoA-I and apoE, showing all bundles of amphipathic α-helices [15]. ApoLp-III is a more simple protein with a single domain, while apoA-I and apoE are each composed of two domains, probably a reflection of a more complex function in vertebrate lipid transport processes [15–18]. Because of its simplicity, apoLp-III is an excellent protein to study the lipid binding properties of apolipoproteins. While it has often been used for preparing nanodisks, a systematic study of the formation of these increasingly popular particles is still lacking. This in contrast to human apoA-I for which this process is known in more detail by the work of Jonas, Pownall, and others [1,7,19]. The present study aims to investigate the factors that affect the transformation of DMPC vesicles into nanodisks. This process, also known as vesicle solubilization, was characterized in detail for apoLp-III from Locusta migratoria, and included the susceptibility to proteases, the effect of phospholipid acyl chain length, and inclusion of cholesterol, sphingomyelin and diacylglycerol in DMPC vesicles. The results show that apoLp-III-induced vesicle solubilization is similar to that observed for apoA-I, indicating that a bundle of amphipathic helices is an attractive protein scaffold to engage in lipid binding activity through insertion of amphipathic α-helices in the PC vesicle bilayer.
2. Materials and methods
2.1. Recombinant protein
ApoLp-III was obtained from a well established recombinant expression system [20]. Proteins expressed in E. coli escaped the bacteria and accumulated in the culture medium from which they were purified by G75 gel filtration chromatography and reversed-phase HPLC (Beckman System Gold) using a semi-preparative Zorbax 300SB column (Agilent Technologies, Santa Clara, CA) which typically yielded 50 mg protein per liter culture medium. The purity of each apoLp-III preparation was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using precast 4–20% gels (Invitrogen, Carlsbad, CA). Proteins were freeze-dried and stored at −80 °C until further use. While the native protein is glycosylated, the recombinant protein does not contain any N-linked carbohydrate [21]. This has no significant effect on the apoLp-III structure as the NMR solution structure of recombinant apoLp-III and the X-ray structure of native (glycosylated) apoLp-III produced very similar high resolution structures [12, 14].
2.2. Isothermal titration calorimetry
1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was dissolved in 2:1 chloroform methanol (v:v) with gentle vortexing. The sample was dried under a stream of argon gas (Praxair, Canada) and placed in vacuum (100 mM Hg) for 12 hours. The lipid film was rehydrated at 40°C in Tris buffer (20 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, pH 7.2) with vortexing and bath sonication. The lipid vesicles underwent extrusion through a 0.1 µM polycarbonate membrane 21 times (Avanti Polar Lipids, Alabaster, AL). The resulting large unilamellar vesicles (LUV) were analyzed using dynamic light scattering (Wyatt Technology Corporation, Santa Barbara, CA) to test for radius and the concentration was determined using the Ames phosphate test [22]. ApoLp-III was dissolved in Tris buffer with the concentration determined using the extinction coefficient at 280 nm. Isothermal titration calorimetry (ITC) measurements were performed with a MicroCal ITC200 titration calorimeter (GE Healthcare, Northhampton, MA). The syringe was loaded with 24.45 mM DMPC (40µL) and the sample cell was filled with a 75 µM apoLp-III solution (200 µL). Tris buffer was used in the reference cell (200 µL). The experiment was performed at 24°C with 20 injections spaced 150 seconds apart in high feedback mode. A pre titration delay of 10 minutes with a reference power of 5 µcal baseline and a filter period of 4 seconds was used. The first injection was 0.2 µL for a duration of 0.4 seconds. The remaining 19 injections were 2 µL for 4 seconds. The data was analyzed using MicroCal Origin 7 fitting to a 1 binding site model.
2.3. Vesicle solubilization
The following lipids were used in our analysis: 1,2-ditridecanoyl-SN-glycero-3-phosphocholine (C13:0-PC), DMPC, 1,2-dipentadecanoyl-sn-glycero-3-phosphocholine (C15:0-PC), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), cholesterol, egg yolk sphingomyelin and 1,2-dimyristoyl-sn-glycerol (DG) and were purchased from Avanti Polar Lipids, Alabaster, AL. Dried pure lipids or their mixtures were rehydrated in phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) at 10 °C above the lipid phase transition temperature, and vigorously vortexed to obtain multilamellar vesicles (MLV). To obtain LUVs, MLVs were extruded fifteen times using various sizes of membranes (Whatman nucleopore track-etch) in the mini-extruder (Avanti Polar Lipids, Alabaster, AL). To measure the vesicle solubilization rate, MLVs or LUVs were incubated at their respective phase transition temperatures. ApoLp-III, equilibrated at the same temperature, was added to the vesicle suspension (1 mL total volume), and the change in light scatter was measured in a spectrophotometer (Shimadzu UV-2401PC equipped with a Peltier controlled cell holder) at 325 nm. To measure the interaction of apoLp-III with DPPC bilayer vesicles, 6-carboxyfluorescein (200 mM, Fluka Biochimica) was trapped into DPPC vesicles by extrusion through 200 nm membranes. Vesicles were separated from free carboxyfluorescein using a Sephadex G-75 column (75 × 2.5 cm) and elution with PBS. Release of the DPPC encapsulated dye was measured in a Fluoromax-2 fluorometer using a 1:1 protein to lipid mass ratio with an excitation wavelength of 480 nm and an emission wavelength of 520 nm.
2.4. Characterization of nanodisks
The resulting nanodisks from LUV-apoLp-III incubations were isolated by FPLC (AKTA purifier, GE healthcare, Waukesha, WI), using a Superdex 200 10/300 GL size exclusion column. PBS was used as the elution buffer with a flow rate of 0.7 mL/min. The nanodisk fraction was collected, and concentrated using 10K spin column concentrators (Microcep, Pall Corp, Ann Arbor, IL). The size of the nanodisks was estimated by native PAGE using 4–20% Tris-glycine pre-cast gels (Invitrogen, Carlsbad, CA). Gels were run for 10 hours at 125 V, and stained with naphthol blue black (Sigma-Aldrich, St. Louis, MO). Transmission electron microscopy (TEM) was used to visualize the nanodisk preparations. Ten µl of a nanodisk solution (containing 28 µg phospholipid and 10 µg apoLp-III) was absorbed to a hydrophilized 300-mesh carbon coated copper formvar film (EM Science), and rinsed once with PBS. Samples were negatively stained for 15 s with 5 µl 2% sodium phosphotungstate, pH 7.4. Electron micrographs were obtained with a Joel JEM 1200 EX-II electron microscope operated at 100 kV. About 75 nanodisks were measured from a preselected grid to determine their dimension.
2.5. Proteolysis analysis
Nanodisks were isolated by size-exclusion chromatography (Superdex-200, GE), and incubated with trypsin (gold mass spec grade, Promega), Endoproteinase Glu-C (sequencing grade, Roche Applied Science), chymotrypsin (sequencing grade, Promega), or elastase (human leukocytes, Sigma-Aldrich). Trypsin incubations were carried out in 50 mM NH4HCO3, pH 8.2; endoproteinase Glu-C in 25 mM NH4HCO3, pH 7.8; chymotrypsin in 100 mM Tris-HCl, 10 mM CaCl2, pH 7.8; and elastase in 50 mM NaH2PO4, pH 6.5. The digests were separated by 16% Tris-tricine SDS-PAGE (Bio-Rad Laboratories). Protein bands in endoproteinase Glu-C digests were blotted to nitrocellulose and N-terminal sequence analysis was carried out at the UC Davis Proteomics Core Facility, providing two sequences: DAAGHV and AEKHQG. The size of the fragments was determined by MALDI-TOF (Applied Biosystems 4800) using sinapinic acid as the matrix (IIRMES facility, CSU Long Beach).
3. Results
3.1. Temperature dependence of DMPC vesicle solubilization
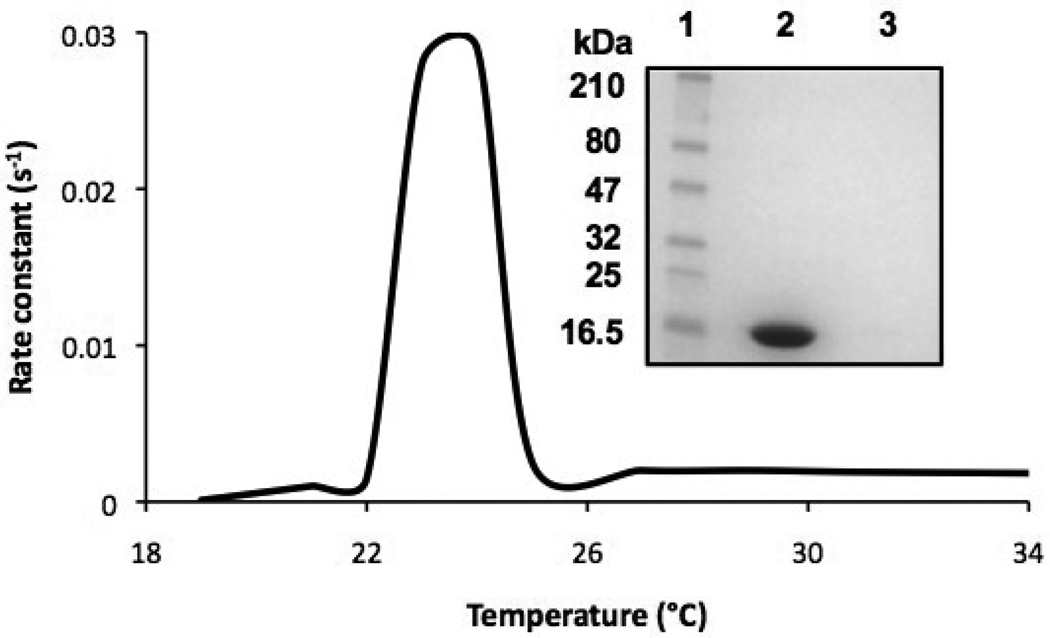
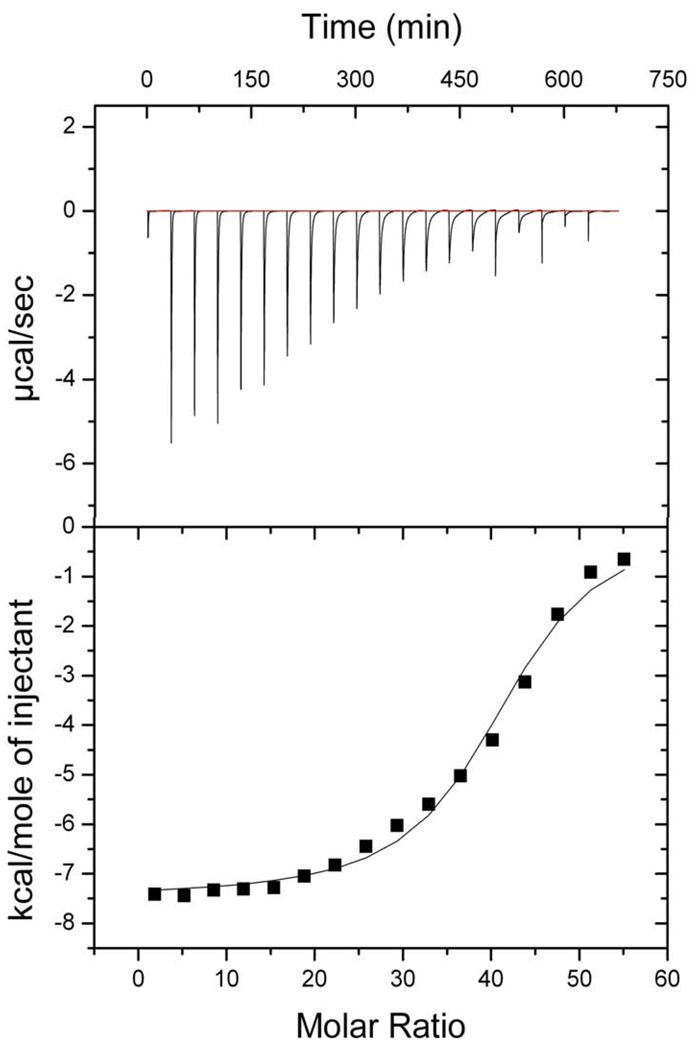
The apolipoprotein-induced spontaneous transformation of DMPC vesicles into nanodisks is a temperature dependent process [7]. ApoLp-III was able to induce the transformation of DMPC vesicles relatively fast when carried out at the narrow temperature range between 23 and 24 °C. The transformation was measured spectrophotometrically at 325 nm and the solubilization of the phospholipid vesicles is evident by a decrease in vesicle light scatter intensity. No significant vesicle solubilization rate was observed at or below 22 or above 25 °C (Figure 1). Since the sample turbidity remained steady outside this narrow temperature range, this raised the question whether apoLp-III associated with the vesicle surface with no transformation into nanodisks. Therefore, apoLp-III and DMPC LUVs were incubated at 4 °C, followed by the separation of LUVs from lipid-free protein by size-exclusion chromatography. The lipid fraction was analyzed by SDS-PAGE and did not contain measurable amounts of protein (Figure 1B), indicating a lack of a stable binding interaction of apoLp-III with the LUVs at 4 °C. In addition, the parameters of the apoLp-III interaction with DMPC SUVs were determined by ITC. The total process was exothermic, with a ΔH of −7.6 • 103 kcal/mol and a ΔS of −5.52 cal/mol/deg at 24 °C (Figure 2). The equilibrium constant of dissociation (KD) was estimated at 4.4 • 10−5 M, however this value does not only result from the binding interaction but also includes the conversion of vesicles into nanodisks. No measurable binding parameters of apoLp-III to DMPC vesicles could be obtained at 4 or at 37 °C, a result similar to the chromatography analysis.
Fig. 1.
Temperature dependence of DMPC solubilization. DMPC vesicles were incubated with apoLp-III at the indicated temperatures, and from the decrease in sample absorbance the initial rate constants were obtained. Inset: SDS-PAGE analysis of DMPC/apoLp-III incubation at 37 °C; lane 1: marker; lane 2: lipid-free fraction, lane 3: lipid-fraction.
Fig. 2.
ITC binding analysis of apoLp-III to DMPC vesicles at 24 °C. Each time, two µL of DMPC (24.45 µM) DMPC was injected into a solution of 200 µL apoLp-III (75 µM) with 150 second time intervals. Top: titration profile of the raw data in which each peak represents the heat released by a single injection after baseline subtraction. Bottom: binding isotherm generated from the integrated heat for each injection and plotted against the molar ratio of lipid to protein.
3.2. Susceptibility to proteases
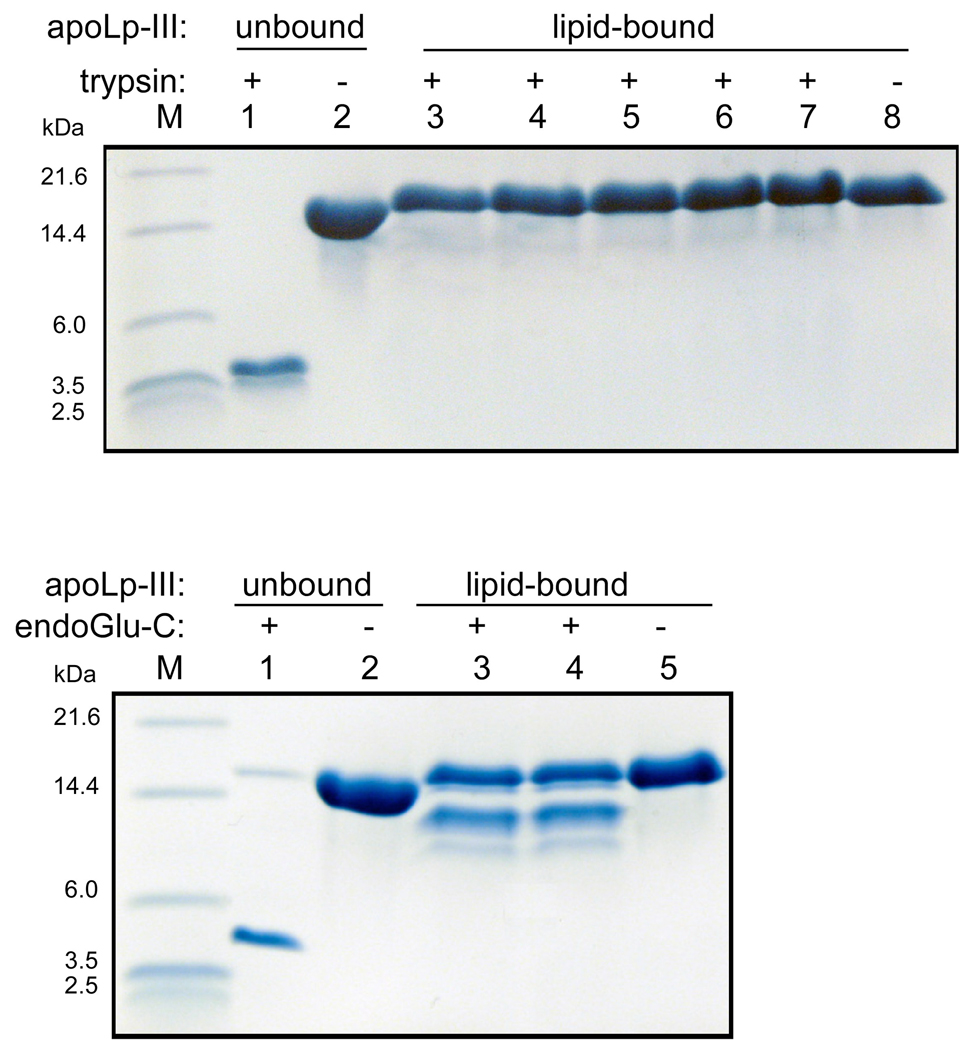
Previous guanidine-HCl denaturation analysis has shown that the midpoint of locust apoLp-III denaturation increased from 0.6 to approximately 3 M, indicating that DMPC nanodisks are exceptionally stable particles [11]. This led us to explore the susceptibility of the nanodisks to protease activity. Four different proteases with various specificities to digest apoLp-III in the lipid-free form and in nanodisks were employed: trypsin, endoproteinase Glu-C (V8 protease), chymotrypsin, and elastase. In this manner the susceptibility to hydrolysis at positively and negatively charged, hydrophobic, and aromatic amino acid residues was analyzed. In the lipid-free form, trypsin fully cleaved apoLp-III into small fragments (enzyme:protein mass ratio of 1:1000); Tris-tricine gels showed the larger products of 4.9 and 4.2 kDa which is close to the expected sizes of 5,341 and 4,275 Da based on the amino acid sequence (Figure 3). However, the protein was completely protected from proteolysis when bound to DMPC, even after a 10- fold increase in the enzyme to protein ratio. Addition of endoproteinase Glu-C to lipid-free apoLp-III also led to a complete digestion, with the largest visible fragment of 5.2 kDa (expected size 5,791 kDa). While DMPC-bound apoLp-III was significantly protected, two protein bands appeared in the 10–14 kDa region. MALDI-TOF and N-terminal amino acid sequence analysis showed that partial hydrolysis at Glu-63 was responsible for the appearance of the 10.6 kDa fragment, while the larger fragment was likely a result of hydrolysis at Glu-126. Nevertheless, the other 11 Glu residues were not susceptible for endoproteinase Glu-C hydrolysis when the protein was bound to DMPC. Chymotrypsin and elastase completely hydrolyzed apoLp-III, and did not produce discrete band on the gel as the peptides were too small to be resolved by Tris-tricine SDS-PAGE (results not shown). On the other hand, incubation of DMPC-bound apoLp-III with these two enzymes did not result in hydrolysis, as the intensity of the intact protein remained unchanged. This result shows that the protein is considerably protected from proteases when in complex with DMPC.
Fig. 3.
Proteolysis of lipid-free and lipid-bound apoLp-III (DMPC nanodisks). Top panel: trypsin incubations of apoLp-III. Lane 1: apoLp-III + trypsin (1:1000 mass ratio); lane 2 apoLp-III. Lanes 3 to 8 contain lipid-bound apoLp-III incubated with trypsin with apoLp-III:trypsin mass ratios of 1:100 (lane 3), 1:200 (lane 4), 1:400 (lane 5), 1:500 (lane 6), 1:1000 (lane 7), or no trypsin (lane 8). Bottom panel shows apoLp-III incubated with endoGlu-C. Lane 1 and 2: apoLp-III in the presence (lane 1) or absence (lane 2) of endoGlu-C (1:100), lane 3–5: lipid-bound apoLp-III with endoGlu-C 1:75 (lane 3), 1:100 (lane 4), and no protease (lane 5).
3.3. Vesicle parameters
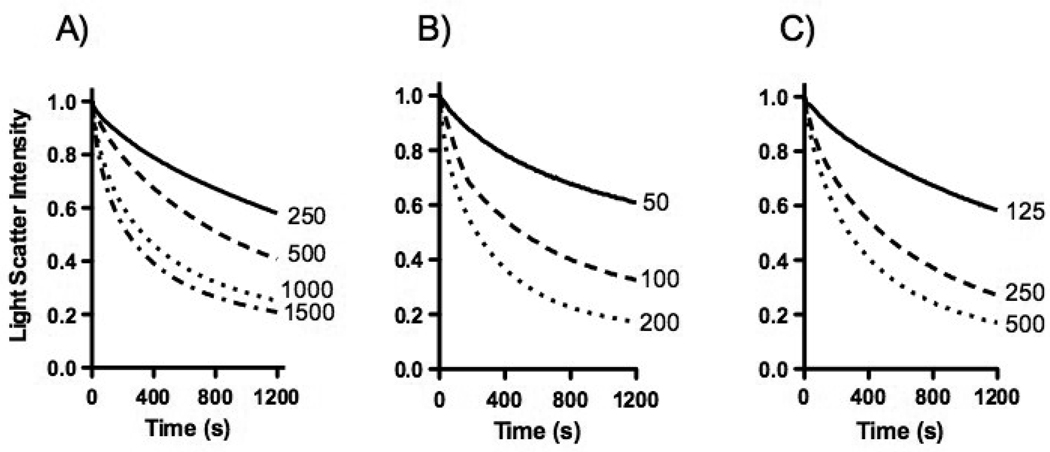
Both MLVs and LUVs were used as the starting point for nanodisk preparations. MLVs were made by vigorously mixing a dried lipid film with buffer, while for LUVs the MLVs were extruded through membranes with narrow pores. LUVs were converted into nanodisks at a faster rate compared to MLVs (not shown). Increasing the protein to lipid ratio, from 1:2 to 3:1 (w/w) resulted in significantly faster solubilization rates (Figure 4A). Next, the solubilization rate of LUVs with different sizes was measured. LUVs prepared by extrusion through 50 nm membranes were converted at a relatively slow rate, while the rate progressively increased using vesicles prepared with 100 nm or 200 nm filters (Figure 4B). Increased concentrations of protein and lipid, maintaining their 2:1 protein to lipid mass ratio, also resulted in substantial faster rates (Figure 4C). Based on these experiments, 500 µg of unilamellar phospholipid vesicles with a diameter of 200 nm mixed with 1000 µg of apoLp-III in a 1 mL total volume was used for subsequent LUV solubilization analysis. The use of these parameters ensures reasonably fast and reproducible solubilization rates.
Fig. 4.
DMPC solubilization. ApoLp-III was mixed with DMPC vesicles and the decrease in sample absorbance monitored for the times indicated. Panel A: 500 µg of DMPC was incubated with 250, 500, 1000 or 1500 µg apoLp-III. Panel B: apoLp-III incubated with 500 µg of DMPC vesicles with a diameter of 50, 100 or 200 nm. Panel C: 125, 250, or 500 µg DMPC incubated with apoLp-III at a 1:2 lipid to protein ratio (mass).
3.4. Effect of phosphatidylcholine acyl chain length
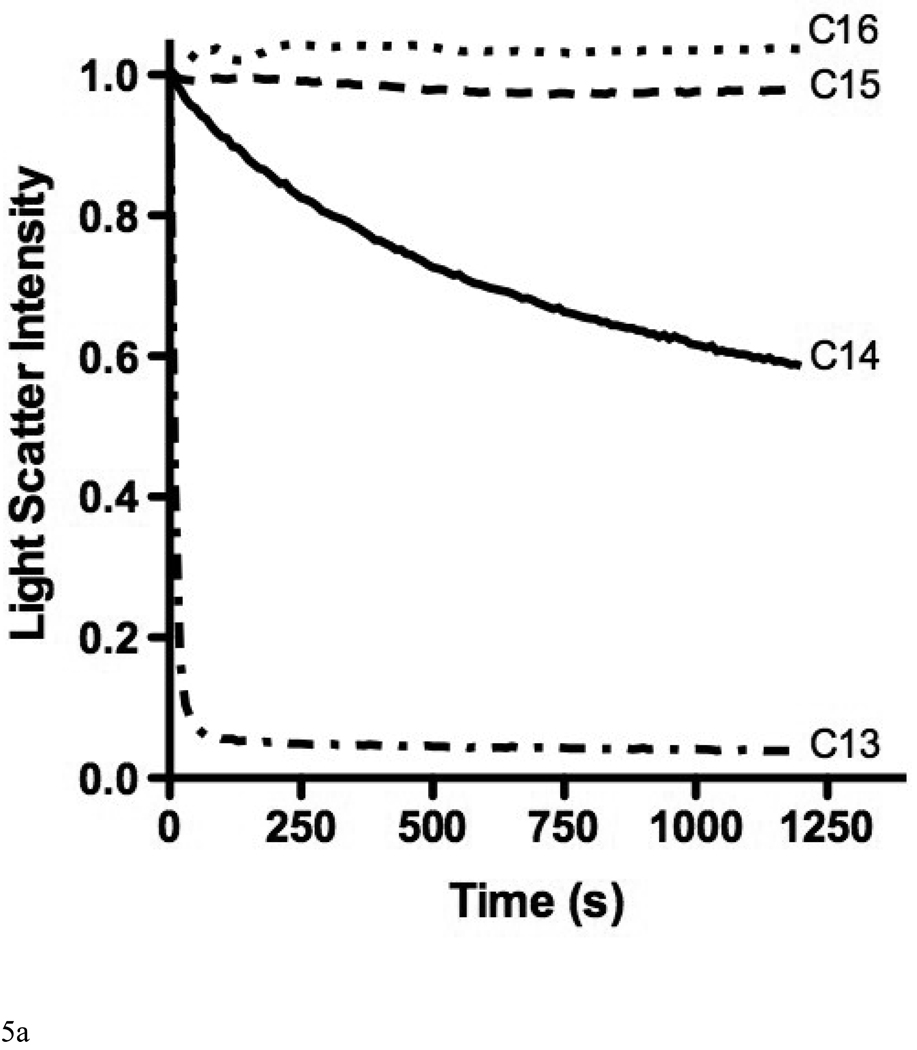
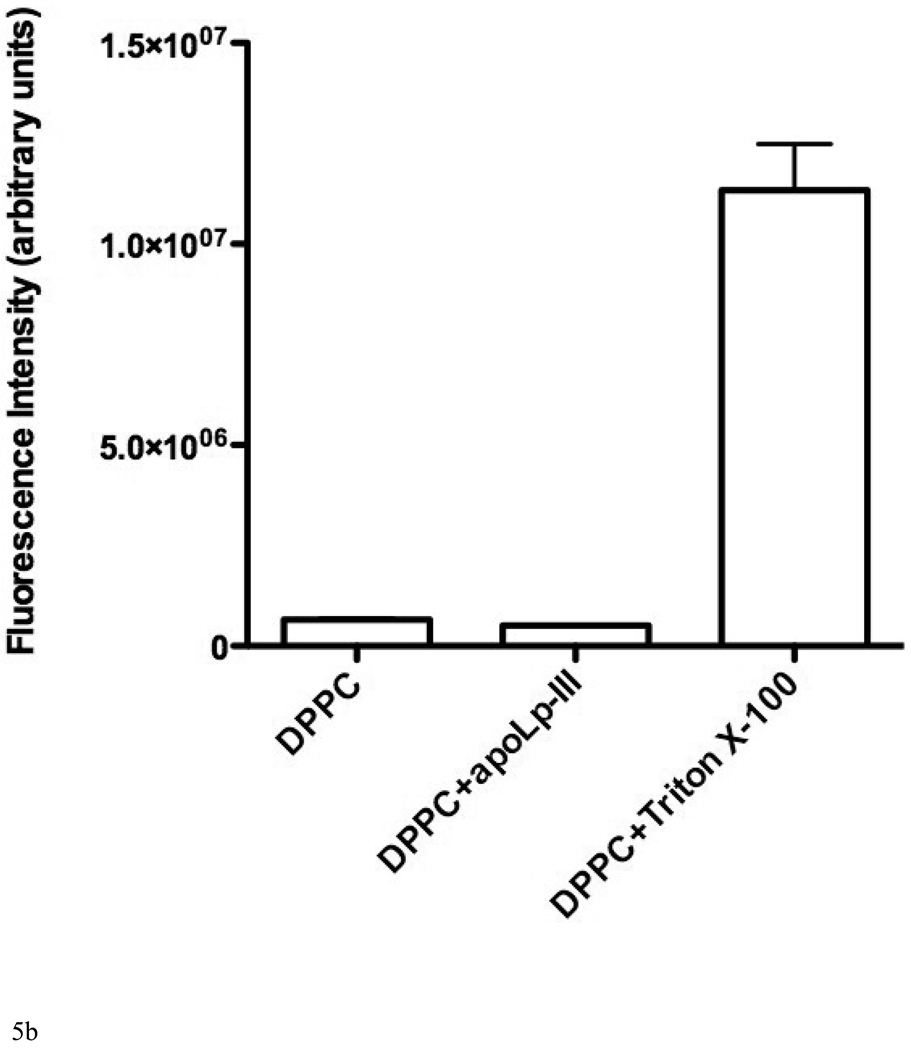
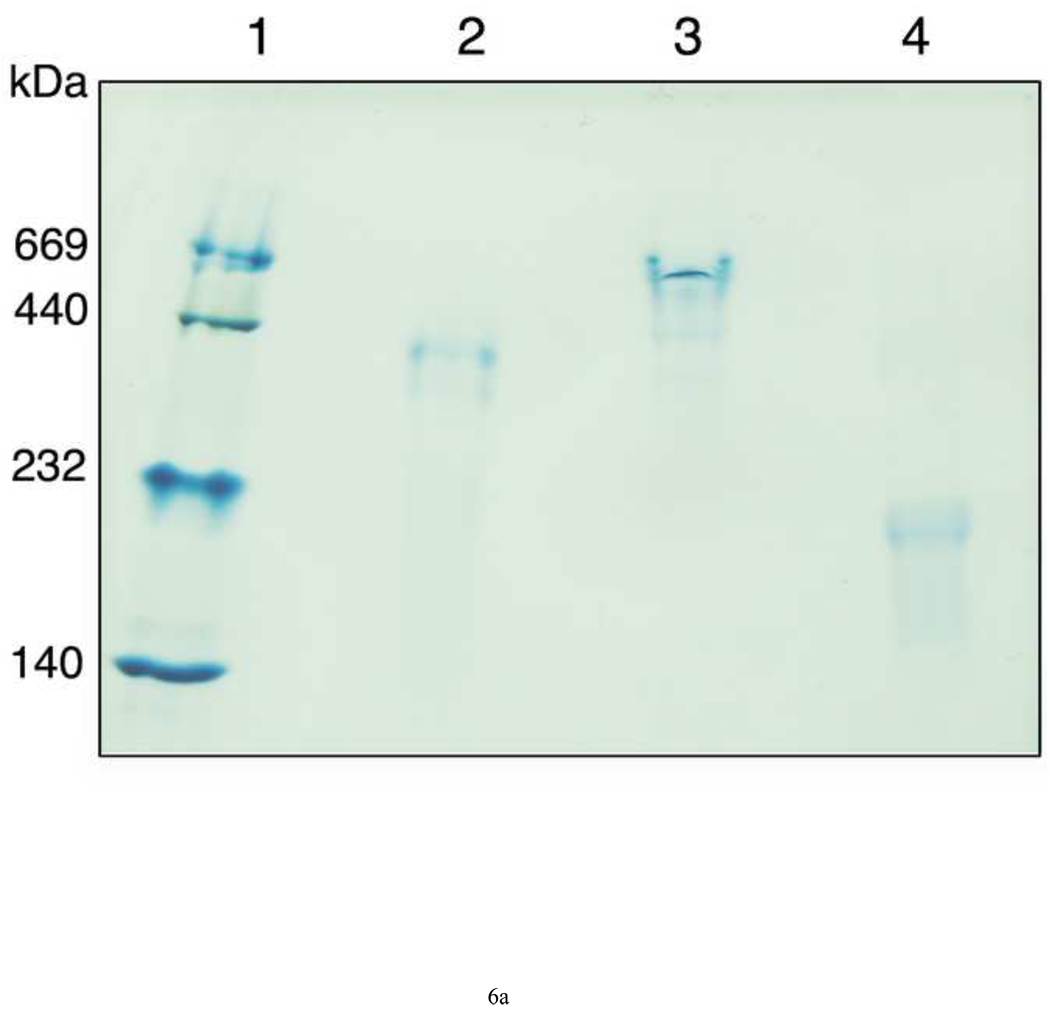
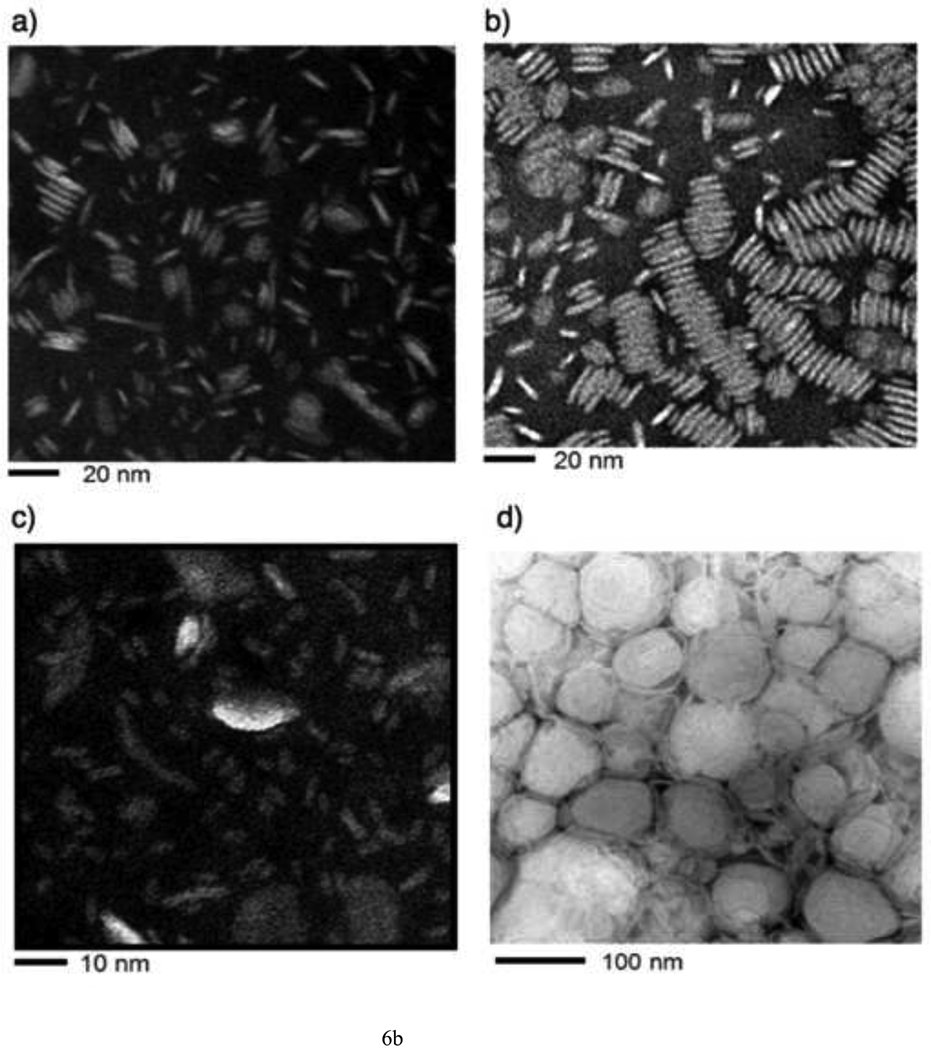
The possibility to employ DPPC LUVs to measure their rate of solubilization was explored as acyl chains with 16 carbons are abundantly found in lipoprotein phospholipids. However, apoLp-III was unable to induce a decrease in sample turbidity of a suspension of DPPC LUVs at the phase transition temperature (Figure 5A). In addition, fluorescein encapsulated in DPPC vesicles could not be released by the addition of apoLp-III, indicating a lack of solubilization (Figure 5B). The inability of apoLp-III to solubilize DPPC vesicles at the phase transition of DPPC (41 °C), is likely not due to protein unfolding as the midpoint of temperature denaturation for apoLp-III is 53 °C [23]. Decreasing the acyl chain length by one carbon (using dipentadecanoyl phosphatidylcholine, phase transition temperature 31 °C) did not result in vesicle solubilization. On the other hand, PC vesicles bearing two saturated acyl chains of 13 carbons (ditridecanoylphosphatidylcholine) were solubilized at a strongly enhanced rate compared to DMPC. The decrease in sample turbidity of C13:0-PC LUVs was accompanied by the formation of discoidal complexes, as shown by native PAGE analysis (Figure 6A). The C13:0-PC nanodisks were substantially smaller compared to that of DMPC, with an apparent molecular mass of 250 kDa compared to 450 kDa for DMPC complexes. Transmission electron microscopic analysis showed a diameter of 11.7 ± 3.1 nm for C13:0-PC and 18.5 ± 5.6 nm for DMPC nanodisks (Figure 6B).
Fig. 5.
A: The effect of acyl chain length of phospholipid vesicles on the solubilization rate. Incubations were carried out at the lipid phase transition temperature. Shown are C13:0-PC, DMPC (C14), C15:0-PC, and DPPC (C16). B: release of carboxyfluorescein from DPPC vesicles after addition of apoLp-III or Triton X-100.
Fig. 6.
A: Native PAGE analysis of nanodisks. Lane 1: marker proteins; lane 2: DMPC nanodisks; lane 3: DMPC/cholesterol 90:10 nanodisks; lane 4: C13:0-PC nanodisks. B: Transmission electron microscopy of DMPC (a), DMPC/cholesterol 90:10 nanodisks (b), C13:0-PC nanodisks (c), and vesicles (d).
3.5. Effect of cholesterol, DG and SM
Cholesterol is abundantly present in HDL [24]. Since nascent HDL has a structure similar to that of apoLp-III nanodisks, the effect of cholesterol in LUVs on the vesicle solubilization rate was investigated. Inclusion of 5 or 10 % of cholesterol (mole %) in DMPC LUVs had a dramatic effect and increased the solubilization rate by 4- and 6-fold, respectively (Figure 7). In contrast, cholesterol contents of 20% and higher prevented the solubilization of LUVs. TEM analysis showed that inclusion of cholesterol in DMPC produced slightly larger nanodisks (diameter of 18.5 ± 5.6 nm versus 22.6 ± 3.1 nm). The positive effect of cholesterol for DMPC LUV solubilization led us to explore the possibility to convert cholesterol enriched DPPC LUVs. However, inclusion of cholesterol in DPPC LUVs did not result in a measurable decrease in sample turbidity, indicating that no significant amounts of nanodisks were formed. Since an increase in DG content in insect lipoproteins is the trigger for apoLp-III binding in vivo, it was tested if DG was able to enhance DMPC solubility rates similar to cholesterol. In addition, SM was used as this is a major HDL lipid component and increased binding was observed in DMPC/SM LUV mixtures (40:60% mol) using differential scanning calorimetric analysis [25]. Surprisingly, addition of neither DG nor SM did result in increased solubility rates. Instead, the ability to form nanodisks was significantly hampered in these lipid mixtures (not shown).
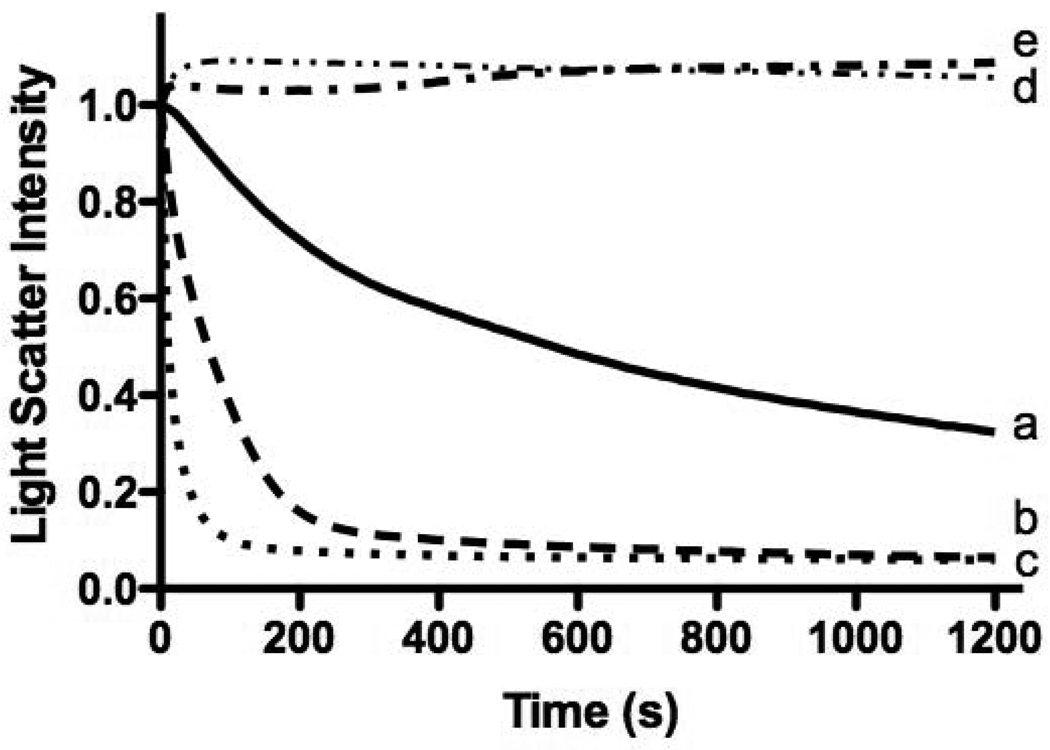
Fig. 7.
Effect of cholesterol on the DMPC vesicles solubilization rate. Shown are pure DMPC (a, solid line), and DMPC vesicles enriched with 5% (b, dashed line) 10% (c, dotted line), 20% (d, dash-dotted line) and 30% (e, double dot-dashed line) cholesterol.
While isolating the lipid fraction of DPPC LUV/apoLp-III incubations to verify the presence of apoLp-III on the vesicle surface, SDS-PAGE analysis indicated the presence of small amounts of apoLp-III. Subsequent TEM analysis surprisingly showed the presence of nanodisks. This prompted us to investigate the ability to solubilize DPPC vesicles in more detail. To detect apoLp-III in the LUV fraction, the concentration of protein and lipid was increased to 10 mg/mL and incubated for several hours. This 10-fold increase in protein and lipid concentration may have driven the formation of nanodisks, and subsequent experiments were carried out using this high concentration. Inclusion of SM (60%) into DPPC vesicles, either LUV or MLV, significantly improved the yield of nanodisks. Of the total amount of protein used, 1.76 ± 0.01 % protein was recovered in the lipid fraction of DPPC LUV incubations. The recovery was improved to 10.50 ± 1.09 % (n=3) when using the SM/DPPC vesicles. The resulting SM/DPPC disks were substantially smaller compared to pure DPPC nanodisks as indicated by native PAGE analysis (~450 kDa for the mixture and ~ 700 kDa for pure DPPC nanodisks, results not shown). Based on the sample turbidity measurements, more than 80% of the vesicles were not converted into nanodisks. Nevertheless, these results demonstrate that apoLp-III is able to solubilize DPPC vesicles spontaneously albeit at a very low yield, and that inclusion of SM improved their formation.
4. Discussion
The ability to form nanodisks has been used extensively in studies investigating the lipid binding properties of apolipoproteins, in particular apoA-I, apoE and apoLp-III [26]. DMPC is often used to produce nanodisks as this synthetic PC is commercially available and inexpensive, a detergent (such as sodium-cholate) is not required, and the rate of LUV solubilization, and thus formation of nanodisks can be readily determined spectrophotometrically at the DMPC phase transition temperature of 24°C. Conversely, a fluorometer can be used when the excitation and emission wavelength are set at identical values to measure right angle light scatter. To follow the conversion of DMPC vesicles into nanodisks, the cuvette temperature needs to be maintained at a constant temperature of 24 ± 0.2 °C as small changes in temperature can lead to a substantial change in solubilization rate, and therefore a peltier controlled cuvette holder or a refrigerated circulating water bath is highly recommended.
The most dramatic effects in apolipoprotein-lipid binding interaction using DMPC liposomes have been observed in apolipoproteins in which the helices have been tethered by disulfide bonds after introduction of closely spaced cysteine residues. The oxidized protein is then maintained in a permanently closed state, unable to undergo the transition to an open conformation. Many of these mutants lost their ability to solubilize DMPC vesicles, however, following cystine reduction, the ability to solubilize the vesicles was restored [27–30]. More subtle changes in lipid binding can also be measured, for which an accurate determination of the rate of solubilization is necessary. Measuring DMPC solubilization rates has been used successfully to demonstrate a molten globule-like state of the apolipoprotein at low pH which facilitates lipid binding [31–33], establish an inverse correlation between lipid binding and apolipoprotein stability for apoE and apoLp-III [17, 23, 34], and used for the lipid binding analysis of truncation mutants of apoA-I and apoA-V [35–37].
For accurate measurement of the solubilization rate, LUVs prepared by extrusion are preferred over MLVs as it is straightforward to produce vesicles with a homogeneous diameter by extrusion, limiting batch to batch variation and the need for vesicle fractionation by size exclusion chromatography to obtain the desired diameter. As shown in the present study, vesicle diameter was inversely correlated with the vesicle solubilization rate. However, one would expect that the increased curvature enhances the strain of the phospholipid bilayers, rendering the bilayers more susceptible for apolipoprotein insertion as reported earlier for the interaction of apoA-I with DPPC vesicles [38]. According to the present data, a protein to lipid ratio of 2:1 (w/w), using 1 mg protein and 0.5 mg of 200 nm LUVs in a total volume of 1 mL, provided relatively quick and reproducible results. Using these conditions approximately 90% of the vesicle population was converted into nanodisks within 30 min. Human apolipoproteins may require other protein to lipid ratios as the intrinsic ability to interact with the phospholipid bilayers may be different. Too much protein may result in extensive solubilization during mixing of LUVs with the protein sample, and initial rate constants will be difficult to obtain. In a scenario where the apolipoprotein is unable to solubilize vesicles at a sufficient rate, cholesterol can be incorporated into the bilayer vesicles. While apoLp-III is monomeric in solution, other apolipoproteins are often present in a multimeric state and this decreases the solubilization rate [39]. Addition of low amounts of guanidine HCl has been used to minimize such protein-protein interactions [9].
Comparison of binding parameters between L. migratoria apoLp-III and other previously studied apolipoproteins is complex, as calorimetry has not been used before to separate the energy from lipid binding and nanodisk formation. ApoA-I binding to pure 1-palmitoyl-2-oleoylphosphatidylcholine mixtures with cholesterol was calculated assuming minimal lipid vesicle perturbation in conditions that did not produce nanodisks [40]. Our data correlates with ΔH values derived for many different plasma apolipoproteins to DMPC whereby the majority reveal a dominant negative enthalpy including apoA-I, apoA-II, apoC-III and apoE [40, 41]. Hence it is likely that the binding of apoLp-III to vesicles is exothermic, however more thermodynamic information about nanodisk formation is required to confirm this. The large negative enthalpy suggests an increase in bonding which includes protein-vesicle and intermolecular protein interactions, as well as van der Waals interactions [40]. ApoLp-III may partition into the DMPC vesicles increasing the van der Waals interactions between hydrophobic faces of the α-helices and the hydrophobic areas of the bilayers yielding a negative ΔH. Such interactions may reduce the lipid mobility and the corresponding reduction in entropy would correlate with our calorimetric finding and the nanodisk model [40].
Boundaries between phospholipid molecules in the gel and liquid state, which occur in the vicinity of the phase transition temperature, likely promote the ability of apoLp-III to solubilize the vesicles, similar as observed for apoA-I [42]. At the phase transition temperature maximum ion permeability [43] and flip-flop [44] have been observed. It appears that pores that develop at such lattice defects form a suitable surface for insertion of the α-helical segments of apolipoproteins, subsequently reorganizing the bilayers vesicles into discoidal particles in a 2nd step. During this binding event, apoLp-III undergoes a dramatic reorganization of its α-helices, switching from a closed helix bundle to an open conformation in which the protein is fully stretched and circumvents the periphery of the discoidal particle [45–46]. This organization is strikingly similar to models proposed for apoA-I and apoE [4–5]. With a diameter of 17 nm, it was calculated that the particle contains four molecules of apoLp-III and ~800 molecules of phospholipid [45]. In addition, nanodisks with other dimensions have been generated by varying the DMPC to apoLp-III ratio, resulting in a different stoichiomtry [47]. A population of nanodisks was generated with a 10.7 nm diameter which contained two apoLp-III molecules. The C13-PC nanodisks produced in our study were similar in diameter which would accommodate approximately two apoLp-III molecules on the disk periphery.
No stable binding interaction of apoLp-III with DMPC was observed below or above the DMPC phase transition temperature. Thus, phospholipids in either an ordered gel or liquid phase are not suitable platforms for apoLp-III binding, emphasizing the need for coexisting phases and the resulting boundaries. At the phase transition temperature, fluctuations of phospholipid clusters between the gel and liquid states result in localized volume changes, causing lattice defects [48]. Additional methylene groups had a strong stabilizing effect on the vesicles: an increase in phospholipid acyl chain length from 14 to 15 or 16 carbons nearly abolished the ability of apoLp-III to spontaneously transform the vesicles into nanodisks. Only at very high lipid and protein concentrations (in the order of 10 to 20 mg mL−1) low amounts of nanodisks were formed with DPPC. This result is similar to apoA-I in which the spontaneous solubilization of DPPC appears to be much slower compared to DMPC with an equilibrium time in the order of days [49]. The tight packing of phosphatidylcholine with acyl chains lengths of 15 or larger in bilayer vesicles results in increased van der Waals interactions, which form a barrier more difficult to overcome and thus minimizes the apolipoprotein-mediated solubilization. On the other hand, C13:0-PC was solubilized about 20-fold faster compared to DMPC. In addition, dilauroylphosphatidylcholine vesicles have been shown to be less stable and are poor permeability barriers to ions, and as the acyl chain length exceeds 14 carbons, the membrane becomes more stable and less permeable [43]. Other studies have shown that the coexistence of gel and fluid domains in membranes promote passive permeation of trapped small molecules, and the rate of flip-flop increases with shorter acyl chain length [50–51]. Thus it is conceivable that the degree of lattice defects increases as the acyl chain length decreases, explaining the order of magnitude faster solubilization rates of C13:0-PC vesicles.
Inclusion of SM into DPPC vesicles resulted in higher yields of the nanodisks. This supports our previous finding that the interaction of apoLp-III was strongest in DMPC/SM (40:60 mol/mol) mixtures, and a less optimal DMPC packing led to surface defects promoting solubilization [24]. This is also in agreement with the enhanced formation of apoA-I containing nanodisks using DPPC/SM mixtures with increasing amounts of SM [52]. Surprisingly, DG inclusion did not promote vesicle solubilization, while in vivo the increased DG content in lipophorins elicits the association of apoLp-III with the lipoprotein surface [53]. It is possible that DG inclusion in the PC bilayers may not result in packing defects but form isolated domains within the PC vesicles. In addition, increased DG content significantly reduces the ability to form stable LUVs, and the DG amount required to form packing defects may not be reached. Inclusion of 10 % cholesterol into DMPC bilayers substantially increased the solubility by apoLp-III. This is similar as observed for apoA-I, for which an optimum of 12.5% cholesterol resulted in the fastest solubilization rate [54]. Cholesterol contents of 20% and higher prevented the nanodisk formation. It has been shown that increased cholesterol incorporation into 14:0-PC bilayers resulted in a strong decrease of the main phase transition as shown in DSC endotherms, which completely disappeared at 50% cholesterol [55]. The absence of a phase transition likely explains the lack of interaction of apoLp-III with PC vesicles with high cholesterol contents. The slightly larger size of cholesterol containing apoLp-III nanodisks is consistent with data published for apoA-I in which cholesterol rich vesicles led to an increase in size of the resulting reconstituted HDL particles [56–57]. The maximum solubilization rates with apoA-I are found at the phase transition of DMPC, and the rate can be modulated by increasing the amount of surface defects such as cholesterol [7, 56–57]. The authors conclude that cholesterol drastically broadens the gel and liquid phases and significantly alters the lipid packing, promoting the binding of apoA-I. The insertion of α-helical segments into these lattice defects are considered as the rate-limiting step. Subsequently, protein/lipid complexes bud off from the vesicle surface and form the characteristic nanodisks as outlined in detail in a model proposed by Segall et al. [9].
ApoLp-III shares many similarities with human apoA-I. Both are bundles of amphipathic α-helices with an up-and-down topology, and the proteins rapidly form nanodisks when incubated with phospholipids at the phase transition temperature. When lipid packing defects were introduced this led to significant increases in vesicle solubilization rates and formation of nanodisks. It appears that the helix bundle motif found in several apolipoproteins seems to be well suited for insertion of one or more helical segments into the lipid surface, enabling the transition from a lipid-free to a more stable lipid-bound form of the protein. The nanodisks made of DMPC and apoLp-III are exceptionally stable and can be stored at 4 °C for months [47]. The lipid-bound apolipoprotein is not only more stable in terms of protein folding, but is also significantly more resistant against protease activity as shown in the present study and by others [58]. This can be exploited when using the nanodisks as a drug delivery vehicle.
Research highlights
-
▪
Apolipophorin III spontaneously solubilizes phosphatidylcholine vesicles
-
▪
ITC shows exothermic character of formation of DMPC nanodisks
-
▪
Nanodisks are well protected from proteases
-
▪
Cholesterol and sphingomyelin stimulate nanodisk formation
-
▪
Nanodisk formation is efficient process for PC with acylchains of 14C or shorter
Acknowledgment
This work was supported by grants of the National Institute of Health (R15 HL077135 and SC3GM089564) to PMMW and the Provost Summer Stipend for Research, Scholarly and Creative Activities to SKL. MHC was supported by scholarships from NSERC, AHFMR and CIHR. The authors would like to acknowledge Dr. Ashraf Elamin, IIRMES, for MALDI-TOF analysis.
Abbreviations
- apoA-I
apolipoprotein A-I
- apoLp-III
apolipophorin III
- DG
1,2-dimyristoyl-sn-glycerol
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DPPC
1,2,-dipalmitoyl-snglycero-3-phosphocholine
- HDL
high density lipoprotein
- ITC
isothermal titration calorimetry
- LUV
large unilamellar vesicle
- MLV
multilamellar vesicle
- PBS
phosphate buffered saline
- PC
phosphatidylcholine
- SM
sphingomyelin
- TEM
transmission electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jonas A. Lipid-binding properties of apolipoproteins. In: Rosseneu M, editor. “Structure and Function of apolipoproteins”. CRC Press; 1992. pp. 217–249. [Google Scholar]
- 2.O’Connor PM, Zysow BR, Schoenhaus SA, Ishida BY, Kunitake ST, Naya-Vigne JM, Duchateau PN, Redberg RF, Spencer SJ, Mark S, Mazur M, Heilbron DC, Jaffe RB, Malloy MJ, Kane JP. Prebeta-1 HDL in plasma of normolipidemic individuals: influences of plasma lipoproteins, age, and gender. J. Lipid Res. 1998;39:670–678. [PubMed] [Google Scholar]
- 3.Raussens V, Goormaghtigh E, Narayanaswami V, Ryan RO, Ruysschaert JM. Alignment of apolipophorin III α-helices in complex with dimyristoylphosphatidylcholine: a unique spatial orientation. J. Biol. Chem. 1995;270:12542–12547. doi: 10.1074/jbc.270.21.12542. [DOI] [PubMed] [Google Scholar]
- 4.Narayanaswami V, Maiorano JN, Dhanasekaran P, Ryan RO, Phillips MC, Lund-Katz S, Davidson WS. Helix orientation of the functional domains in apolipoprotein E in discoidal high density lipoprotein particles. J. Biol. Chem. 2004;279:14273–14279. doi: 10.1074/jbc.M313318200. [DOI] [PubMed] [Google Scholar]
- 5.Davidson WS, Thompson TB. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- 6.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 7.Surewicz WK, Epand RM, Pownall HJ, Hui S-W. Human apolipoprotein A-I forms thermally stable complexes with anionic but not zwitterionic phospholipids. J. Biol. Chem. 1986;261:16191–16197. [PubMed] [Google Scholar]
- 8.Wientzek M, Kay CM, Oikawa K, Ryan RO. Binding of insect apolipophorin III to dimyristoylphosphatidylcholine vesicles. Evidence for a conformational change. J. Biol. Chem. 1994;269:4605–4612. [PubMed] [Google Scholar]
- 9.Segall ML, Dhanasekaran P, Baldwin F, Anantharamaiah GM, Weisgraber KH, Phillips MC, Lund-Katz S. Influence of apoE domain structure and polymorphism on the kinetics of phospholipid vesicle solubilization. J. Lipid Res. 2002;43:1688–1700. doi: 10.1194/jlr.m200157-jlr200. [DOI] [PubMed] [Google Scholar]
- 10.Weers PMM, Ryan RO. Apolipophorin III: Role model apolipoprotein. Insect Biochem. Mol. Biol. 2006;36:231–240. doi: 10.1016/j.ibmb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Weers PMM, Kay CM, Oikawa K, Wientzek M, Van der Horst DJ, Ryan RO. Factors affecting the stability and conformation of Locusta migratoria apolipophorin III. Biochemistry. 1994;33:3617–3624. doi: 10.1021/bi00178a019. [DOI] [PubMed] [Google Scholar]
- 12.Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5-Å resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc. Natl. Acad. Sci. USA. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan D, Zheng Y, Yang D, Wang J. NMR solution structure and dynamics of an exchangeable apolipoprotein, Locusta migratoria apolipophorin III. J. Biol. Chem. 2003;278:21212–21220. doi: 10.1074/jbc.M208486200. [DOI] [PubMed] [Google Scholar]
- 15.Narayanaswami V, Kiss RS, Weers PMM. The helix bundle: a reversible lipid binding motif. Comparative Biochemistry Physiology A. Mol. Integr. Physiol. 2010;155:123–133. doi: 10.1016/j.cbpa.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 2004;43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 19.Pownall HJ, Massey JB. Mechanism of association of human plasma apolipoproteins with dimyristoylphosphatidylcholine: effect of lipid clusters on reaction rates. Biophys. J. 1982;37:177–179. doi: 10.1016/S0006-3495(82)84659-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weers PMM, Wang J, Van der Horst DJ, Kay CM, Sykes BD, Ryan RO. Recombinant locust apolipophorin III: characterization and NMR spectroscopy. Biochim. Biophys. Acta. 1998;1393:99–107. doi: 10.1016/s0005-2760(98)00063-0. [DOI] [PubMed] [Google Scholar]
- 21.Hård K, Van Doorn JM, Thomas-Oates JE, Kamerling JP, Van der Horst DJ. Structures of the Asn-linked oligosaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate-linked 2-aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry. 1993;32:766–775. doi: 10.1021/bi00054a005. [DOI] [PubMed] [Google Scholar]
- 22.Ames BN. Assay of inorganic phosphates, total phosphate and phosphatase. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 23.Weers PMM, Cabrera J, Abdullahi EW, Hsu TC. Role of buried polar residues in helix bundle stability and lipid binding of apolipophorin III: destabilization by threonine 31. Biochemistry. 2005;44:8810–8816. doi: 10.1021/bi050502v. [DOI] [PubMed] [Google Scholar]
- 24.Jonas A, Phillips MC. Lipoprotein structure. In: Vance DE, Vance JE, editors. Biochemistry of Lipids. Amsterdam: Elsevier BV; 2008. pp. 485–506. Lipoproteins and Membranes. [Google Scholar]
- 25.Chiu MC, Wan C-PL, Weers PMM, Prenner EJ. Apolipophorin III interaction with model membranes composed of phosphatidylcholine and spingomyelin using differential scanning calorimetry. Biochim. Biophys Acta. 2009;1788:2160–2168. doi: 10.1016/j.bbamem.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chromy BA, Arroyo E, Blanchette CD, Bench G, Benner H, Cappuccio JA, Coleman MA, Henderson PT, Hinz AK, Kuhn EA, Pesavento JB, Segelke BW, Sulchek TA, Tarasow T, Walsworth VL, Hoeprich PD. Different apolipoproteins impact nanolipoprotein particle formation. J. Am. Chem. Soc. 2007;129:14348–14354. doi: 10.1021/ja074753y. [DOI] [PubMed] [Google Scholar]
- 27.Narayanaswami V, Wang J, Kay CM, Scraba DG, Ryan RO. Disulfide bond engineering to monitor conformational opening of apolipophorin III during lipid binding. J. Biol. Chem. 1996;271:26855–26862. doi: 10.1074/jbc.271.43.26855. [DOI] [PubMed] [Google Scholar]
- 28.Lu B, Morrow JA, Weisgraber KH. Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid. J. Biol Chem. 2000;275:20775–20781. doi: 10.1074/jbc.M003508200. [DOI] [PubMed] [Google Scholar]
- 29.Soulages JL, Arrese EL. Interaction of the α-helices of apolipophorin III with phospholipid acyl chains in discoidal lipoprotein particle: a fluorescence quenching study. Biochemistry. 2001;40:14279–14290. doi: 10.1021/bi010949d. [DOI] [PubMed] [Google Scholar]
- 30.Leon LJ, Idangodage H, Wan C-PL, Weers PMM. Apolipophorin III: lipopolysaccharide binding requires helix bundle opening. Biochem. Biophys. Res. Commun. 2006;348:1328–1322. doi: 10.1016/j.bbrc.2006.07.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soulages JL, Bendavid OJ. The lipid binding activity of the exchangeable apolipoprotein apolipophorin-III correlates with the formation of a partially folded conformation. Biochemistry. 1998;37:10203–10210. doi: 10.1021/bi980622l. [DOI] [PubMed] [Google Scholar]
- 32.Weers PMM, Kay CM, Ryan RO. Conformational changes of an exchangeable apolipoprotein, apolipophorin III from Locusta migratoria at low pH: correlation with lipid binding. Biochemistry. 2001;40:7754–7760. doi: 10.1021/bi010410f. [DOI] [PubMed] [Google Scholar]
- 33.Morrow JA, Hatters DM, Lu B, Hochtl P, Oberg KA, Rupp B, Weisgraber KH. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J. Biol. Chem. 2002;277:50380–55038. doi: 10.1074/jbc.M204898200. [DOI] [PubMed] [Google Scholar]
- 34.Weers PMM, Narayanaswami V, Ryan RO. Modulation of the lipid binding properties of the N-terminal domain of human apolipoprotein E3. Eur. J. Biochem. 2001;268:3728–3735. doi: 10.1046/j.1432-1327.2001.02282.x. [DOI] [PubMed] [Google Scholar]
- 35.Ji Y, Jonas A. Properties of an N-terminal proteolytic fragment of apolipoprotein A-I in solution and in reconstituted high density lipoproteins. J. Biol. Chem. 1995;270:11290–11297. doi: 10.1074/jbc.270.19.11290. [DOI] [PubMed] [Google Scholar]
- 36.Beckstead JA, Block BL, Bielicki JK, Kay CM, Oda MN, Ryan RO. Combined N-and C-terminal truncation of human apolipoprotein A-I yields a folded, functional central domain. Biochemistry. 2005;44:4591–4599. doi: 10.1021/bi0477135. [DOI] [PubMed] [Google Scholar]
- 37.Beckstead JA, Wong K, Gupta V, Wan CP, Cook VR, Weinberg RB, Weers PMM, Ryan RO. The C terminus of apolipoprotein A-V modulates lipid-binding activity. J. Biol. Chem. 2007;282:15484–15489. doi: 10.1074/jbc.M611797200. [DOI] [PubMed] [Google Scholar]
- 38.Wetterau JR, Jonas A. Effect of dipalmitoylphosphatidylcholine vesicle curvature on the reaction with human apolipoprotein A-I. J. Biol. Chem. 1982;257:10961–10966. [PubMed] [Google Scholar]
- 39.Pownall HJ, Pao Q, Hickson D, Sparrow JT, Kusserow SK, Massey JB. Kinetics and mechanism of association of human plasma apolipoproteins with dimyristoylphosphatidylcholine: effect of protein structure and lipid clusters on reaction rates. Biochemistry. 1981;20:6630–6635. doi: 10.1021/bi00526a017. [DOI] [PubMed] [Google Scholar]
- 40.Arnulphi C, Jin L, Tricerri MA, Jonas A. Enthalpy-driven apolipoprotein A-I and lipid bilayer interaction indicating protein penetration upon lipid binding. Biochemistry. 2004;43:12258–12264. doi: 10.1021/bi036118k. [DOI] [PubMed] [Google Scholar]
- 41.Rosseneu M. Isothermal calorimetry of apolipoproteins. Methods Enzymol. 1986;128:365–375. doi: 10.1016/0076-6879(86)28080-5. [DOI] [PubMed] [Google Scholar]
- 42.Pownall HJ, Massey JB, Kusserow SK, Gotto AM. Kinetics of lipid-protein interactions: interaction of apolipoprotein A-I from human plasma high density lipoproteins with phosphatidylcholines. Biochemistry. 1978;17:1183–1188. doi: 10.1021/bi00600a008. [DOI] [PubMed] [Google Scholar]
- 43.Blok MC, Van der Neut-Kok ECM, Van Deenen LLM, De Gier J. The effect of chain length and lipid phase transition on the selective permeability properties of liposomes. Biochim. Biophys. Acta. 1975;406:187–196. doi: 10.1016/0005-2736(75)90003-6. [DOI] [PubMed] [Google Scholar]
- 44.John K, Schreiber S, Kubelt J, Hermann A, Muller P. Transbilayer movement of phospholipids at the main phase transition of lipid membranes: implications for rapid flip-flop in biological membranes. Biophys. J. 2002;83:3315–3323. doi: 10.1016/S0006-3495(02)75332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahoo D, Weers PMM, Ryan RO, Narayanaswami V. Lipid triggered molecular switch of apoLp-III helix bundle to an extended helix conformation. J. Mol. Biol. 2002;321:201–214. doi: 10.1016/s0022-2836(02)00618-6. [DOI] [PubMed] [Google Scholar]
- 46.Garda HA, Arrese EL, Soulages JL. Structure of apolipophorin III in discoidal lipoproteins, Interhelical distances in the lipid-bound state and conformational change upon binding to lipid . J. Biol. Chem. 2002;277:19773–19782. doi: 10.1074/jbc.M110089200. [DOI] [PubMed] [Google Scholar]
- 47.Fischer NO, Blanchette CD, Segelke BW, Corzett M, Chromy BA, Kuhn EA, Bench G, Hoeprich PD. Isolation, characterization, and stability of discretely-sized nanolipoprotein particles assembled with apolipophorin-III. PLoS One. 2010. 2010;5:e11643. doi: 10.1371/journal.pone.0011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swaney JB, Chang BC. Thermal dependence of apolipoprotein A-I-phospholipid recombination. Biochemistry. 1980;19:5637–5644. doi: 10.1021/bi00565a028. [DOI] [PubMed] [Google Scholar]
- 49.Jonas A, Mason WR. Interactions of dipalmitoyl- and dimyristoylphosphatidylcholines and their mixtures with apoA-I. Biochemistry. 1981;20:3801–3805. doi: 10.1021/bi00516a020. [DOI] [PubMed] [Google Scholar]
- 50.Clerc SG, Thompson TE. Permeability of dimyristoyl phosphatidylcholine/dipalmitoyl phosphatidylcholine bilayer membranes with coexisting gel and liquid-crystalline phases. Biophys. J. 1995;68:2333–2341. doi: 10.1016/S0006-3495(95)80415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howman R, Powell HJ. Transbilayer diffusion of phospholipids: dependence on headgroup structure and acyl chain length. Biochim. Biophys. Acta. 1988;938:155–156. doi: 10.1016/0005-2736(88)90155-1. [DOI] [PubMed] [Google Scholar]
- 52.Fukuda M, Nakano M, Sriwongsitanont S, Ueno M, Kuroda Y, Handa T. Spontaneous reconstitution of discoidal HDL from sphingomyelin-containing model membranes by apolipoprotein A-I. J. Lipid Res. 2007;48:822–889. doi: 10.1194/jlr.M600495-JLR200. [DOI] [PubMed] [Google Scholar]
- 53.Van der Horst DJ, Van Hoof D, Van Marrewijk WJA, Rodenburg KW. Alternative lipid mobilization: the insect shuttle system. Mol. Cell. Biochem. 2002;239:113–119. [PubMed] [Google Scholar]
- 54.Pownall HJ, Massey JB, Kusserow SK, Gotto AM., Jr Kinetics of lipid-protein interactions: effect of cholesterol on the association of human plasma high-density apolipoprotein A-I with L-alpha-dimyristoylphosphatidylcholine. Biochemistry. 1979;18:574–579. doi: 10.1021/bi00571a004. [DOI] [PubMed] [Google Scholar]
- 55.McMullen TPW, Lewis RNAH, McElhaney RN. Comparative differential scanning calorimetric and FTIR and 31P-NMR spectroscopic studies of the effects of cholesterol and androstenol. Biophys. J. 1994;66:741–752. doi: 10.1016/s0006-3495(94)80850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Massey JB, Pownall HJ. Role of oxysterol structure on the microdomain-induced microsolubilization of phospholipid membranes by apolipoprotein A-I. Biochemistry. 2005;44:14376–14384. doi: 10.1021/bi051169y. [DOI] [PubMed] [Google Scholar]
- 57.Massey JB, Pownall HJ. Cholesterol is a determinant of the structures of discoidal high density lipoproteins formed by the solubilization of phospholipid membranes by apolipoprotein A-I. Biochim. Biophys. Acta. 2008;1781:245–253. doi: 10.1016/j.bbalip.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massey JB, Hickson-Dick DL, Gotto AM, Jr, Pownall HJ. Kinetics of trypsin hydrolysis as a probe of the structure of human plasma apolipoprotein A-II. Biochim. Biophys. Acta. 1989;999:121–127. doi: 10.1016/0167-4838(89)90208-2. [DOI] [PubMed] [Google Scholar]