Abstract
During metazoan development, Wnt molecules are secreted from Wnt-producing cells, diffuse to target cells, and determine cell fates; therefore, Wnt secretion is tightly regulated. However, the molecular mechanisms controlling Wnt diffusion are not fully elucidated. The specific chondroitin sulfate (CS) structure synthesized by chondroitin-4-O-sulfotransferase-1 (C4ST-1) binds to Wnt-3a with high affinity (Nadanaka, S., Ishida, M., Ikegami, M., and Kitagawa, H. (2008) J. Biol. Chem. 283, 27333–27343). In this study we tested whether Wnt signaling regulates sulfation patterns of cell-associated CS chains by suppressing expression of C4ST-1 to trigger release of Wnt molecules from Wnt-producing cells. C4ST-1 expression was dramatically reduced in L cells that stably expressed Wnt-3a (L-Wnt-3a cells) and had CS with low affinity for Wnt-3a. Forced expression of C4ST-1 in L-Wnt-3a cells inhibited diffusion of Wnt-3a due to structural alterations in CS chains mediated by C4ST-1. Furthermore, sustained Wnt signaling negatively regulated C4ST-1 expression in a cell-autonomous and non-cell autonomous fashion. These results demonstrated that C4ST-1 is a key downstream target of Wnt signaling that regulates Wnt diffusion from Wnt-producing cells.
Keywords: Chondroitin Sulfate, Extracellular Matrix, Glycosaminoglycan, Proteoglycan Structure, Signal Transduction, Sulfotransferase, Wnt Pathway
Introduction
Many developmental and disease-related processes are mediated by Wnt proteins secreted by specific Wnt-producing cells to regulate cellular programs in surrounding tissues (1). Upon secretion from Wnt-producing cells, Wnt molecules generate a concentration gradient with positional information that instructs developing cells to adopt particular fates (2), and the formation of Wnt concentration gradients might require freely diffusible proteins that can traverse the extracellular space. However, biochemical analyses have shown that secreted Wnt proteins are hydrophobic and sticky (3, 4). Thus, Wnt proteins are tightly associated with the surface of cells and the extracellular matrix and are unlikely to diffuse freely through the aqueous extracellular environment. Previous studies showed that secreted wingless (Wg),2 the Drosophila homologue of mammalian Wnt-1, could bind to glycosaminoglycan (GAG) moieties of proteoglycans (PGs) with high affinity and that the extracellular diffusion of Wg molecules was restricted by binding to the surface of cells and to extracellular matrix PGs (4). Therefore, we hypothesized that binding of Wnt to GAG chains might control Wnt release from Wnt-producing cells.
GAG is attached as a side chain to PGs found on the cell surface and in the extracellular matrix, and it modulates signals that initiate differentiation during development and those that maintain homeostasis in adults. Previous studies indicate that PGs are critical modulators of wingless, hedgehog, and decapentaplegic morphogenic gradients and FGF signaling (5, 6). GAGs bind to a large variety of proteins on the cell surface and in the extracellular matrix, and the interactions between various signaling molecules and GAG moieties of PGs are one molecular basis for the function of PGs as signaling modulators. In addition, it is thought that the interactions between proteins and GAG chains are regulated by the fine structures of GAGs. Linear sulfated GAGs are classified based on structural units chondroitin sulfate (CS) and heparan sulfate (HS), which are composed of disaccharide units [-GlcUA-GalNAc-]n and [-GlcUA-GlcNAc-]n, respectively. During synthesis of the GAG backbone, sulfotransferases catalyzed various modifications such as sulfation. “Sugar codes” specified by specific sulfation patterns are thought to define specificity and affinity of protein-sugar interactions. Thus, we have been studying a sugar code to understand an intrinsic role for sugar in cell function.
Our previous study showed that Wnt-3a binds to a specific structure containing E-disaccharide units (GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)) with high affinity (7). In addition, CS chains containing E-disaccharide units concentrate Wnt-3a molecules on the surface of Wnt-producing cells and enhance Wnt-3a signaling (7). Furthermore, we demonstrated that chondroitin-4-O-sulfotransferase (C4ST-1) is involved in the biosynthesis of E-disaccharide units (8). Based on these observations, we hypothesized that the level of C4ST-1 controls the E-disaccharide units on the cell surface and, therefore, modulates the association and dissociation of Wnt-3a and the cell surface. In this study, we found that C4ST-1 gene expression is negatively regulated by Wnt signaling. Therefore, we hypothesize that Wnt molecules secreted from the Wnt-producing cells down-regulate C4ST-1 gene expression in an autocrine manner, leading to structural changes in CS chains on the surface of the Wnt-producing cells and trigger the release of Wnt molecules from these cells. Here, we propose a model detailing how Wnt molecules diffuse from Wnt-producing cells.
EXPERIMENTAL PROCEDURES
Materials
Purified recombinant mouse Wnt-3a (carrier-free) was obtained from R&D Systems (Minneapolis, MN). Anti-Wnt-3a antibody (#2391), the anti-β-catenin antibody (#610154), and the monoclonal anti-actin antibody (clone AC-40) (#A3853) were purchased from Cell Signaling Technology (Boston, MA), BD Transduction Laboratories, and Sigma, respectively. Recombinant N-glycosidase F from Escherichia coli (EC 3.5.1.52) and chondroitinase ABC from Proteus vulgaris (EC 4.2.2.4) were purchased from Roche Diagnostics and Seikagaku Corp. (Tokyo, Japan), respectively. Mouse L fibroblasts were kindly provided by Dr. Frank Tufaro (Allera Health Products, Inc., St. Petersburg, FL), and L cells overexpressing Wnt-3a (CRL-2647), C2C12 cells (CRL-1772), human colon carcinoma LoVo cells (CCL-229), WiDr cells (CCL-218), and human hepatocellular carcinoma HepG2 cells (HB-8065) were purchased from American Type Culture Collection (ATCC) (Manassas, VA). RCM-1 (JCRB0256) and human embryonic fibroblasts, OUMS-36 (JCRB1006.0), were purchased from RIKEN BioResource Center Cell Bank (JCRB) (Tsukuba, Japan). HCT-8 (HRT-18) (catalogue no. 86040306) was obtained from the European Collection of Cell Cultures (Salisbury, England). The inhibitor of glycogen synthase kinase-3β, SB216763 (3-(2,4-dichlorophenyl)-4-(1-methyl-1H-indol-3-yl)-1H-pyrrole-2,5-dione), and the inhibitor of Wnt signaling, XAV939 (3,5,7,8-tetrahydro-2-[4-(trifluoromethyl)phenyl]-4H-thiopyrano[4,3-d]pyrimidin-4-one), were purchased from TOCRIS (Ellisville, MO) and Cayman (Ann Arbor, MI), respectively.
Plasmid Construction
Mouse Wnt-3a cDNA was obtained by reverse transcription-coupled polymerase chain reaction using total RNA isolated from L cells and the primers 5′-CCCAAGCTTGCGATGGCTCCTCTCGGATA-3′ and 5′-GGAATTCTTGCAGGTGTGCACGTCATAGACACG-3′. The resultant fragments were inserted into the HindIII and EcoRI sites of p3xFLAG CMV14 (Sigma) to express Wnt-3a protein tagged with the FLAG epitope at the C terminus. pIRESneo3-mWnt-3a-3xFLAG was constructed by inserting the NheI-NotI fragment of a PCR product amplified using p3xFLAG CMV14-mWnt-3a as a template with primers 5′-GCTAGCATGGCTCCTCTCGGATACCT-3′ and 5′-GCGGCCGCCTTGTCATCGTCATCCTTGTA-3′ into the NheI and NotI sites of pIRESneo3 (Clontech).
Cell Culture and Stable Transfection
L-Wnt-3a cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, and 400 μg/ml G418, and maintained in a 5% CO2 incubator at 37 °C. L, C2C12, human embryonic fibroblasts, LoVo, and HepG2 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate, and RCM-1 and HCT-8 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate. All cells were maintained in a 5% CO2 incubator at 37 °C.
The expression plasmid (pcDNA3.1/zeo(−)-mC4ST-1) was transfected into L-Wnt-3a cells using FuGENETM 6 transfection reagent (Roche Diagnostics) according to the manufacturer's instructions. Transfectants were cultured in the presence of 400 μg/ml G418 and 400 μg/ml zeocin (Invitrogen). Zeocin-resistant clones were then picked up and propagated for experiments. C2C12 cells were transfected with pIRESneo3-mWnt-3a-3xFLAG. After selection with 700 μg/ml (clone 1) or 800 μg/ml (clone 2) G418 (Invitrogen), surviving colonies were used for experiments.
Co-culture Conditions of HeLa Cells or Human Embryonic Fibroblasts in the Presence of L Cells or L-Wnt-3a Cells
On day 0, L or L-Wnt-3a cells (2.6 × 104 cells) were inoculated with HeLa cells (2.6 × 104 cells) in a 6-cm dish. On day 5, cells were passaged at a ratio of 1:20. On day 10, total RNA was isolated from the co-cultured cells after cells were washed with PBS. Human embryonic fibroblasts (passage 27) (1 × 104 cells) were inoculated with L or L-Wnt-3a cells (1.0 × 104 cells) in a 3.5-cm dish and cultured for the indicated times without passage. To evaluate the expression level of hC4ST-1 in HeLa cells and human embryonic fibroblasts, real-time PCR was carried out using primer pairs specific for the human C4ST-1 and the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes (see supplemental Fig. S1A); human C4ST-1 forward primer (5′-GTGGGGAGAGGGAGAGAATCATG-3′) and reverse primer (3′-CAAGAACGACCCATCCTATTTCC-3′) and human GAPDH forward primer (5′-TGCTGGGGAGTCCCTGCCACA-3′) and (reverse primer 5′-GGTACATGACAAGGTGCGGCTC-3′).
Conditioned Media
The conditioned media recovered from confluent cultures of L-Wnt-3a cells were centrifuged, sterilized by filtration through 0.2-μm filters, and mixed at 1:2 ratio with normal growth medium before addition to cells.
On day 0, mouse L-Wnt-3a cells and L-Wnt-3a-C4ST-1 transfectants were seeded at a density of 0.5 × 106 cells per 3.5-cm dish and cultured in DMEM containing 10% fetal bovine serum, 400 μg/ml G418, and 400 μg/ml zeocin. On day 1, the growth medium in the cultures was replaced with 1 ml of fresh growth medium without G418 and zeocin. On day 3, conditioned medium was recovered from confluent cultures, centrifuged at 10,000 rpm for 5 min and sterilized by filtration through 0.2-μm filters. The conditioned medium corresponding to 250 μg of cellular proteins was used in the luciferase reporter assay using C2C12 cells.
Luciferase Reporter Assay
One day before transfection, C2C12 cells were seeded on a 24-well culture plate. The reporter plasmid, pTCF7wt-luc (1 μg) (9), and the reference plasmid, pRL-TK (harboring thymidine kinase promoter just upstream of Renilla luciferase; Promega) (0.1 μg), were transfected into C2C12 cells using Lipofectamine 2000 (Invitrogen). After 12 h, cells were treated with conditioned medium prepared from L-Wnt-3a cells for activation of Wnt signal pathway and cultured for an additional 12 h. Cells were washed with PBS and lysed in 100 μl of passive lysis buffer (Promega). Firefly luciferase and Renilla luciferase activities were measured with 5 μl of cell lysate using the Dual-Luciferase Reporter Assay System (Promega). A Lumat LB9507 (EG&G Beltold) was used in the linear range. “Relative activity” was defined as the ratio of firefly luciferase activity to Renilla luciferase activity.
Inhibition of the Wnt Signal Pathway Using XAV939
On day 0, L or L-Wnt-3a cells (1 × 103 cells) were seeded on a 6-cm dish and treated with 5 μm XAV939 or DMSO as a vehicle. The incubation medium was replaced with fresh growth medium containing XAV939 or DMSO at 12-h intervals for 7 days.
Real-time PCR Analysis
Total RNA was extracted from cells by the guanidine phenol method using TRIzol reagent (Invitrogen) according to the manufacturer's protocols. Aliquots of 1 μg of total RNA were digested with 2 IU of RQ1 RNase-free DNase (Promega) for 30 min at 37 °C and then incubated for 10 min at 65 °C with stop solution (Promega). For reverse transcription, 0.75 μg of total RNA were treated with Moloney murine leukemia virus reverse transcriptase (Invitrogen) using random primers (nonadeoxyribonucleotide mixture; pd (N)9) (Takara bio Inc., Shiga, Japan). Quantitative real-time PCR was conducted using a FastStart DNA Master plus SYBR Green I in a LightCycler 1.5 (Roche Applied Science) according to the manufacturer's protocols. The housekeeping genes (GAPDH and β-actin) were used as internal controls for quantification. The expression level of C4ST-1 mRNA was normalized to that of the GAPDH or β-actin transcripts; mouse C4ST-1 forward primer (5′-ACCTCGTGGGCAAGTATGAG-3′) and reverse primer (5′-TCTGGAAGAACTCCGTGGTC-3′); mouse GAPDH forward primer (5′-CATCTGAGGGCCCACTG-3′) and reverse primer (5′-GAGGCCATGTAGGCCATGA-3′); mouse β-actin forward primer (5′-AGAGGGAAATCGTGCGTGAC-3′) and reverse primer 5′-CAATAGTGATGACCTGGCCGT-3′).
Northern Blotting
Poly(A)+ RNA was purified from L and L-Wnt-3a cells using the Quick Prep Micro mRNA purification kit (GE Healthcare). Aliquots of 1 μg of poly(A)+ RNA were subjected to 1.2% agarose gel electrophoresis containing 2.2 m formaldehyde, transferred to a Hybond N+ membrane (GE Healthcare), and hybridized with radiolabeled cDNA probe specific to either mC4ST-1 or mGAPDH. A specific cDNA probe to C4ST-1 was prepared from pCMV-mC4ST-1 (8) by double-digestion with SacII and SalI. cDNA probes (25 ng) were labeled with Redivue [α-32P]CTP (6000 Ci/mmol) (GE Healthcare) using the RediprimeTMII Random Prime Labeling System (GE Healthcare), and poly(A)+ RNA that had been transferred to membranes were hybridized with radiolabeled cDNA probe in ULTRAhyb solution (Ambion) overnight at 42 °C according to the manufacturer's protocols.
Immunoblotting
Cells grown in 60-mm dishes were treated with 2 ml of conditioned media for the indicated periods. When cells were treated with recombinant mouse Wnt-3a, cells grown in 35-mm dishes were washed twice with serum-free DMEM and incubated in 1 ml of serum-free DMEM containing recombinant mouse Wnt-3a. To measure β-catenin accumulation in the cytosol, cells were treated with Wnt-3a, washed with PBS, harvested, lysed in 200 μl of PBS containing 0.5 mm EDTA, 0.5% Nonidet P-40, 10 μm Z-Leu-Leu-Leu-H (Peptide Institute, Inc., Osaka, Japan), and protease inhibitor mixture (Nacalai Tesque, Kyoto, Japan) and incubated at 4 °C for 5 min. The lysates were centrifuged at 1,000 × g for 5 min, and the resulting supernatants were removed from the pellets and further centrifuged at 10,000 rpm for 10 min. The supernatants resulting from the second centrifugation were mixed with 100 μl of 3× SDS loading buffer (187.5 mm Tris-HCl, pH 6.8, containing 6% (w/v) SDS, 30% glycerol, and 0.03% (w/v) phenol red) and boiled for 5 min. Alternatively, cells were solubilized in 0.1% saponin lysis buffer (25 mm Hepes containing 75 mm potassium acetate) for 30 min at 4 °C (10) and centrifuged at 15,000 rpm for 5 min. To detect cellular Wnt-3a, cells were lysed in 1% Triton X-100 lysis buffer (20 mm Tris-HCl, pH 7.4, containing 1 mm EDTA, 0.15 m NaCl, 10% glycerol, 10 μm Z-Leu-Leu-Leu-H, and protease inhibitor mixture) for 30 min on ice and centrifuged at 10,000 rpm for 5 min. The protein concentration of each sample was determined using a BCA protein assay kit (Thermo Fisher Scientific, Inc., Waltham, MA) according to the manufacturer's protocol. Each sample was subjected to SDS-PAGE (7.5% gel), transferred to Hybond-ECL nitrocellulose filters (GE Healthcare), and analyzed using the procedure as described in Yoshida et al. (11) with an ECL AdvanceTM Western blotting detection kit (GE Healthcare) according to the manufacturer's protocol. To detect secreted Wnt-3a proteins, conditioned medium obtained from L-Wnt-3a cells and L-Wnt-3a-C4ST-1 transfectants was centrifuged at 2000 rpm for 5 min, and supernatants were analyzed by SDS-PAGE under non-reducing conditions. Cell lysates (8 μg of proteins) prepared using 0.1% saponin lysis buffer were digested with 1 IU of N-glycosidase F (Roche Diagnostics) at 37 °C overnight.
Immunoprecipitation
After C2C12 cells overexpressing FLAG-tagged Wnt-3a proteins were cultured for 3 days, the conditioned medium was collected and centrifuged at 10,000 rpm for 5 min. Cells were lysed in 1% Triton X-100 lysis buffer (20 mm Tris-HCl, pH 7.4, containing 1 mm EDTA, 0.15 m NaCl, 10% glycerol, 10 μm Z-Leu-Leu-Leu-H, and a protease inhibitor mixture) for 30 min on ice and then clarified by centrifugation at 10,000 rpm for 5 min. Anti-FLAG M2 affinity gel (Sigma) was added to the medium and cell lysate and incubated overnight at 4 °C with rotating. The beads were collected by brief centrifugation and washed with the lysis buffer three times. Immunoprecipitated materials were eluted by boiling for 5 min in 1× Laemmli sample buffer in the absence of dithiothreitol and then subjected to immunoblotting using anti-FLAG rabbit polyclonal antibody (Sigma).
Inhibition Assay
Recombinant Wnt-3a (5 ng (0.125 pmol)) was preincubated with CS chains (50 ng as GlcUA) prepared from each cell line in 1 ml of serum-free DMEM for 30 min at room temperature and then added to L cells cultured in 6-cm dishes. L cells were washed with serum-free DMEM before adding recombinant Wnt-3a (rWnt-3a)-CS complexes. After 2 h, accumulation of β-catenin in the cytosol was quantified using immunoblotting. Similar competition assays using CS-derived tetrasaccharides with various sulfation patterns as the inhibitor were carried out. GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-O-p-methoxyphenyl (E-E sequence) was chemically synthesized (12). GalNAc(4-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-O-p-methoxyphenyl (A-E sequence) and GalNAc(6-O-sulfate)-GlcUA-GalNAc(6-O-sulfate)-GlcUA-O-p-methoxyphenyl (C-C sequence) were obtained by the complete digestion of E-E sequence using chondro-6-sulfatase (50 mIU) and chondro-4-sulfatase (50 mIU), respectively. Each tetrasaccharide (125 pmol) was preincubated with rWnt-3a (0.125 pmol) for 30 min at room temperature and then added to L cells. After 2 h, cytosolic β-catenin was quantified using immunoblotting.
Human embryonic fibroblasts treated with or without recombinant Wnt-3a or SB216763 were homogenized by passing through a 23-gauge needle in PBS containing protease inhibitor mixture and centrifuging at 9000 rpm for 10 min. The resultant supernatants were analyzed by immunoblotting.
Isolation and Purification of GAGs
Cells were homogenized in acetone, extracted with acetone three times, and air-dried thoroughly. The dried materials were digested with heat-activated actinase E (10% by weight of dried materials) in 0.1 m borate-sodium, pH 8.0, containing 10 mm CaCl2 at 55 °C for 48 h. The samples were adjusted to 5% v/v in trichloroacetic acid and centrifuged. The resulting supernatants were extracted with diethyl ether three times to remove the trichloroacetic acid. The aqueous phase was evaporated to dryness, dissolved in 50 mm pyridine acetate, pH 5.0, and subjected to gel filtration on a PD-10 column (GE Healthcare) using 50 mm pyridine acetate, pH 5.0, as an eluent. The flow-through fractions were collected and evaporated to dryness. The dried materials were dissolved in 300 mm phosphate buffer containing 150 mm NaCl and applied to an anion-exchange Micro SpinColumnTM (Harvard Apparatus, Holliston, MA). After washing with 300 mm phosphate buffer containing 150 mm NaCl, the bound GAGs were eluted by 300 mm phosphate buffer containing 1.5 m NaCl and then desalted using a PD-10 column as described above.
Disaccharide Composition Analysis of CS and HS
Purified GAGs were digested with 5 mIU of chondroitinase ABC or a mixture of 0.5 mIU of heparinase and 0.5 mIU of heparitinase at 37 °C for 2 h. Reactions were terminated by boiling for 1 min. The digests were derivatized with a fluorophore 2-aminobenzamide and then analyzed by high performance liquid chromatography (HPLC) as reported previously (13).
RESULTS
The Expression Levels of C4ST-1 Decrease Dramatically in Wnt-producing Cells
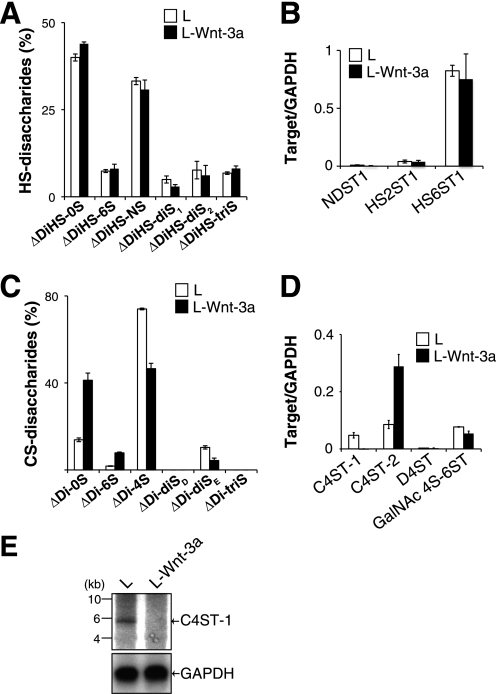
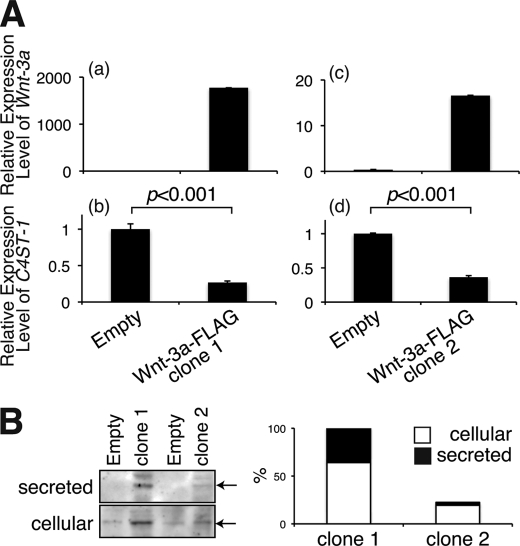
We found that ∼80% of the Wnt-3a protein from mouse L cells expressing high amounts of Wnt-3a protein (L-Wnt-3a cells) was secreted into the culture medium, contrary to previous findings in which secreted Wnt-3a molecules tightly associated with the cell surface and extracellular matrix. Based on this observation and our previous finding that Wnt-3a could bind to GAG chains with high affinity depending on the specific sulfation patterns of the GAGs (7), we hypothesized that structural alterations of cellular GAG chains might occur in L-Wnt-3a cells. Thus, we analyzed the structures of GAG chains associated with L cells and L-Wnt-3a cells. As shown in Fig. 1A, disaccharide composition of HS in L-Wnt-3a cells was comparable with that of L cells. Consistent with this finding, the expression levels of the biosynthetic enzymes involved in the sulfation of HS were not significantly affected by Wnt-3a signaling (Fig. 1B). In contrast, the CS chains of L-Wnt-3a cells have less 4-O-sulfation than those of L cells (Fig. 1C and supplemental Table S1). In addition, the E-disaccharide unit (ΔDi-diSE), which may be involved in modulation of Wnt-3a signaling, was also reduced in L-Wnt-3a cells relative to L cells (Fig. 1C and supplemental Table S1). Therefore, we investigated the expression levels of sulfotransferases involved in the sulfation of CS chains. To date, three sulfotransferases, C4ST-1, C4ST-2, and D4ST, involved in the 4-O-sulfation of GalNAc residues in CS/dermatan sulfate have been reported (14). In addition, GalNAc4S-6ST was implicated in the 6-O-sulfation of 4-O-sulfated GalNAc residues and production of E-disaccharide units (8, 15). Therefore, we measured the expression levels of these genes using real-time PCR. There was a sharp decrease in the expression of C4ST-1 in L-Wnt-3a cells relative to the expression in L cells; in contrast, levels of D4ST-1 and GalNAc4S-6ST mRNA in L-Wnt-3a cells were indistinguishable from those in L cells (Fig. 1D). C4ST-2 mRNA was expressed at elevated levels in L-Wnt-3a cells compared with L cells (Fig. 1D). Northern blot analysis confirmed that C4ST-1 expression was depressed in L-Wnt3a cells (Fig. 1E). In addition, we established two C2C12 cell clones (clones 1 and 2) stably expressing Wnt-3a tagged with FLAG epitope to exclude the possibility that C4ST-1 expression was down-regulated in L-Wnt-3a cells due to clonal selection of cell lines. As shown in Fig. 2A, C4ST-1 expression levels were also suppressed in the two C2C12 cell clones expressing Wnt-3a, corresponding to the expression levels of Wnt-3a. Moreover, secretion of Wnt-3a molecules into the culture medium appeared to be negatively correlated with C4ST-1 expression (Fig. 2, A and B).
FIGURE 1.
The expression of C4ST-1 gene was suppressed in L-Wnt-3a cells. A, the disaccharide composition of HS chains produced in L and L-Wnt-3a cells was analyzed. GAG chains isolated from L and L-Wnt-3a cells were digested with a mixture of heparitinase and heparinase, labeled with the fluorophore 2-aminobenzamide, and analyzed by HPLC. The percentage of different HS disaccharides is shown. Results are the means ± S.E. for two experiments. ΔDiHS-0S, ΔHexUAα1–4GlcNAc; ΔDiHS-6S, ΔHexUAα1–4GlcNAc(6-O-sulfate); ΔDiHS-NS, ΔHexUAα1–4GlcN(2-N-sulfate); ΔDiHS-diS1, ΔHexUAα1–4GlcN(2-N-sulfate,6-O-sulfate); ΔDiHS-diS2, ΔHexUA(2-O-sulfate)α1–4GlcN(2-N-sulfate); ΔDiHS-triS, ΔHexUA(2-O-sulfate)α1–4GlcN(2-N-sulfate,6-O-sulfate). B, NDST1 (AF049894), HS2ST1 (AF060178), and HS6ST1 (NM_015818.2) mRNA expression levels in L (open bars) and L-Wnt-3a cells (closed bars) were measured using quantitative real-time PCR. Transcript levels of each gene are expressed relative to GAPDH mRNA levels ± S.D. Two independent experiments were run for each data set. C, the disaccharide composition of CS chains produced in L and L-Wnt-3a cells was analyzed as described in A. The percentage of different CS disaccharides is shown. ΔDi-0S, ΔHexUAα1–3GalNAc; ΔDi-6S, ΔHexUAα1–3GalNAc(6-O-sulfate); ΔDi-4S, ΔHexUAα1–3GalNAc(4-O-sulfate); ΔDi-diSD, ΔHexUA(2-O-sulfate)α1–3GalNAc(6-O-sulfate); ΔDi-diSE, ΔHexUAα1–3GalNAc(4-O-sulfate,6-O-sulfate); ΔDi-triS, ΔHexUA(2-O-sulfate)α1–3GalNAc(4-O-sulfate,6-O-sulfate). D, C4ST-1 (NM_021439.2), C4ST-2 (NM_021528), D4ST (BC043700), and GalNAc4S-6ST (NM_029935.5) mRNA expression levels in L (open bars) and L-Wnt-3a cells (closed bars) were analyzed using quantitative real-time PCR as described in B. E, poly(A)+ RNA was isolated from L and L-Wnt-3a cells and analyzed by Northern blot hybridization using DNA probes specific for C4ST-1 and GAPDH.
FIGURE 2.
The expression of C4ST-1 gene was suppressed in C2C12 cells expressing Wnt-3a. A, two cell lines of C2C12 cells expressing FLAG-tagged Wnt-3a proteins were established and named clone 1 (panels a and b) and clone 2 (panels c and d). C4ST-1 mRNA expression levels in these clones were examined by real-time PCR as described under “Experimental Procedures.” Panels a and c and panels b and d indicate the expression levels of Wnt-3a and C4ST-1, respectively. B, cellular and secreted FLAG-tagged Wnt-3a proteins of C2C12 cells expressing FLAG-tagged Wnt-3a, clones 1 and 2, were immunoprecipitated with anti-FLAG M2 affinity gel and then analyzed by immunoblotting using anti-FLAG antibody. Empty indicates C2C12 cells carrying a pIRES-neo3 empty vector. The total amount of Wnt-3a protein in clone 2 was expressed relative to that in clone 1 (right panel).
Specific Sugar Modifications Catalyzed by C4ST-1 Control Diffusion of Wnt-3a Molecules
We next established L-Wnt-3a cell clones stably transfected with C4ST-1 cDNA to examine whether down-regulation of C4ST-1 enhanced the release of Wnt-3a into the culture medium. Eight stably transfected clones were obtained, and the expression levels of C4ST-1, GalNAc4S-6ST, and Wnt-3a were examined by real-time PCR. C4ST-1 expression had little effect on the level of GalNAc4S-6ST or Wnt-3a expression (Fig. 3A). In addition, because Komekado et al. (16) reported that N-glycosylation of Wnt-3a is needed for secretion of an active Wnt-3a, we confirmed that overexpression of C4ST-1 had no effect on the N-glycosylation of Wnt-3a using two independently transfected clones, 3 and 11 (Fig. 3B).
FIGURE 3.
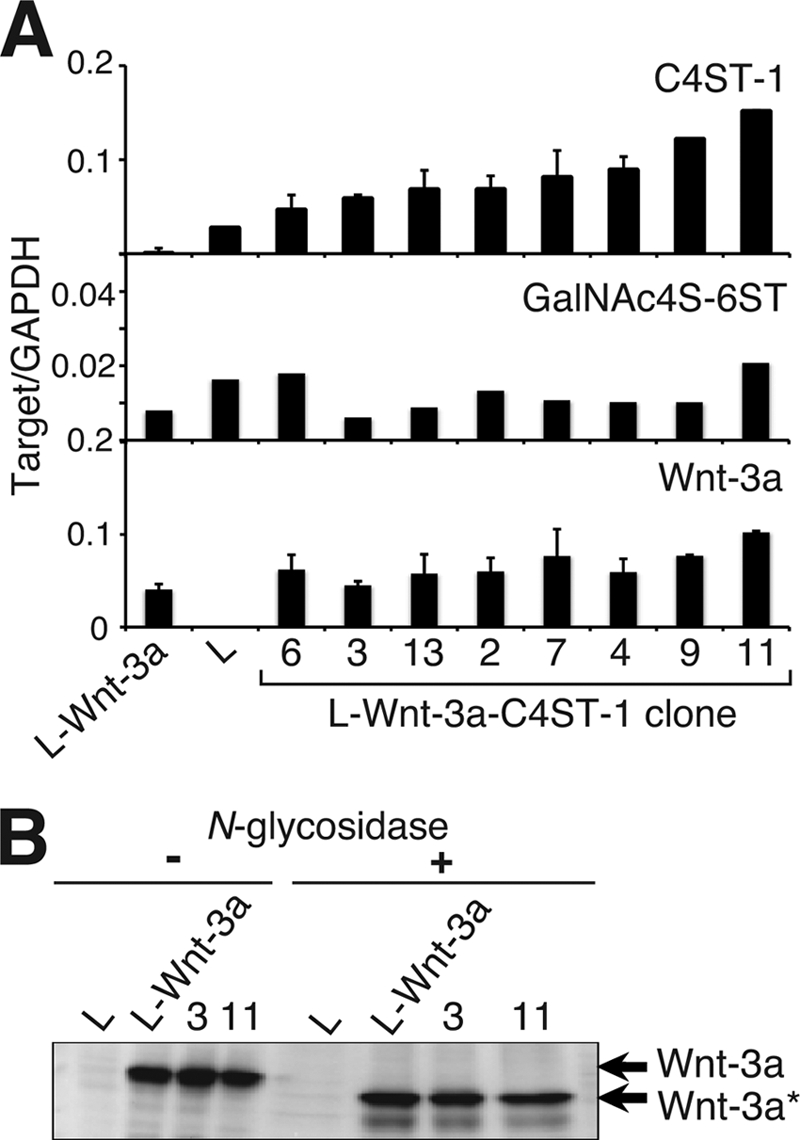
Forced expression of C4ST-1 in L-Wnt-3a cells hardly affected GalNAc4S-6ST and Wnt-3a mRNA levels or Wnt-3a N-glycosylation. A, the mRNA levels of C4ST-1, GalNAc4S-6ST, and Wnt-3a in parental L cells, L-Wnt-3a cells, and L-Wnt-3a-C4ST-1 clones were analyzed using real-time PCR as described in Fig. 1B. B, cell lysates prepared from L, L-Wnt-3a cells, and L-Wnt-3a-C4ST-1 clone 3 and clone 11 were digested with N-glycosidase F and analyzed using immunoblots probed with anti-Wnt-3a antibody. Wnt-3a* indicates Wnt-3a proteins without N-glycosylation.
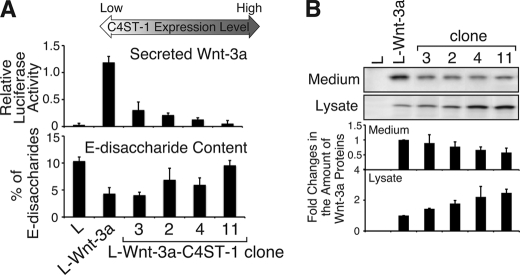
We then assayed the amount of Wnt-3a in the medium of each transfectant clone using C2C12 cells transiently transfected with pTCF7wt-luc reporter vector carrying seven repeats of the TCF binding consensus sequence (9) (supplemental Fig. S2). Conditioned medium prepared from each transfectant clone was used to stimulate C2C12 cells carrying the reporter vector for Wnt signals. A decline in the amount of Wnt-3a secreted into the medium, as determined by stimulation of C2C12 cells, was accompanied by an increase in C4ST-1 expression levels in Wnt-3a-producing L cells (Fig. 4A and supplemental Fig. S3), whereas the amount of Wnt-3a protein bound to the Wnt-3a-producing L cells increased (Fig. 4B). As shown in Fig. 3, we confirmed that a decrease in secreted Wnt-3a protein was not caused by reduction in the expression level of the Wnt-3a gene or defects in glycosylation of Wnt-3a. In addition, the amount of E-disaccharide units increased approximately linearly with the expression level of C4ST-1 (correlation coefficient: 0.75) (Fig. 4A and supplemental Fig. S3 and Table S1). The level of E-disaccharide units was apparently controlled by the level of C4ST-1, but not GalNAc4S-6ST, gene expression (Fig. 3A), even though E-disaccharide units are synthesized via sequential sulfation by C4ST-1 and GalNAc4S-6ST. Secretion of Wnt-3a declined exponentially with increasing C4ST-1 expression (correlation coefficient, 0.98) and E-disaccharide unit content (correlation coefficient, 0.69) (supplemental Fig. S3), implying that a sugar motif containing E-disaccharide units provided a recognition site for Wnt-3a molecules. These results suggested that C4ST-1 regulated the secretion of Wnt-3a into the medium by producing E-disaccharide units.
FIGURE 4.
C4ST-1 expression levels are positively correlated with E-disaccharide content and negatively correlated with Wnt-3a secretion. A, the amount of secreted Wnt-3a (top panel) and the content of E-disaccharide units in cellular CS chains (bottom panel) in L, L-Wnt-3a, and L-Wnt-3a-C4ST-1 transfectants (clone 2, clone 3, clone 4, and clone 11) were examined. Four L-Wnt-3a-C4ST-1 clones were ranked in order of increasing levels of C4ST-1 mRNA. Secretion of Wnt-3a into the medium was measured as described under “Experimental Procedures.” E-disaccharide content was determined based on HPLC analysis of CS chains isolated from each cell type as described under “Experimental Procedures.” B, Wnt-3a proteins in medium and cell lysates were analyzed using immunoblots probed with anti-Wnt-3a antibody. The amount of Wnt-3a protein in L and L-Wnt-3a-C4ST-1 clones was expressed relative to that in L-Wnt-3a cells. The average of the amount of Wnt-3a protein from two independent experiments is presented with S.D.
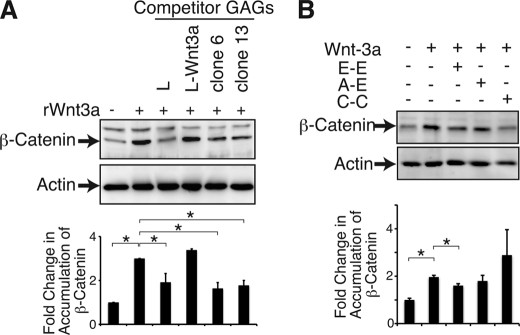
Next, we investigated whether the CS chains produced by L-Wnt-3a-C4ST-1 cells bound more Wnt-3a than L-Wnt-3a cells. CS chains isolated from L, L-Wnt-3a, and L-Wnt-3a-C4ST-1 cells were used as competitors in competitive inhibition assays because the binding activity of CS chains to Wnt-3a were positively correlated with their ability to inhibit Wnt-3a signaling (7). β-Catenin accumulation in the cytosol was used as a quantitative indicator of canonical Wnt signaling because degradation of cytosolic β-catenin is inhibited by activated Wnt signaling. More Wnt-3a bound to CS chains synthesized by L-Wnt-3a-C4ST-1 cells than to the CS chains synthesized by L-Wnt-3a cells (Fig. 5A). This result suggested that CS binding affinity for Wnt-3a was closely associated with the expression level of C4ST-1. To further examine whether Wnt-3a bound to the E-disaccharide unit formed by C4ST-1, we carried out inhibition assays using three chemically synthesized tetrasaccharide sequences including the E-E-containing sequence GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-O-p-methoxyphenyl. Preincubation of Wnt-3a with the E-E-containing sequence significantly inhibited accumulation of β-catenin in the cytosol, whereas GalNAc(4-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-O-p-methoxyphenyl (A-E sequence) and GalNAc(6-O-sulfate)-GlcUA-GalNAc(6-O-sulfate)-GlcUA-O-p-methoxyphenyl (C-C sequence) did not dramatically block accumulation of β-catenin in response to Wnt-3a (Fig. 5B). We also examined the binding of Wnt-3a to the E-E-containing sequence by competitive inhibition experiments using the BIAcore system (supplemental Fig. S4). As reported (7), Wnt-3a molecules bound strongly to chondroitin sulfate E (CS-E) with a high E-disaccharide unit content. The interaction between CS-E and Wnt-3a was inhibited by the E-E sequence but not the A-E or C-C sequence (supplemental Fig. S4). These results indicated that Wnt-3a might preferentially recognize tandem E-disaccharide units. This observation was consistent with those in supplemental Fig. S3 that showed that an increase in the number of E-disaccharide unit was correlated with an exponential decreased in secretion of Wnt-3a. Thus, C4ST-1 generated sequentially arranged E disaccharide motif (E-E sequence) within cellular CS chains to capture Wnt-3a molecules on the surface of Wnt-3a-producing cells.
FIGURE 5.
Wnt-3a preferentially binds to E-disaccharide units. A, L cells were treated with rWnt-3a preincubated with or without competitor GAGs prepared from the indicated cells. Accumulated β-catenin in the cytosol was examined by immunoblotting. Each β-catenin band was calculated relative to that of each actin band and normalized with the value of L cells in the absence of Wnt-3a. The means from two independent experiments with the S.D. (error bars) are plotted. *, p < 0.05. B, L cells were treated with rWnt-3a preincubated with or without chemically synthesized tetrasaccharides carrying various sulfation patterns. E-E, GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-O-p-methoxyphenyl; A-E, GalNAc(4-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-O-p-methoxyphenyl; C-C, GalNAc(6-O-sulfate)-GlcUA-GalNAc(6-O-sulfate)-GlcUA-O-p-methoxyphenyl. The expression level of β-catenin was also determined as in A. The means from two independent experiments are plotted with the S.D. (error bars). *, p < 0.05.
Sustained Wnt-3a Signaling Is Required for Down-regulation of C4ST-1
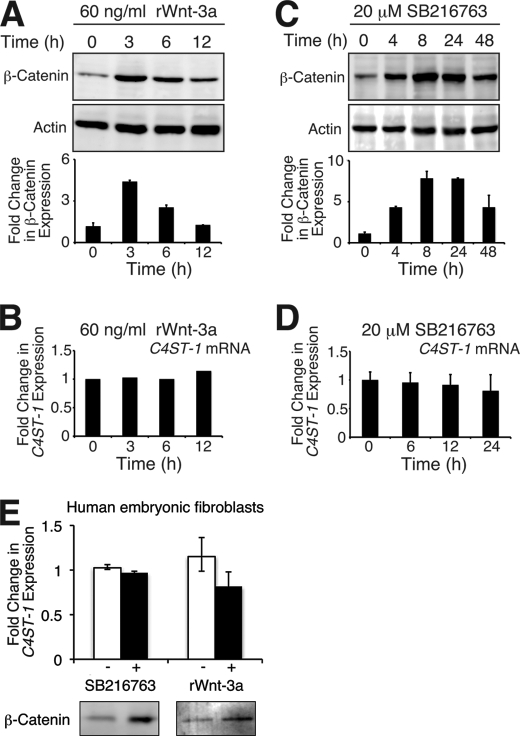
We investigated whether transient Wnt-3a signals were sufficient for down-regulation of C4ST-1. After L cells were treated with recombinant Wnt-3a for the indicated defined period, accumulation of β-catenin was examined. Three hours after treatment of L cells with recombinant Wnt-3a, accumulation of β-catenin increased by <5-fold and then gradually decreased to background levels (Fig. 6A). In the meantime, C4ST-1 mRNA expression levels were not changed significantly by adding recombinant Wnt-3a (Fig. 6B). Wnt-3a signaling was also transiently activated using an inhibitor of glycogen synthase kinase 3, SB216763. The activation of Wnt-3a signals via SB216763 reached maximal levels between 8 and 24 h of exposure and then reduced (Fig. 6C). Although C4ST-1 was not down-regulated by transient Wnt-3a signals (Fig. 6, B and D), C4ST-1 mRNA levels were down-regulated in L-Wnt-3a cells (Fig. 1, D and E). The accumulation of β-catenin induced by Wnt-3a signaling in L-Wnt-3a cells did not differ greatly from β-catenin accumulation observed in L cells transiently activated with recombinant Wnt-3a or SB216763 (Fig. 6, A and C, and supplemental Fig. S5). In addition, we confirmed the effect of transient Wnt-3a signals on the expression of C4ST-1 using one of primary cells, human fibroblasts, which are thought to be more sensitive to the stimulation of Wnt than established cell lines. As shown in Fig. 6E, transiently activated Wnt pathway by recombinant Wnt-3a and SB216763 had little effect on the expression levels of C4ST-1 in human fibroblasts. Taken together, the observation suggested that sustained Wnt-3a signaling was required for down-regulation of C4ST-1.
FIGURE 6.
Transient Wnt signaling has little effect on the accumulation of C4ST-1 mRNA. A and C, L cells were incubated in the presence of either 60 ng/ml rWnt-3a or 20 μm SB216763 for the indicated periods. Accumulation level of cytosolic β-catenin was measured as in Fig. 5A. The means from two independent experiments are plotted with the S.D. (error bars). B and D, total RNA was extracted from cells treated as described in A and C and subjected to quantitative real-time PCR. The expression level of C4ST-1 was calculated relative to that of GAPDH and normalized with the value at 0 h. E, human embryonic fibroblasts were treated with 60 ng/ml rWnt-3a for 6 h or 20 μm SB216763 for 12 h and then analyzed by immunoblotting. Accumulation level of cytosolic β-catenin was measured as in Fig. 5A. The means from two independent experiments are plotted with the S.D. (error bars).
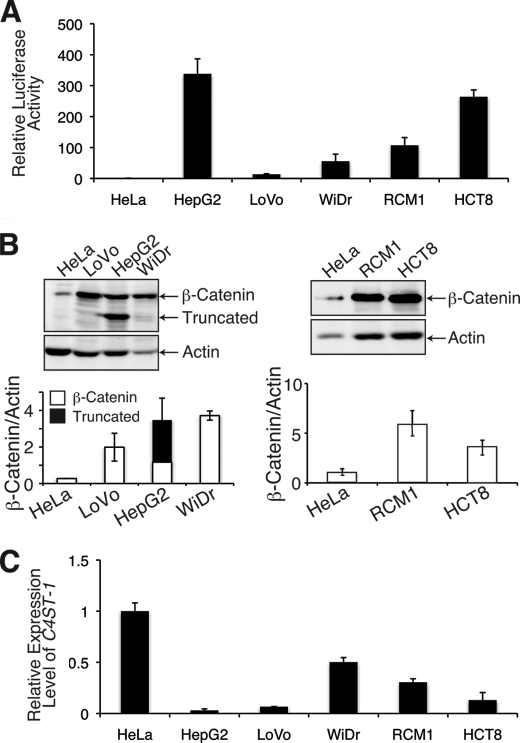
We further examined whether sustained Wnt signaling down-regulates C4ST-1 gene expression using gastrointestinal and liver cancer cell lines that have constitutively active Wnt/β-catenin signaling. We examined four human colorectal cancer cell lines, LoVo, WiDr, HCT8, and RCM1 cells, and one human liver carcinoma cell line, HepG2. As reported previously (17), the activity of a pTCF7-luc reporter in HepG2, LoVo, WiDr, HCT8, and RCM1 cells was higher than the activity in HeLa cells that do not have constitutive Wnt/β-catenin signaling (Fig. 7A). In addition, the levels of cytosolic β-catenin were increased by the activated Wnt signaling in gastrointestinal and liver cancer cell lines (Fig. 7B). A truncated form of β-catenin generated by deletion of amino acids 25–140 was observed in HepG2 cells (Fig. 7B) (18). Moreover, C4ST-1 mRNA levels were lower in the five cell lines that have constitutive Wnt/β-catenin signaling than in HeLa cells (Fig. 7C). Therefore, C4ST-1 gene expression was down-regulated by constitutive activation of Wnt/β-catenin signaling.
FIGURE 7.
C4ST-1 gene expression is dramatically suppressed in tumor cell lines with constitutive Wnt signaling. A, HeLa, HepG2, LoVo, WiDr, HCT8, and RCM1 cells were transfected with pTCF7wt-luc reporter vector together with the pRL-TK reference vector. The relative luciferase activity was measured in duplicate, and the averages from two independent experiments are presented with the S.D. (error bars). Experiments were independently repeated twice. B, accumulated levels of β-catenin in the cytosol was measured as described in Fig. 5A. The averages from two independent experiments are presented with the S.D. (error bar). Truncated type β-catenin generated by deletion of amino acids 25–140 was detected in addition to the full-length type β-catenin in HepG2 cells. C, C4ST-1 expression levels in the six cancer cell lines were investigated by real-time PCR as described in Fig. 1B.
Secreted Wnt-3a Molecules Act on Target Cells to Down-regulate C4ST-1 Gene Expression
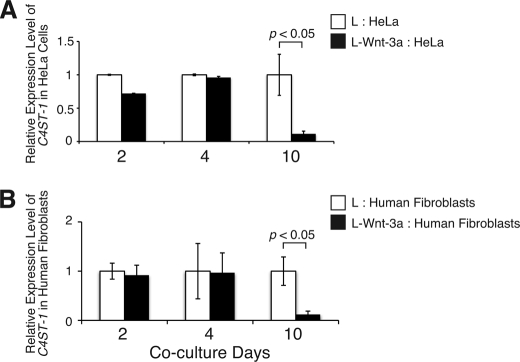
To investigate whether Wnt-3a can control the expression of C4ST-1 in a paracrine manner, L-Wnt-3a cells were co-cultured for 2, 4, and 10 days with HeLa cells, which increase expression level of β-catenin in response to Wnt-3a (supplemental Fig. S1B). If Wnt-3a molecules secreted from L-Wnt-3a cells function as paracrine signals, the level of C4ST-1 mRNA in the co-cultured HeLa cells should decrease. Co-culture with L-Wnt-3a cells for 10 days suppressed C4ST-1 mRNA expression in HeLa cells; however, the level of C4ST-1 mRNA expression was not affected in HeLa cells co-cultured with L-Wnt-3a cells for only 2 or 4 days (Fig. 8A). In addition, human embryonic fibroblasts were co-cultured with L-Wnt-3a cells for the indicated periods. Co-culture with L-Wnt-3a cells for 4 days was not sufficient to down-regulate the expression of C4ST-1, whereas C4ST-1 mRNA levels were significantly decreased by co-culture with L-Wnt-3a cells for 10 days (Fig. 8B). Thus, these results suggested that sustained Wnt stimuli were also required for down-regulation of C4ST-1 when Wnt-3a acted as a paracrine signal.
FIGURE 8.
Wnt-3a can suppress the expression level of C4ST-1 in a non-cell autonomous manner. HeLa cells (A) and human embryonic fibroblasts (B) were cultured with L or L-Wnt-3a cells for the indicated periods, and C4ST-1 gene expression was measured by real-time PCR as described in Fig. 1B. Sustained Wnt stimuli resulted in reduced C4ST-1expression in both HeLa cells and human fibroblasts. Experiments were independently repeated twice.
C4ST-1 Is an Indirect Target of the Wnt Signal Pathway
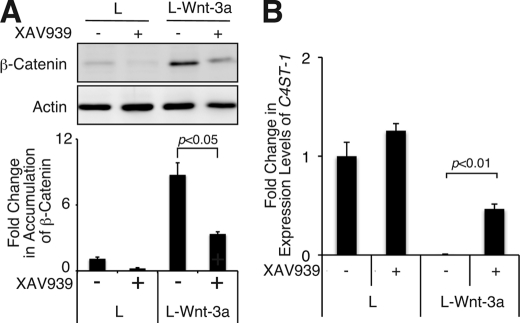
We investigated whether the expression of C4ST-1 could be recovered by blocking Wnt signaling. Recently, it has been reported that a small molecule, XAV939, selectively inhibits β-catenin-mediated transcription because it stimulates β-catenin degradation by stabilizing axin. Treatment of L-Wnt-3a cells with XAV939 decreased the expression level of β-catenin (Fig. 9A), indicating that Wnt signaling was inhibited in L-Wnt-3a cells. C4ST-1 expression levels were measured using real-time PCR (Fig. 9B). The C4ST-1 expression level in L-Wnt-3a cells increased by ∼70-fold after treatment for 7 days with 5 μm XAV939, which was restored to about half that of L cells (Fig. 9B). These results indicated that Wnt signaling down-regulated the expression of C4ST-1. Thus, sustained Wnt signaling was apparently required for decreased expression of C4ST-1.
FIGURE 9.
Sustained inhibition of Wnt signaling can recover C4ST-1 expression. A, L and L-Wnt-3a cells were incubated in the presence or absence of 5 μm XAV939 for 7 days, and cytosolic β-catenin accumulation was examined in each cell and treatment type. The means from two independent experiments are plotted with the S.D. (error bars). B, C4ST-1 mRNA levels in each cell from each treatment described in A were analyzed by real-time PCR as in Fig. 1B. Experiments were independently repeated twice.
DISCUSSION
Here we showed that a negative feedback loop acts in Wnt-producing cells. Sustained Wnt-3a signals resulted in repression of C4ST-1, a positive regulator of Wnt signaling (7). C4ST-1 is the first of the sulfotransferases involved in the biosynthesis of CS targeted and is negatively regulated by Wnt signaling. Thus, suppression of the expression of C4ST-1 induces structural alteration of CS chains, resulting in decreased affinity between CS chains and Wnt-3a molecules and the release of Wnt-3a molecules from the surface of Wnt-producing cells. Therefore, we suggest that the negative regulation of C4ST-1 by Wnt-3a has two biological implications; first, it acts as a feedback inhibitor of sustained Wnt signaling that tightly controls the strength of Wnt signaling. Second, it functions as a trigger for the diffusion of Wnt-3a molecules from Wnt-producing cells.
It has been shown that the strength of Wnt/β-catenin signaling determines whether mesenchymal progenitor cells differentiate into osteoblasts or chondrocytes (19). During endochondral ossification, canonical Wnt signaling is first down-regulated to form the cartilage and later up-regulated to form endochondral bone. Aberrant up-regulation of Wnt signaling inhibits chondrogenesis, whereas low levels of Wnt signaling are insufficient for ossification. C4ST-1 activity is also tightly controlled during chondrogenesis. Previous studies using a gene-trap mutation in the C4ST-1 gene showed that C4ST-1 is required for morphogenesis of bones formed by endochondral ossification (20). Because C4ST-1 is a positive regulator of Wnt signaling and a target gene negatively controlled by Wnt signaling, it can participate in a negative feedback loop to tightly control the strength of Wnt signaling. Thus, mutual regulation between Wnt signaling and C4ST-1 might determine chondrocyte versus osteoblast differentiation.
Secreted Wnt proteins are poor candidates for long range signaling because they are hydrophobic and mostly found associated with cell membranes and the extracellular matrix. Nevertheless, a body of cell biological evidence demonstrates that they effectively diffuse. How these sticky proteins diffuse to distant cells is currently unclear. Recent studies show that packaging of Wnt molecules into lipoprotein particles is needed for release from Wnt-producing cells (21). Lipoproteins particles have been proposed to act as vehicles for the intracellular movement of lipid modified proteins. Wnt proteins are lipid-modified proteins anchored in the cell membrane by lipid moieties. Therefore, it is thought that lipoproteins function as intracellular carriers for lipid-modified Wnt proteins. In addition, Wnt molecules have a high affinity for CS chains expressed on the surface of cells and in the extracellular matrix (4, 7). A decrease in the affinity between CS chains and Wnt proteins is needed for release of Wnt molecules from Wnt-producing cells and for the long-range action of Wnt molecules. In this study we showed that down-regulation of C4ST-1 decreases the affinity between CS chains and Wnt proteins and triggers release of Wnt molecules from Wnt-producing cells. Additionally, C4ST-1 regulated the association and dissociation of Wnt-3a molecules and CS chains by controlling the level of GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA (E-E sequences) that strongly binds to Wnt-3a proteins (Fig. 5). PGs and GAGs regulate the distribution of morphogens; for example, a glycosylphosphatidylinositol (GPI)-anchored PG, glypican, is essential for Wg long range diffusion, and Notum, an enzyme that cleaves GPI-anchored proteins, regulates distribution of Wg mainly by induction of the release of glypicans from the cell surface (22, 23). In addition, the complete loss of HS or PGs in model organisms including Drosophila melanogaster and mice generated using genetic techniques results in an abnormal distribution of Wg and Wnt (24–27). Here we demonstrated that the expression level of a specific sugar structure produced by C4ST-1 mediated the release of Wnt-3a molecules from the cell surface.
CS and HS chains are composed of repeating disaccharide units, GlcUA-GalNAc and GlcUA-GlcNAc, respectively. This disaccharide backbone structure is acted upon sequentially by a series of modifying enzymes such as sulfotransferases and epimerases. Differences in the sulfation and epimerization of the core disaccharide units give rise to a variety of structural units. We hypothesize that the differential arrangement of these units forms a sugar code that determines the specificity and the strength of binding affinity in protein-sugar interactions. A comprehensive study of mutations in each of the HS biosynthetic enzymes in Caenorhabditis elegans provides the first hint that specific modification patterns encode specific functions. Mutants lacking any of HS-modifying enzymes encoded by hse-5 (C5-epimerase), hst-2 (2-O-sulfotransferase), or hst-6 (6-O-sulfotransferase) have very specific defects in nervous system development (28). Some axons require all three modifying enzymes for their guidance, others require only hse-5 or hst-2, and many require none of these enzymes. Moreover, the same neuron could need different modifications for different guidance decisions. These results imply that specific sugar codes formed by sulfotransferases function as a determinant for guidance. In this study, we identified the E-E sequence, GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)-GlcUA, formed by C4ST-1 as a sugar code for the diffusion and signaling of Wnt-3a.
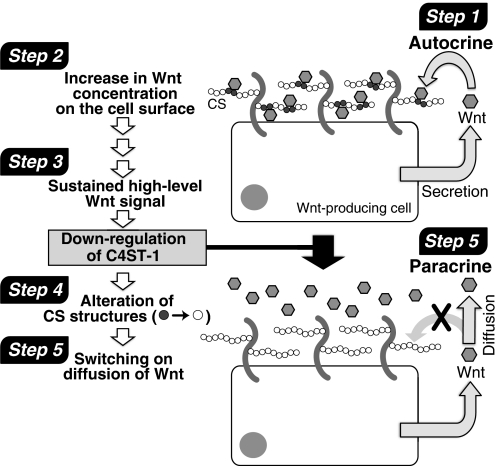
As shown in Fig. 10, continuous Wnt/β-catenin signaling in L-Wnt-3a cells depressed the expression of the C4ST-1 gene, caused structural alteration of CS chains, and resulted in the diffusion of Wnt-3a molecules from Wnt-producing cells. Continuous Wnt/β-catenin signaling was found to ubiquitously depress the expression of the C4ST-1 gene. C4ST-1 gene expression was dramatically suppressed in human hepatocarcinoma HepG2 cells and human colon adenocarcinoma LoVo, WiDr, RCM-1, and HCT-8 cells, where Wnt/β-catenin signaling was highly activated (Fig. 7). Therefore, we suggest that many cells share a common mechanism for negative regulation of C4ST-1 by sustained Wnt/β-catenin signaling. In addition, the E-disaccharide unit content in HepG2 cells (1.6%), LoVo cells (2.5%), and WiDr cells (0.21%) was lower than that of HeLa cells (3.0%) and correlated positively with the expression levels of C4ST-1 (data not shown). These results indicated that continuous Wnt/β-catenin signaling acted ubiquitously on the CS biosynthetic pathway and created a microenvironment that maximized long range Wnt activity. In addition, Wnt-3a can also act in a paracrine manner and down-regulate C4ST-1 in the target cells as shown in Fig. 8. These results suggest that Wnt-3a has effects on the CS biosynthesis in the target cells and causes changes in extracellular matrix surrounding the target cells. These alterations in the microenvironment are likely of great importance, for example, in understanding how niches control stem cell fate.
FIGURE 10.
A model of C4ST-1-mediated regulation of Wnt-3a diffusion from Wnt-producing. Step 1, Wnt acts in an autocrine manner in Wnt-producing cells. Step 2, secreted Wnt molecules are caught by CS chains expressing on the cell surface or in extracellular matrix. Step 3, sustained Wnt signaling down-regulates C4ST-1 gene expression. Step 4, down-regulation of C4ST-1 alters sulfation patterns of cell-surface and extracellular-matrix CS chains (gray circles and white circles indicate CS disaccharide structures with a high and low affinity to Wnt-3a, respectively); these modifications result in altered, lower affinity binding interactions between CS chains and Wnt proteins. Step 5, eventually, diffusion of Wnt molecules is switched on, and Wnt can act as a paracrine signal.
Interestingly, association of C4ST-1 with tumorigenesis has been reported. For example, de-regulation of the C4ST-1 gene causes B-cell chronic lymphocytic leukemia (29), and decreases in C4ST mRNA levels are correlated with advanced tumor stage of colorectal cancer (30). Therefore, it is important to understand how Wnt/β-catenin signaling down-regulates the expression of C4ST-1. We are now searching for cis-regulatory sites in the C4ST-1 gene that are responsive to suppression mediated by Wnt-3a signaling. C4ST-1 is not a direct transcriptional target of canonical Wnt signaling because it was not suppressed by transient Wnt signaling (Fig. 6). Therefore, C4ST-1 repression may be a multistep process mediated through histone modifications. In this regard, it is reported that C4ST-1 shows a highly specific temporal and spatial expression pattern during development and is up-regulated by various growth factor signaling containing BMP2 signaling (31). Furthermore, cycloheximide experiments showed that C4ST-1 is an indirect target of BMP2, BMP7, and TGFβ signaling (31). In addition, it has been reported that bone formation and homeostasis are propelled by integrated cooperative effects of canonical Wnt and BMP pathways (32) and that C4ST-1 is controlled by BMP pathways during chondrogenesis (20). Therefore, it is suggested that C4ST-1 is regulated in a multistep event by Wnt signaling as well as BMP signaling.
Supplementary Material
Acknowledgments
We are especially grateful to Prof. Frank Tufaro (Allera Health Products, Inc., St. Petersburg, FL) for kind provision of the L mutant cell lines and to Prof. Kunitada Shimotohno (Chiba Institute of Technology, Chiba, Japan) and Assistant Prof. Makoto Hijikata (Institute for Virus Research, Kyoto University, Kyoto, Japan) for the kind gift of the reporter vector, pTcf7wt-luc. We thank Miho Ishida for construction of pcDNA3.1/zeo(−)-mC4ST-1 expression vector and Masami Ikegami for establishment of L-Wnt-3a stably expressing C4ST-1 cells.
This work was supported by the Scientific Research Promotion Fund from the Japan Private School Promotion Foundation and Grant-in-aid for Scientific Research-B 21390025 (to H. Kitagawa), and Grant-in-aid for Young Scientists (B) 20790081 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- Wg
- wingless
- BMP
- bone morphogenetic protein
- rWnt-3a
- recombinant Wnt-3a
- C4ST-1
- chondroitin 4-O-sulfotransferase-1
- CS
- chondroitin sulfate
- E disaccharide unit
- GlcUA-GalNAc(4-O-sulfate,6-O-sulfate)
- GAG
- glycosaminoglycan
- GalNAc4S-6ST
- N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase
- GlcUA
- glucuronic acid
- ΔHexUA
- unsaturated hexuronic acid
- HS
- heparan sulfate
- PG
- proteoglycan.
REFERENCES
- 1. Bartscherer K., Boutros M. (2008) EMBO Rep. 9, 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashe H. L., Briscoe J. (2006) Development 133, 385–394 [DOI] [PubMed] [Google Scholar]
- 3. Smolich B. D., McMahon J. A., McMahon A. P., Papkoff J. (1993) Mol. Biol. Cell 4, 1267–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reichsman F., Smith L., Cumberledge S. (1996) J. Cell Biol. 135, 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 6. Haltiwanger R. S., Lowe J. B. (2004) Annu. Rev. Biochem. 73, 491–537 [DOI] [PubMed] [Google Scholar]
- 7. Nadanaka S., Ishida M., Ikegami M., Kitagawa H. (2008) J. Biol. Chem. 283, 27333–27343 [DOI] [PubMed] [Google Scholar]
- 8. Uyama T., Ishida M., Izumikawa T., Trybala E., Tufaro F., Bergström T., Sugahara K., Kitagawa H. (2006) J. Biol. Chem. 281, 38668–38674 [DOI] [PubMed] [Google Scholar]
- 9. Ueda Y., Hijikata M., Takagi S., Takada R., Takada S., Chiba T., Shimotohno K. (2002) Int. J. Cancer 99, 681–688 [DOI] [PubMed] [Google Scholar]
- 10. Soriano S., Kang D. E., Fu M., Pestell R., Chevallier N., Zheng H., Koo E. H. (2001) J. Cell Biol. 152, 785–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 12. Tamura J., Neumann K. W., Kurono S., Ogawa T. (1997) Carbohydr. Res. 305, 43–63 [DOI] [PubMed] [Google Scholar]
- 13. Kitagawa H., Kinoshita A., Sugahara K. (1995) Anal. Biochem. 232, 114–121 [DOI] [PubMed] [Google Scholar]
- 14. Kusche-Gullberg M., Kjellén L. (2003) Curr. Opin Struct. Biol. 13, 605–611 [DOI] [PubMed] [Google Scholar]
- 15. Ohtake S., Kondo S., Morisaki T., Matsumura K., Kimata K., Habuchi O. (2008) Biochim. Biophys. Acta 1780, 687–695 [DOI] [PubMed] [Google Scholar]
- 16. Komekado H., Yamamoto H., Chiba T., Kikuchi A. (2007) Genes Cells 12, 521–534 [DOI] [PubMed] [Google Scholar]
- 17. Ikenoue T., Ijichi H., Kato N., Kanai F., Masaki T., Rengifo W., Okamoto M., Matsumura M., Kawabe T., Shiratori Y., Omata M. (2002) Jpn. J. Cancer Res. 93, 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de La Coste A., Romagnolo B., Billuart P., Renard C. A., Buendia M. A., Soubrane O., Fabre M., Chelly J., Beldjord C., Kahn A., Perret C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8847–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Day T. F., Guo X., Garrett-Beal L., Yang Y. (2005) Dev. Cell 8, 739–750 [DOI] [PubMed] [Google Scholar]
- 20. Klüppel M., Wight T. N., Chan C., Hinek A., Wrana J. L. (2005) Development 132, 3989–4003 [DOI] [PubMed] [Google Scholar]
- 21. Neumann S., Coudreuse D. Y., van der Westhuyzen D. R., Eckhardt E. R., Korswagen H. C., Schmitz G., Sprong H. (2009) Traffic 10, 334–343 [DOI] [PubMed] [Google Scholar]
- 22. Han C., Yan D., Belenkaya T. Y., Lin X. (2005) Development 132, 667–679 [DOI] [PubMed] [Google Scholar]
- 23. Traister A., Shi W., Filmus J. (2008) Biochem. J. 410, 503–511 [DOI] [PubMed] [Google Scholar]
- 24. Bellaiche Y., The I., Perrimon N. (1998) Nature 394, 85–88 [DOI] [PubMed] [Google Scholar]
- 25. The I., Bellaiche Y., Perrimon N. (1999) Mol. Cell 4, 633–639 [DOI] [PubMed] [Google Scholar]
- 26. Takei Y., Ozawa Y., Sato M., Watanabe A., Tabata T. (2004) Development 131, 73–82 [DOI] [PubMed] [Google Scholar]
- 27. Inatani M., Irie F., Plump A. S., Tessier-Lavigne M., Yamaguchi Y. (2003) Science 302, 1044–1046 [DOI] [PubMed] [Google Scholar]
- 28. Bülow H. E., Hobert O. (2004) Neuron 41, 723–736 [DOI] [PubMed] [Google Scholar]
- 29. Schmidt H. H., Dyomin V. G., Palanisamy N., Itoyama T., Nanjangud G., Pirc-Danoewinata H., Haas O. A., Chaganti R. S. (2004) Oncogene 23, 6991–6996 [DOI] [PubMed] [Google Scholar]
- 30. Kalathas D., Theocharis D. A., Bounias D., Kyriakopoulou D., Papageorgakopoulou N., Stavropoulos M. S., Vynios D. H. (2009) Oncol. Rep. 22, 369–375 [PubMed] [Google Scholar]
- 31. Klüppel M., Vallis K. A., Wrana J. L. (2002) Mech. Dev. 118, 77–89 [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez-Carballo E., Ulsamer A., Susperregui A. R., Manzanares-Cespedes C., Sanchez-Garcia E., Bartrons R., Rosa J. L., Ventura F. (2010) J. Bone Miner. Res., in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.