Abstract
Neuronal, cardiac, and skeletal muscle action potentials are produced and conducted through the highly regulated activity of several types of voltage-gated ion channels. Voltage-gated potassium (Kv) channels are responsible for action potential repolarization. Glycans can be attached to glycoproteins through N- and O-linkages. Previous reports described the impact of N-glycans on voltage-gated ion channel function. Here, we show that sialic acids attached through O-linkages modulate gating of Kv2.1, Kv4.2, and Kv4.3. The conductance-voltage (G-V) relationships for each isoform were shifted uniquely by a depolarizing 8–16 mV under conditions of reduced sialylation. The data indicate that sialic acids modulate Kv channel activation through apparent electrostatic mechanisms that promote channel activity. Voltage-dependent steady-state inactivation was unaffected by changes in sialylation. N-Linked sialic acids cannot be responsible for the G-V shifts because Kv4.2 and Kv4.3 cannot be N-glycosylated, and immunoblot analysis confirmed Kv2.1 is not N-glycosylated. Glycosidase gel shift analysis suggested that Kv2.1, Kv4.2, and Kv4.3 were O-glycosylated and sialylated. To confirm this, azide-modified sugar residues involved specifically in O-glycan and sialic acid biosynthesis were shown to incorporate into all three Kv channel isoforms using Cu(I)-catalyzed cycloaddition chemistry. Together, the data indicate that sialic acids attached to O-glycans uniquely modulate gating of three Kv channel isoforms that are not N-glycosylated. These data provide the first evidence that external O-glycans, with core structures distinct from N-glycans in type and number of sugar residues, can modulate Kv channel function and thereby contribute to changes in electrical signaling that result from regulated ion channel expression and/or O-glycosylation.
Keywords: Carbohydrate Chemistry, Cell Surface Protein, Glycosylation, Membrane Proteins, Potassium Channels, Membrane Potential, O-Glycosylation, Sialic Acids, Voltage-gated Ion Channels
Introduction
Precise gating of voltage-gated ion channels is a crucial component in producing proper neuronal, cardiac, and skeletal muscle action potential (AP)2 waveform and duration. The orchestrated activities of voltage-gated potassium (Kv) channels are responsible for the repolarizing phases of the AP, with regulated or aberrant changes in Kv channel activity often modifying AP repolarization (1). Such AP remodeling is observed throughout physiological processes, including development, aging, and synaptic plasticity (2–4). AP remodeling also may produce pathological consequences such as long QT syndrome, deafness, and epilepsy (5–8).
Ion channels undergo significant post-translational modification, including glycosylation, which comprises upwards of 30% of the total channel mass (9). Ion channel activity can be modulated by glycosylation through isoform-specific mechanisms (10–17). Such changes in ion channel activity would lead to altered AP waveforms and thereby modulate excitability.
Glycans are attached to channel proteins through N- and O-linkages (18, 19). Several studies detailed the isoform-specific effects of N-glycans on Kv (13–17, 20) and voltage-gated Na+ (Nav) channel functions (10–12). Negatively charged sialic acids (SA) are typically the terminal residues of glycoprotein glycan structures and were shown to impact gating of various voltage-gated ion channels differentially (10–17, 20, 21). Such studies were possible because an external consensus site for N-glycosylation has been identified.
On the other hand, little is known about the role of O-glycosylation in voltage-gated ion channel activity. The enzymes regulating O-glycosylation are complex, and O-glycosylation does not have a recognized consensus sequence (22). The most prevalent form of extracellular O-glycosylation is the mucin-type, in which a GalNAc is bound to hydroxyl groups of serine or threonine side chains (19, 23).
All previous efforts to study the impact of glycosylation on Kv channel function concentrated on determining the effects of N-glycosylation or generic sialylation (13–17, 20, 21). To our knowledge, no one has reported a role for external O-glycans in Kv channel function. Kv4.2 and Kv4.3 are members of the Shal subfamily of Kv channels and are rapidly activating and inactivating (A-type) channels (1). The putative external sequences of Kv4.2 and Kv4.3 do not contain potential N-glycosylation sites; therefore, neither isoform can be N-glycosylated. Kv2.1 is a delayed rectifier with one N-glycosylation site located on the S3-S4 linker. Based on recent structural models, this site may be inaccessible to the glycosylation machinery of the cell (24, 25); consistently, brain Kv2.1 was shown to contain no N-glycans (26).
In this study, we investigated whether and how sialic acids modulate gating of Kv2.1, Kv4.2, and Kv4.3, and we probed for the presence of sialylated O-glycans using two independent methods, Click chemistry and glycosidase treatment. The data show for the first time that Kv channel isoforms are uniquely modulated by sialic acid residues attached to the channel through O-linkages.
EXPERIMENTAL PROCEDURES
CHO Cell Culture and Transfection
Pro5 and Lec2 cells were grown in minimal medium and transfected with channel cDNA as described previously (13, 27). Briefly, the cells were plated at 25–50% confluence on 35-mm dishes 24 h prior to transfection with 1 ml of Opti-MEM (Invitrogen) containing 8 μl of Lipofectamine (Invitrogen), 0.25 μg of enhanced GFP, and 2.5 μg of channel cDNA (rat) and incubated at 37 °C in a 5% CO2 humidified incubator. 24 h post-transfection, the media were replaced with growth medium, consisting of α-minimum essential medium (Invitrogen) with (Pro5) and without (Lec2) ribo- and deoxyribonucleosides supplemented with 10% fetal bovine serum (FBS; Hyclone) and penicillin/streptomycin (Mediatech). Cells were incubated at 37 °C for another 48 h prior to commencing electrophysiological recordings.
Whole Cell Recordings in CHO Cells
The Pro5/Lec2 expression system previously was used successfully to determine the effects of sialic acids on channel gating (10–17, 21). Whole cell current recordings were performed using pulse protocols, solutions, whole cell patch clamp techniques, and data analyses as described previously (13, 14). All experiments were conducted at room temperature, ∼22 °C. Drummond capillary tubes were pulled into electrodes with a resistance of 1–2 megohm using a Model P-97 Sutter electrode puller. Series resistance was compensated 95–98%. The extracellular solution was (in mm) 65 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 155 sucrose, 5 glucose, 10 Hepes (pH 7.3); and the intracellular solution used was (in mm) 70 KCl, 65 KF, 5 NaCl, 1 MgCl2, 10 EGTA, 5 glucose, 10 Hepes (pH 7.3). The extracellular low divalent cation solution was similar to the extracellular solution listed above, with the exception of a 0.2 mm concentration of CaCl2 and a 0.1 mm concentration of MgCl2. To ensure complete dialysis of the intracellular solution, data were collected at least 10 min after attaining whole cell configuration.
Pulse Protocols
The steady-state and kinetic gating parameters were examined through the use of standard pulse protocols and solutions previously described by our laboratory (13, 14).
G-V Relationship
Cells were held at −120 mV, stepped to more depolarized potentials (−100 to +40 mV in 10-mV increments) for 100 ms (Kv4.2 and Kv4.3) and 200 ms (Kv2.1), and returned to the holding potential. Consecutive pulses were stepped every 1.5 s, and the data were leak-subtracted using the P/4 method, stepping negatively from the holding potential. Steady-state whole cell conductance values (G) were determined using Ohm's law (G = I/(V − Ek)), where I is the measured peak current at a test potential, V. Ek is the predicted K+ Nernst equilibrium potential (+84 mV) for the set of intra- and extracellular solutions used. The maximum conductance generated by each cell was used to normalize the data for each cell to its maximum conductance by fitting the data to a single Boltzmann distribution (Equation 1, solving for maximal conductance). These single Boltzmann distributions were used to determine the average Va ± S.E. and Ka ± S.E. values listed in Table 1. The normalized data were averaged with those from the other cells, and the resulting average G-V curve was fit via least squares using the following Boltzmann relationship,
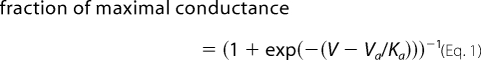 |
where V is the membrane potential; Va is the voltage of half-activation, and Ka is the slope factor.
TABLE 1.
Measured gating parameters for Kv4.2, Kv4.3, and Kv2.1
Data are the means ± S.E. Va indicates voltage of half-activation; Ka indicates Boltzmann activation slope factor; Vi indicates voltage of half-inactivation; and Ki indicates Boltzmann inactivation slope factor. Significance was tested using a two-tailed Student's t test to compare gating parameters between Pro5 (+SA) and Lec2 (−SA) cells. * indicates significance (p < 0.005); # indicates not significant (p > 0.05).
| Isoform ± SA | n | Va | Ka | Vi | Ki |
|---|---|---|---|---|---|
| mV | mV | mV | mV | ||
| Kv4.2 +SA | 8 | −20.7 ± 1.7 | 15.6 ± 0.9 | −66.2 ± 2.8 | −7.8 ± 0.6 |
| Kv4.2 −SA | 11 | −4.5 ± 2.2* | 16.9 ± 0.6# | −67.2 ± 2.6# | −6.7 ± 0.4# |
| Kv4.3 +SA | 10 | −16.4 ± 2.2 | 13.3 ± 0.6 | −51.3 ± 2.1 | −5.9 ± 0.9 |
| Kv4.3 −SA | 13 | −8.3 ± 1.3* | 14.2 ± 0.5# | −57.3 ± 2.5# | −6.1 ± 0.4# |
| Kv2.1 +SA | 14 | −12.9 ± 1.9 | 13.6 ± 0.3 | −39.3 ± 4.9 (n = 3) | −14.4 ± 0.9 |
| Kv2.1 −SA | 12 | −3.7 ± 1.8* | 13.3 ± 0.4# | −36.4 ± 5.1# (n = 3) | −12.8 ± 1.0# |
Steady-state Inactivation Curves (hinf)
To determine the steady-state channel availability for the fast-inactivating Kv4.2 and Kv4.3 isoforms, cells were prepulsed from −120 to +20 mV (10-mV increments) for 1000 ms, then stepped to +60 mV for 100 ms, and returned to the holding potential (−120 mV). To discern the effects of glycosylation on Kv2.1 steady-state slow inactivation, cells were pre-pulsed from −110 mV to +40 mV (10 mV increments) for 5 s, stepped to +30 mV for 500 ms, and returned to the holding potential (−120 mV) for 15 s, similar to that described previously (28, 29).
For both protocols, the maximum current generated by each cell was used to normalize the data for each cell to its maximum current by fitting the data to a single Boltzmann distribution (Equation 2, solving for maximal current), from which the mean Vi ± S.E. and Ki ± S.E. values listed in Table 1 were determined.
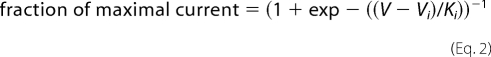 |
where V is the membrane potential; Vi is the voltage of half-inactivation, and Ki is the slope factor.
Recovery from Fast Inactivation (for Kv4.2 and Kv4.3)
Cells were held at −120 mV and stepped to +60 mV for 100 ms and then stepped to a −140-mV recovery potential for various time intervals (10–200 ms in 10-ms increments). The membrane potential then was stepped to +60 mV for 100 ms. Peak currents measured during the +60-mV depolarizations were compared, and the fractional peak current that remained during the second depolarization was plotted as a function of the recovery pulse duration, representative of the fraction of channels recovered from inactivation during the recovery interval.
Kv2.1 Channel Mutagenesis
Kv2.1 vector construction and mutagenesis were performed similar to that described previously (13, 14). A Kv2.1 plasmid encoding the 857-amino acid form of the channel protein was utilized throughout this study. The asparagine residue located at site 283 was mutated to glutamine. Generally, the cDNA containing the α-subunit open reading frame was inserted into pcDNA3.1. Using the Stratagene QuikChange IIXL site-directed mutagenesis kit, mutagenesis was completed, and the constructs were sequenced.
Whole Cell Homogenization
Cells were rinsed with cold PBS and incubated for 5 min in ice-cold sodium pyrophosphate buffer with protease inhibitors as described previously (13, 14). Cells were then homogenized using manual tissue grinders. The homogenates were centrifuged for 10 min at 1000 × g in a Beckman bench-top centrifuge. The supernatant was centrifuged in a Beckman ultracentrifuge for 1 h at 100,000 × g after which the pellet was resuspended in an appropriate volume of sodium pyrophosphate buffer containing protease inhibitors. The lysates were then stored at −80 °C. Protein levels were determined using the Pierce BCA protein assay kit.
Deglycosylation of Homogenates
Several sets of glycosidases were used to remove N- and O-glycans and sialic acids. For experiments shown in Fig. 3, two different N-glycanase enzymes were used, PNGase-F (5 units/10 μg of protein, Sigma) and N-glycanase (5 milliunits/10 μg of protein for Kv1.4; 5 milliunits/2 μg of protein for Kv2.1, Glyko). Lysates were treated for 3 h at 37 °C. For removal of sialic acids and O-glycans, as done in Fig. 6 and Table 2, lysates were treated for 3 h at 37 °C with at least 1 milliunit/10 μg of protein O-glycanase and with at least 4 milliunits/10 μg of protein sialidase A (both from Glyko). Sialic acids must be removed from O-glycans in order for O-glycanase to work properly, and therefore, the O-glycanase-treated samples were treated with sialidase A for 1.5 h at 37 °C prior to O-glycanase treatment.
FIGURE 3.
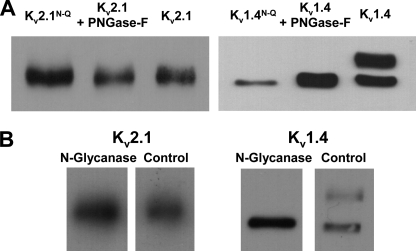
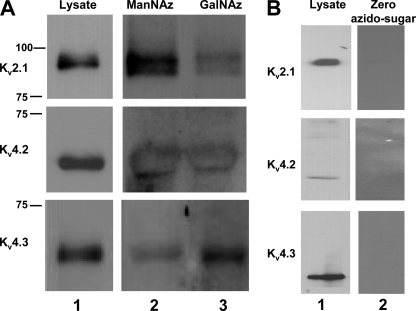
Kv2.1 is not likely N-glycosylated. Immunoblots of Kv2.1 and Kv1.4 ± N-glycosylation. Lysates isolated from transfected Pro5 cells. A, N-glycosylation site mutant and PNGase-treated lysates. Kv2.1 (left) and Kv1.4 (right). Lane 1, N-glycosylation site mutant; lane 2, PNGase-F treated; lane 3, untreated control. B, N-glycanase-treated. Lane 1, N-glycanase-treated; lane 2, untreated control. Kv2.1, 1.5–2.0 μg of protein loaded/lane. Kv1.4, 10 μg of protein loaded/lane.
FIGURE 6.
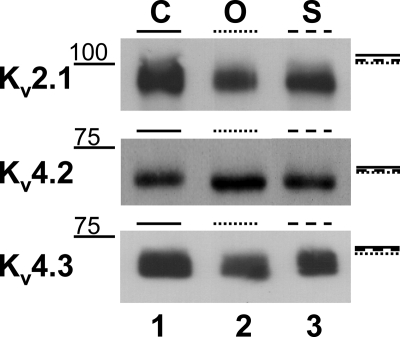
Glycosidase gel shift analyses suggest that Kv2.1, Kv4.2, and Kv4.3 are O-glycosylated and sialylated. Immunoblots of Kv2.1, Kv4.2, and Kv4.3 Pro5 lysates are shown. Lane 1, untreated control (C); lane 2, O-glycanase-treated (O); lane 3, sialidase treated (S). Molecular weight markers noted on left side of blot. Lines along blot right side demarcate the top of the control (solid), O-glycanase-treated (dotted), and sialidase-treated (dashed) bands. 2 (Kv2.1) to 25 (Kv4.2) μg of protein were loaded/lane. See Table 2 for mean Mr ± S.E. for 4–8 blots/isoform.
TABLE 2.
Electrophoretic mobility of Kv2.1, Kv4.2, and Kv4.3 ± sialidase and O-glycanase treatments
Data are the mean Mr ± S.E. for Kv2.1, Kv4.2, and Kv4.3 whole Pro5 cell lysates. Significance was tested using a two-tailed Student's t-test to compare untreated controls with treated lysates. * indicates significance (p < 0.005), or between treated lysates, # indicates significance (p < 0.05).
| Isoform | n | Untreated control | Sialidase-treated | O-Glycanase-treated |
|---|---|---|---|---|
| kDa | kDa | kDa | ||
| Kv2.1 | 6 | 105.2 ± 0.5 | 101.0 ± 0.6* | 98.9 ± 0.4*# |
| Kv4.2 | 4 | 65.6 ± 0.1 | 63.4 ± 0.5* | 61.6 ± 0.5*# |
| Kv4.3 | 8 | 69.0 ± 0.8 | 66.5 ± 0.7* | 64.2 ± 0.8*# |
Immunoblots
Immunoblot analysis was performed as described previously (10, 13, 14). Briefly, cell homogenates (2–25 μg/lane) were combined with 1 volume of 2× sample buffer (10% glycerol, 5% 2-mercaptoethanol, 3% SDS, and 12.5% upper Tris buffer) and denatured for 3 min in boiling water. Samples were then run on a 6.0–6.5% SDS-polyacrylamide gel for 90 min at 75–110 mV and transferred onto a nitrocellulose membrane using a semi-dry transfer cell (Bio-Rad). Monoclonal anti-Kv2.1, anti-Kv4.2, and anti-Kv4.3 antibodies (1:500–1000 dilution; NeuroMab) were utilized to detect Kv2.1, Kv4.2, and Kv4.3, respectively. After incubation with primary antibody, the blot was treated with a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:25,000 dilution; Jackson ImmunoResearch) and visualized using an enhanced chemiluminescence kit (Pierce).
Click-iT Glycoprotein Labeling and Detection
Click-iT glycoprotein metabolic labeling reagents and detection kits were utilized for these reactions (Invitrogen). Pro5 cells were transfected with each of the Kv channel isoforms and with the expression vector pcDNA3.1 alone, as described previously (10, 11, 13, 14). Eight hours post-transfection, the cells were re-plated in the presence of the tetraacetylated azido sugar of interest at a concentration of 2.5 million cells/100-mm dish and incubated for 48–72 h at 37 °C. Tetraacetylated azido-modified sugars were incorporated into protein glycan structures through the permissive characteristics of oligosaccharide synthesis. N-Azidoacetylgalactosamine (Ac4GalNAz) was metabolically integrated into O-glycosylation structures through the GalNAc salvage pathway. N-Azidoacetylmannosamine (Ac4ManNAz) was utilized to incorporate a modified sugar into the sialic acid biosynthesis pathway. The cells were then harvested and lysed. Azide-modified glycoprotein samples were labeled with a biotin-alkyne and precipitated (30, 31). The precipitated sample was resolubilized and incubated on streptavidin-bound Dynabeads (Invitrogen). Because of the strong interaction between streptavidin and biotin, this step allowed for the isolation of only those proteins with the modified sugar incorporated into their glycosylation structures. Immunoblot analysis was performed on the samples, and channel-specific antibodies were used to probe for channels of interest (1:100–1000 dilution) as described above. Three different controls were used. 1) Samples from the first wash following incubation of the biotinylated-modified glycoprotein samples with streptavidin-bound beads were run on the gel. Little to no signal was detected in the wash samples upon incubation with channel-specific antibody, indicating that biotin-labeled Kv channel proteins bound to streptavidin specifically and with apparent high efficiency. 2) The pcDNA3.1-transfected Pro5 cell line was treated identically to the Kv channel transfectants. No bands were observed for any of the pcDNA3.1-transfected blots. 3) Cells were not incubated with the azido-modified sugars, but all other steps of the process were completed. As shown in Fig. 6, no bands were observed. The three controls combined with the positive bands observed in the experimental samples indicate that azido-modified sugars incorporated specifically into each of the three Kv channel isoforms. The experiment was repeated at least five times for each Kv channel isoform, with nearly identical results.
Data Analysis
Electrophysiological data were analyzed using Pulse/PulseFit (HEKA) and Sigma Plot (SSPS Inc.) as described previously (10–14, 32, 33). Glycosidase gel shift data were analyzed using the Quantity One one-dimensional gel analysis software (Bio-Rad), as described previously (32).
RESULTS
Several Kv channel isoforms, including Kv2.1, Kv4.2, and Kv4.3, either do not contain N-glycosylation consensus sequences (Kv4.2 and Kv4.3) or are not likely N-glycosylated (Kv2.1) (26). One previous report showed that the cardiac transient outward current (Ito), which is generated in the mouse primarily by the activity of Kv4.2 and Kv4.3, was altered under conditions of reduced sialylation (21). To question whether sialic acids attached to channel O-glycans modulate the function of Kv channels that are not N-glycosylated, these three isoforms were expressed in two Chinese hamster ovary (CHO) cell lines that differentially sialylate glycoproteins. Pro5 cells are fully sialylating (+SA), whereas Lec2 cells are essentially nonsialylating (−SA) due to a deficiency in the CMP-sialic acid transporter (34). This deficiency serves as a model for a form of congenital disorders of glycosylation (CDG), CDG type IIf (35). The Pro5/Lec2 cell system has been used by several investigators, including our laboratory, to question how sialic acids modulate ion channel gating (10–16, 21). Channel sialylation was reduced through a second independent mechanism, enzymatic desialylation through sialidase treatment. Enzymatic desialylation of Lec2 cells was used as a negative control. As shown previously by our laboratory and others, channels expressed in Lec2 cell lines (± sialidase treatment) and fully glycosylated channels treated with sialidase showed nearly identical gating characteristics (15, 21, 36, 37). Kv channel O-glycosylation and sialylation were confirmed through two independent methods, O-glycosidase gel shift analysis and Click chemistry.
Sialic Acids Modulate Kv4.2 and Kv4.3 Activation
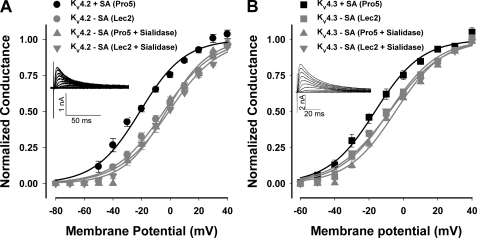
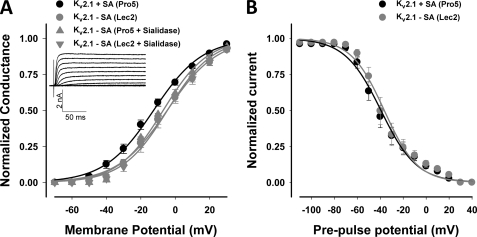
Kv4.2 and Kv4.3, members of the Shal subfamily of Kv channels, are rapidly activating and inactivating (A-type) channels. Neither Kv4.2 nor Kv4.3 contains an external N-glycosylation site (1). Fig. 1 shows the G-V relationships measured for both isoforms as expressed in Pro5 and Lec2 cells and following sialidase treatment. For all three conditions of reduced sialylation, note that the G-V relationships and the voltages of half-activation (Va) for Kv4.2 and Kv4.3 were shifted significantly, linearly, and nearly identically (among conditions of reduced sialylation) to more depolarized potentials compared with control (by 16 and 8 mV, respectively; Fig. 1 and Table 1). These data indicate that channels expressed in the Lec2 cell line behaved similarly to those following enzymatic desialylation, consistent with previous work, demonstrating the efficacy of the Lec2 cell line in mimicking the desialylated protein state (15, 21, 36, 37). Additionally, the data indicate that sialic acids impact Kv4.2 and Kv4.3 gating, causing a leftward shift in voltage-dependent activation.
FIGURE 1.
Sialic acids modulate Kv4.2 and Kv4.3 activation. Conductance-voltage (G-V) relationships for Kv4.2 (A) and Kv4.3 (B) under conditions of full (Pro5) and reduced sialylation (Lec2, sialidase-treated Pro5 and Lec2) are shown. Data are the mean normalized peak conductances ± S.E. and are fit to single Boltzmann relationships (lines). Insets, typical whole cell Kv4.2 and Kv4.3 current traces. Black lines/symbols, +sialic acids (Pro5, n = 8–10; circles, Kv4.2; squares, Kv4.3); Gray lines/symbols, −sialic acids (Lec2, n = 11–13 (circles, Kv4.2; squares, Kv4.3); sialidase-treated Pro5 (triangles) or Lec2 (inverted triangles), n = 3–6).
Kv4.2 and Kv4.3 Fast Inactivation and Recovery from Fast Inactivation Were Not Altered by Changes in Channel Sialylation
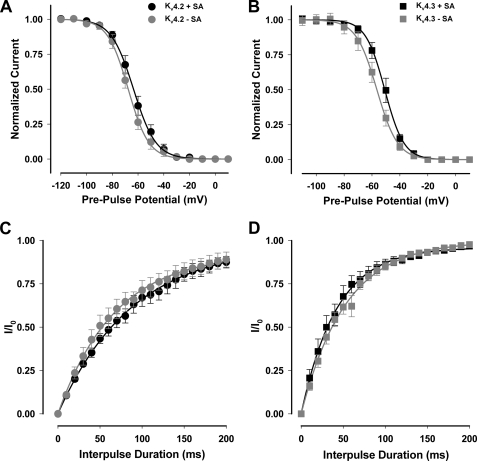
Kv4.2 and Kv4.3 are rapidly inactivating channels that undergo N-type fast inactivation. To determine whether sialic acids modulate fast inactivation, we measured steady-state inactivation and recovery from fast inactivation for the two Kv4 isoforms ± sialic acids. As shown in Fig. 2 and Table 1, Kv4.2 and Kv4.3 channel sialylation had no measurable effect on steady-state fast inactivation or recovery from fast inactivation.
FIGURE 2.
Kv4.2 and Kv4.3 fast inactivation and recovery from fast inactivation are not affected by sialic acids. Fast inactivation and recovery from fast inactivation for Kv4.2 (circles) and Kv4.3 (squares) are expressed under conditions of full and reduced sialylation. Data are means ± S.E. A and B, steady-state channel availability (inactivation). Curves are single Boltzmann distribution fits to the data (lines). C and D, recovery from fast inactivation at a −140 mV recovery potential. Curves are single exponential fits to the data. Black lines/symbols, +sialic acids (Pro5); gray lines/symbols, −sialic acids (Lec2). A and C, Kv4.2. B and D, Kv4.3. n = 8–13.
Kv2.1 Is Not Likely N-Glycosylated
Kv2.1 is a delayed rectifier channel that produces a slowly inactivating current and contains one N-glycosylation site located on the S3-S4 linker (1). Previously, Kv2.1 was shown to not be N-glycosylated in the brain (26) or in COS cells (38). To determine whether Kv2.1 at site 283N is N-glycosylated in CHO cells, a Kv2.1 N-glycosylation mutant (Kv2.1N283Q) was generated by mutating the asparagine residue that initiates the potential N-glycosylation consensus sequence to a glutamine. In addition, fully glycosylated Kv2.1 lysates were treated with two N-glycanases, PNGase-F and N-glycanase. These enzymes were used to remove the full N-glycan structures, as previously performed by our laboratory (13, 14, 33). Immunoblot analyses showed that the electrophoretic mobilities (Mr) of untreated Kv2.1, Kv2.1N283Q, and Kv2.1 treated with each N-glycanase were nearly identical, consistent with Kv2.1 having no N-glycans attached to the channel protein as expressed in CHO cells (Fig. 3). Conversely, note the large shift in Mr of Kv1.4 following PNGase-F and N-glycanase treatment; this was used as an example of the efficacy of removing channel N-glycans by enzymatic treatment, as Kv1.4 is a Kv channel isoform previously shown to be N-glycosylated (14, 26). There are several possible explanations for the lack of glycosylation of the potential Kv2.1 N-glycosylation site. A likely possibility is that the N-glycosylation site, located on the S3-S4 linker, is not accessible to the glycosylation machinery (as suggested by the Kv channel crystal structures (24, 25)). Thus, addition of N-glycans would be prevented.
Sialic Acids Modulate Kv2.1 Activation
To determine whether sialic acids impact Kv2.1 channel function, channel gating was studied under conditions of full and reduced sialylation. Under all three conditions of reduced sialylation, Kv2.1 activated at more depolarized potentials than the fully sialylated channel (Fig. 4A and Table 1). The G-V relationship and Va for the less sialylated Kv2.1 were shifted linearly by a significant depolarizing ∼9 mV compared with fully sialylated channels. The data indicate that a stronger depolarization is required to activate Kv2.1 in the absence of sialic acids (Fig. 4 and Table 2).
FIGURE 4.
Sialic acids modulate Kv2.1 activation but do not affect slow inactivation. G-V (A) and steady-state inactivation (B) relationships for Kv2.1 under conditions of full and reduced sialylation. Data are the means ± S.E. and are fit to single Boltzmann relationships (lines). Inset (A), typical whole cell Kv2.1 current traces. Black lines/circles, +sialic acids (Pro5, n = 14); gray lines/symbols, −sialic acids (Lec2 (circles), n = 12; sialidase-treated Pro5 (triangles) or Lec2 (inverted triangles), n = 3–4).
Sialic Acids Do Not Affect Kv2.1 Slow Inactivation
Kv2.1 does not undergo fast N-type inactivation but does slowly inactivate. Thus, we measured the voltage dependence of steady-state slow inactivation for Kv2.1 in Pro5 and Lec2 cells (Fig. 4B and Table 1). There was no significant difference measured in the steady-state inactivation curves or in the voltages of half-inactivation (Vi) under conditions of full and reduced sialylation. These data are consistent with that previously reported by our laboratory for Kv1.5 (14). However, Kv2.1 steady-state inactivation does not show U-type inactivation, as reported previously by some laboratories for Kv2.1 expressed in Xenopus oocytes and HEK cells (39–42). Instead, the steady-state inactivation curves shown in Fig. 4B are similar to that reported by others in recent studies using comparable pulse protocols to measure Kv2.1 inactivation in other mammalian cells (29, 43–47).
Electrostatic Mechanisms Account for the Effect of Sialic Acids on Gating of Three Kv Channel Isoforms
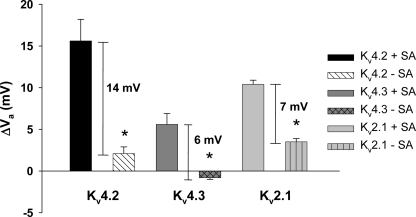
Sialic acids are negatively charged residues at physiological pH that may contribute to the external negative surface potential, impacting voltage-dependent channel gating. With increased negative surface charge, the channel would activate at less depolarized potentials, i.e. if sialic acids contribute to a negative surface potential, then channels with greater levels of sialic acids would be predicted to activate at less depolarized potentials than channels with reduced levels of sialylation. The presence of external divalent cations will screen this surface potential and limit the impact of the negatively charged sialic acid residues on channel gating (48, 49). Here, the Va for Kv2.1, Kv4.2, and Kv4.3 as expressed under conditions of full and reduced sialylation were measured and compared at two different divalent cation concentrations. The data show the Va for each isoform as expressed under conditions of full sialylation was significantly more sensitive to changes in external divalent concentrations than were the Va values for the isoforms expressed in Lec2 cells (Fig. 5). These data are consistent with an electrostatic mechanism by which sialic acids alter the activation of Kv4.2, Kv4.3, and Kv2.1.
FIGURE 5.
Activation of three Kv channel isoforms is modulated through apparent electrostatic interactions. The Va values for Kv2.1, Kv4.2, and Kv4.3 expressed under conditions of full and reduced sialylation were measured and compared at normal and low (10-fold reduction) external divalent cation concentrations. Data are mean hyperpolarizing shifts in Va with a 10-fold reduction in external divalent concentration (ΔVa) ± S.E. Note the significantly larger ΔVa in the presence of sialic acids. Solid bars, +sialic acid (Pro5); patterned bars, −sialic acid (Lec2). n = 3–4. Significance (*) tested using two-tailed Student's t test comparing Va shifts measured in Pro5 versus Lec2 cells (p < 0.0005).
Kv2.1, Kv4.2, and Kv4.3 Channels Are O-Glycosylated and Sialylated
Data shown here indicate that sialic acids attached to three Kv channel isoforms uniquely impact channel activation. Despite a number of studies investigating whether and how N-glycans modulate Kv channel function, the putative effects of extracellular O-glycans on voltage-gated ion channel function have not been defined. Kv4.2 and Kv4.3 do not contain N-glycosylation sites, and we show here that Kv2.1 is not N-glycosylated as expressed in CHO cells (Fig. 3). Thus, the data suggest it is likely that sialic acids attached to O-glycans are responsible for the observed effects on channel gating.
To question whether Kv2.1, Kv4.2, and Kv4.3 are O-glycosylated and sialylated, we employed previously used methods of glycosidase gel shift analysis (10, 14, 32, 33). Fig. 6 shows typical blots for the three isoforms under control and O-glycosidase- and sialidase-treated conditions. Following repeated experiments (n = 4–8 for each isoform, see Table 2), the data show a significant shift in O-glycosidase-treated samples compared with untreated control and sialidase-treated lysates, suggesting that each isoform is O-glycosylated and sialylated.
Although the gel shifts observed following O-glycosidase and sialidase treatments are significant, they are small. This is not surprising given the relatively small size of O-glycan structures, i.e. O-glycans typically consist of only a few sugar residues; one would expect to observe only a small shift in Mr following removal of the O-glycan structure. In addition, the complete removal of glycans using glycosidase (particularly O-glycanase) treatment is difficult. Thus, we employed a second independent method to question whether the Kv channel isoforms were O-glycosylated and sialylated using Click chemistry (30, 31, 50, 51). Click chemistry previously was used effectively to identify the appearance and presence of O-glycosylation during zebra fish development (52). Tetraacetylated azido-modified sugars, Ac4GalNAz (O-glycosylation) or Ac4ManNAz (sialylation), were incorporated into protein glycan structures and bound with biotin-alkyne for Pro5 cells transfected individually with each Kv channel isoform. Because of the strong interaction between streptavidin and biotin, the biotinylated samples were incubated with streptavidin-bound magnetic beads to isolate and precipitate only those proteins with the modified sugar incorporated into the protein glycan structures. The streptavidin precipitates were then run on 6.0–6.5% SDS-polyacrylamide gels, and antibodies specific for each Kv channel isoform were used to visualize O-glycosylated and sialylated channels. Immunoblot analysis of the Ac4GalNAz and Ac4ManNAz samples detected bands at the appropriate molecular weights for Kv2.1, Kv4.2, and Kv4.3 (Fig. 7), as described under “Experimental Procedures.” The three controls, along with the positive banding data shown in Fig. 7, indicate that the O-glycosylation and sialic acid-specific azido-modified sugar residues incorporated into all three Kv channel isoforms. This set of experiments was repeated at least six times per isoform, with similar results. These data corroborate the glycosidase gel shift analysis shown in Fig. 6 and suggest that each Kv channel isoform tested is O-glycosylated and sialylated.
FIGURE 7.
Click-iT chemistry confirmed the presence of sialylated O-glycans attached to Kv2.1, Kv4.2, and Kv4.3. A, immunoblots of Kv2.1, Kv4.2, and Kv4.3 Pro5 lysates. Lane 1, untreated whole cell lysates; lane 2, Ac4ManNAz-labeled (sialic acid) samples; lane 3, Ac4GalNAz-labeled (O-glycosylated) samples. Molecular weight markers are noted to the left of lane 1. B, control treatment in the absence of azido-modified sugars. Lane 1, untreated whole cell lysates. Lane 2, biotin-treated, streptavidin-precipitated samples. Top, Kv2.1. Middle, Kv4.2. Bottom, Kv4.3.
DISCUSSION
Sialylated O-Glycans Modulate Kv2.1, Kv4.2, and Kv4.3 Gating Uniquely through Apparent Electrostatic Mechanisms
We examined the role of sialic acids attached to Kv channel O-glycans on the gating of rat Kv2.1, Kv4.2, and Kv4.3. Figs. 1 and 4 show that a reduction in sialic acids causes a significant and depolarizing shift in the G-V relationship that is unique for each isoform. Furthermore, the data indicate that sialic acids modulate the activity of each isoform through apparent electrostatic mechanisms. Based on the surface potential theory, negative charges on the outer surface of the membrane (e.g. sialic acids) generate a surface potential. A depolarization would be sensed by the channel gating mechanism with increases in external negative surface charge, and thus, channel activation would require a smaller depolarization (48, 49). Extracellular divalent cations should act to limit the effects of negatively charged sialic acid residues, effectively reducing the negative surface potential, and cause a shift in the Va to more depolarized potentials. The greater the surface potential, the larger the predicted shift in the Va with changes in extracellular divalent cation concentration because there are effectively more negative surface charges that can be screened by divalent cations. We found that the Va for channels expressed in the fully sialylating Pro5 cells were more sensitive to changes in extracellular divalent cation concentration than channels expressed in the Lec2 cells (−SA, Fig. 6). Thus, sialic acids modulate Kv2.1, Kv4.2, and Kv4.3 activation through apparent electrostatic mechanisms (48, 49). Furthermore, changes in sialylation shifted the G-V curve for each isoform nearly linearly; there was no significant difference in the Boltzmann slope factors (Ka values) with changes in sialylation (Figs. 1 and 4; Table 1). This suggests that changes in Kv2.1, Kv4.2, and Kv4.3 sialylation primarily alter the negative surface potential, with little to no effect on the stability of channels among functional states. These data are consistent with our recent findings for Kv1.5 (14).
Channel availability for each isoform and recovery from fast inactivation for Kv4.2 and Kv4.3 were not altered by channel sialylation levels (Fig. 2 and Table 1). Some recent studies showed that the surface charge effects of sialic acids on channel voltage-dependent gating typically and similarly affected all voltage-dependent gating mechanisms (13, 16), i.e. a shift in fast inactivation voltage similar to the observed linear shift in channel activation voltage is consistent with an electrostatic mechanism. In this study, we show that sialic acids shift only the G-V relationship for each isoform, with no effect on inactivation or recovery from fast inactivation (for Kv4.2 and Kv4.3). The data also indicate that sialic acids modulate activation voltage by contributing to the negative surface potential (Fig. 5). Together, these data suggest that sialic acids affect channel gating through electrostatic mechanisms, but do not confer a uniform effect on all voltage-dependent gating mechanisms. One likely explanation is that sialic acids impose a variable effect on the electric field sensed by the channel gating mechanisms, such that only activation is affected. As reviewed by Hille (1), contribution of local surface potentials to voltage-dependent gating may vary with each gating step and between forward and reverse gating kinetics. Our recent work on Kv1.5 showed a similar phenomenon (14). Thus, an inhomogeneous effect of sialic acids on the local electric field could account for the variable effect of reduced sialylation among voltage-dependent gating characteristics such as channel activation, inactivation, and recovery from fast inactivation with changes in channel sialylation.
A single previous report indicated that sialic acids impact gating of cardiac Ito and Kv4.3 (21). Specifically, sialic acids were shown to modulate Ito of primarily isolated adult ventricular myocytes. Ito in the mouse is composed largely of Kv4.2 and Kv4.3, neither of which can be N-glycosylated. Thus, this previous report strongly suggests that Kv4.3 and likely Kv4.2 are sialylated in vivo. However, the study did not determine whether the channel sialic acids were attached through N- or O-linkages. Our data are in general agreement with the previous report with respect to Kv4.3 gating in vitro; Kv4.3 channel activation voltage was shifted to depolarized potentials under conditions of reduced sialylation. The preceding report indicated a small, but significant, shift in channel inactivation voltage that we did not observe. There are several possible reasons for the relatively minor differences between our data and that previously described, including the use of different external and internal solutions to record K+ currents. It was shown in the previous study that sialolipids contribute at most 3–4 mV to Kv4.3 channel gating. We measure a much larger sialic acid-dependent shift for Kv4.3 (∼8 mV) and Kv2.1 gating (∼9 mV), with an even greater shift observed for Kv4.2 gating (∼16 mV). Together, these data indicate that most of the effect of sialic acids on gating of these three Kv channel isoforms is caused by channel sialic acids attached to O-glycans.
Although Kv4.2 and Kv4.3 are homologous proteins, changes in channel sialylation have a much greater impact on Kv4.2. This suggests that O-linked sialic acids attached to Kv4.2 and Kv4.3 impact channel activation differently. Apparently, Kv4.2 and Kv4.3 O-linked sialic acids are different in number and/or location relative to the channel activation mechanisms. Further studies are required to determine how specific changes in the number and/or location of O-linked sialic acids impact Kv4 channel activation.
In general, N- and O-glycan structures are very diverse, even the core structures for N- and O-glycans are distinct from one another. N-Glycans tend to be significantly larger and more branched than O-glycans, which are typically a few residues in length. The smaller (shorter) O-glycans would likely be more rigid structures located closer to the membrane surface. Sialic acids typically are attached to the terminus of both N- and O-glycans, which would likely place sialic acids attached to N- and O-glycans at different locations relative to the membrane, channel, and electric field. Thus, the sialic acids attached to the end of the shorter O-glycan structures likely affect channel gating differently than sialic acid residues terminally located on a much longer and branched N-glycan. This becomes more apparent when comparing the data showing changes in Kv channel gating with reduced glycosylation and sialylation. A spectrum of effects was observed, ranging from no measurable effect to ∼20-mV shifts on activation only (14), to large effects on several (or all) voltage-dependent gating parameters (13, 15–17), to apparent changes in stability of the channel among functional states (13, 16). These variable effects alone suggest that the location (in three dimensions) of the sialic acids relative to the channel gating mechanism likely contributes to the measured impact of sialic acids on channel gating.
Here, the impact of O-linked sialic acids on Kv gating is variable in magnitude for two Kv4 isoforms (Kv4.2 versus Kv4.3), but the mechanism appears to be similar. However, the magnitude and mechanism by which N-linked sialic acids modulate orthologous Kv channel gating appear to be strictly isoform-specific, as we and others have published previously (13–17). Together, there is abundant evidence that sialic acids attached to N- and O-glycans can, and do, modulate Kv channel isoforms through isoform-specific mechanisms that include the following: the magnitude of an electrostatic effect, the impact of the electrostatic effect on gating, and/or the contribution to channel state stability.
None of the Kv channel isoforms investigated in this study is N-glycosylated. Our data indicate that all three isoforms are O-glycosylated and sialylated (Figs. 6 and 7). Here, we show that Kv channel sialic acids attached to O-glycans modulate channel gating through isoform-specific mechanisms. No previous reports linked Kv channel extracellular O-glycosylation to channel gating. Thus, the data presented in this study reflect a novel finding that Kv channel gating can be modulated by channel sialic acids attached to O-glycans through isoform-specific mechanisms.
Physiological and Pathophysiological Implications
Normal gating of voltage-gated ion channels is essential for proper excitable cell function. Changes in ion channel function can lead to disorders such as long QT syndrome and epilepsy (5, 6, 8). Because channel glycosylation and sialylation modulate Kv channel gating through isoform-specific mechanisms, aberrant changes in Kv channel isoform expression, distribution, and glycosylation may play a role in the pathogenesis of such disorders.
Aberrant sialylation is involved in CDGs. CDGs are autosomal dominant disorders in which afflicted individuals present with severe developmental delay (53–57). CDGs are caused by mutant or missing glycogenes, primarily glycosyltransferases, resulting in glycoproteins with relatively modest reductions in the levels and types of attached sugars. Currently, there are ∼30 documented forms of CDGs, each of which are caused by the dysfunction of one gene product involved in N- or O-linked protein glycosylation (53, 57). Nearly all CDGs impact multiple organ systems, with prominent effects on neuromuscular and cardiovascular systems, resulting in hypotonia, seizures, and cardiomyopathies. Although the exact mutated glycosylation structure varies among CDGs, all CDG patients suffer from reduced protein sialylation. It is intriguing to consider whether the patient's reduced glycoprotein sialylation may be responsible for changes in ion channel activity and potentially contribute to symptoms.
Chagas disease afflicts ∼18 million people and is caused following infection by the parasite Trypanosoma cruzi. Mortality rate is ∼30% of the total cases, with nearly all of these patients experiencing heart failure preceded by ventricular tachycardia (58–62). Interestingly, T. cruzi release a neuraminidase shown to reduce the level of cardiomyocyte sialylation (63). Individuals suffering from Chagas disease experience arrhythmias and conduction anomalies more frequently than those with nonchagasic dilated cardiomyopathies (62, 64). A recent study measured mouse ECG as a function of time post-infection using two strains of T. cruzi to infect (64). The data indicated that ∼60% of infected mice (compared with ∼5% of control) showed some conduction abnormality. Could the reduced cardiomyocyte sialylation of chagasic patients lead to altered cardiac ion channel behavior and ultimately to the arrhythmias and heart failure observed in these patients?
Kv4.2 and Kv4.3 are responsible for producing a rapidly activating and inactivating cardiac current, Ito, involved in early action potential repolarization (phase 1). A previous report indicated that changes in murine ventricular Ito following neuraminidase treatment used to remove sialic acids may contribute to the extended AP duration and the increase in number of early after depolarizations observed (21). Our recent work showed that regulated changes in channel sialylation were sufficient to modulate action potential waveform, including duration (32). Increased action potential duration would be predicted if Kv channel activity was limited by reduced sialylation. With a rightward shift in the G-V relationship and no shift in the inactivation-voltage relationship, the window current, defined as the portion under the overlapping steady-state activation and inactivation curves, would be right-shifted and smaller. This would lead to an overall reduction in voltage-dependent K+ current.
Furthermore, Kv2.1, which produces a slowly inactivating current, aids in regulating excitability in cortical and hippocampal pyramidal neurons by acting as a suppressor during periods of hyperexcitability (65, 66). Reductions in Kv2.1 sialylation may limit Kv2.1 activity, disrupt this suppressive role, and thereby increase the frequency and/or duration of hyperexcitability in these neurons.
Summary
Here, the data show that reduced sialic acid levels attached to channel O-glycans cause unique depolarizing shifts in activation voltage for three Kv channel isoforms that are not N-glycosylated. An electrostatic mechanism is apparently responsible for these effects, with O-linked sialic acids contributing to the external negative surface potential. Steady-state inactivation and recovery from fast inactivation were not impacted significantly by reduced sialylation; this would lead to further reductions in Kv channel activity, likely extending the action potential. Mechanistically, this suggests an inhomogeneous influence of Kv channel sialic acids on the electric field sensed by the activation, inactivation, and recovery gating mechanisms. Together, the data indicate that channel sialic acids attached to O-glycans modulate gating of Kv channels that are not N-glycosylated, and this modulation is unique for each isoform. This is the first study to report direct effects of O-glycans on voltage-gated ion channel gating, suggesting that sialylated Kv channel O-glycans enhance channel activity. Such modulation is relevant to changes in action potential repolarization that occur as ion channel expression, distribution, and O-glycosylation are regulated.
Acknowledgments
We thank Dr. Stephen Korn and Dr. Jeanne M. Nerbonne for generously providing us with the Kv2.1 and Kv4.2/Kv4.3 cDNAs, respectively.
This work was supported by an American Heart Association predoctoral fellowship (to T. A. S.), an American Heart Association grant-in-aid (to E. S. B.), and Bridge and Challenge grants from the James and Esther King Biomedical Research Program (to E. S. B.).
- AP
- action potential
- Kv
- voltage-gated potassium
- Nav
- voltage-gated sodium
- SA
- sialic acid
- Click
- Cu(I)-catalyzed cycloaddition
- G-V
- conductance-voltage
- Ac4GalNAz
- tetraacetylated N-azidoacetylgalactosamine
- Ac4ManNAz
- tetraacetylated modified N-azidoacetylmannosamine
- CDG
- congenital disorders of glycosylation
- PNGase
- peptide:N-glycosidase.
REFERENCES
- 1. Hille B. (ed) (2001) Ion Channels of Excitable Membranes, 3rd Ed., pp. 25–60, 131,–158, 646–660, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 2. Cerbai E., Barbieri M., Mugelli A. (1996) Circulation 94, 1674–1681 [DOI] [PubMed] [Google Scholar]
- 3. Plotnikov A. N., Sosunov E. A., Patberg K. W., Anyukhovsky E. P., Gainullin R. Z., Shlapakova I. N., Krishnamurthy G., Danilo P., Jr., Rosen M. R. (2004) Circulation 110, 489–495 [DOI] [PubMed] [Google Scholar]
- 4. Zhang J. F., Robinson R. B., Siegelbaum S. A. (1992) Neuron 9, 97–103 [DOI] [PubMed] [Google Scholar]
- 5. Abriel H., Cabo C., Wehrens X. H., Rivolta I., Motoike H. K., Memmi M., Napolitano C., Priori S. G., Kass R. S. (2001) Circ. Res. 88, 740–745 [DOI] [PubMed] [Google Scholar]
- 6. Clancy C. E., Kass R. S. (2005) Physiol. Rev. 85, 33–47 [DOI] [PubMed] [Google Scholar]
- 7. Kubisch C., Schroeder B. C., Friedrich T., Lütjohann B., El-Amraoui A., Marlin S., Petit C., Jentsch T. J. (1999) Cell 96, 437–446 [DOI] [PubMed] [Google Scholar]
- 8. Smart S. L., Lopantsev V., Zhang C. L., Robbins C. A., Wang H., Chiu S. Y., Schwartzkroin P. A., Messing A., Tempel B. L. (1998) Neuron 20, 809–819 [DOI] [PubMed] [Google Scholar]
- 9. Schmidt J. W., Catterall W. A. (1987) J. Biol. Chem. 262, 13713–13723 [PubMed] [Google Scholar]
- 10. Bennett E. S. (2002) J. Physiol. 538, 675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson D., Montpetit M. L., Stocker P. J., Bennett E. S. (2004) J. Biol. Chem. 279, 44303–44310 [DOI] [PubMed] [Google Scholar]
- 12. Johnson D., Bennett E. S. (2006) J. Biol. Chem. 281, 25875–25881 [DOI] [PubMed] [Google Scholar]
- 13. Johnson D., Bennett E. S. (2008) Pflugers Arch. 456, 393–405 [DOI] [PubMed] [Google Scholar]
- 14. Schwetz T. A., Norring S. A., Bennett E. S. (2010) Biochim. Biophys. Acta 1798, 367–375 [DOI] [PubMed] [Google Scholar]
- 15. Thornhill W. B., Wu M. B., Jiang X., Wu X., Morgan P. T., Margiotta J. F. (1996) J. Biol. Chem. 271, 19093–19098 [DOI] [PubMed] [Google Scholar]
- 16. Watanabe I., Wang H. G., Sutachan J. J., Zhu J., Recio-Pinto E., Thornhill W. B. (2003) J. Physiol. 550, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe I., Zhu J., Sutachan J. J., Gottschalk A., Recio-Pinto E., Thornhill W. B. (2007) Brain Res. 1144, 1–18 [DOI] [PubMed] [Google Scholar]
- 18. Kornfeld R., Kornfeld S. (1985) Annu. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 19. Van den Steen P., Rudd P. M., Dwek R. A., Opdenakker G. (1998) Crit. Rev. Biochem. Mol. Biol. 33, 151–208 [DOI] [PubMed] [Google Scholar]
- 20. Zhu J., Watanabe I., Poholek A., Koss M., Gomez B., Yan C., Recio-Pinto E., Thornhill W. B. (2003) Biochem. J. 375, 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ufret-Vincenty C. A., Baro D. J., Santana L. F. (2001) Am. J. Physiol. Cell Physiol. 281, C464–CC474 [DOI] [PubMed] [Google Scholar]
- 22. Tian E., Ten Hagen K. G. (2009) Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanisch F. G. (2001) Biol. Chem. 382, 143–149 [DOI] [PubMed] [Google Scholar]
- 24. Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 25. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 26. Shi G., Trimmer J. S. (1999) J. Membr. Biol. 168, 265–273 [DOI] [PubMed] [Google Scholar]
- 27. Stanley P., Caillibot V., Siminovitch L. (1975) Cell 6, 121–128 [DOI] [PubMed] [Google Scholar]
- 28. McEwen D. P., Li Q., Jackson S., Jenkins P. M., Martens J. R. (2008) Mol. Pharmacol. 73, 678–685 [DOI] [PubMed] [Google Scholar]
- 29. Bocksteins E., Raes A. L., Van de Vijver G., Bruyns T., Van Bogaert P. P., Snyders D. J. (2009) Am. J. Physiol. Cell Physiol. 296, C1271–C1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tornøe C. W., Christensen C., Meldal M. (2002) J. Org. Chem. 67, 3057–3064 [DOI] [PubMed] [Google Scholar]
- 31. Laughlin S. T., Agard N. J., Baskin J. M., Carrico I. S., Chang P. V., Ganguli A. S., Hangauer M. J., Lo A., Prescher J. A., Bertozzi C. R. (2006) Methods Enzymol. 415, 230–250 [DOI] [PubMed] [Google Scholar]
- 32. Montpetit M. L., Stocker P. J., Schwetz T. A., Harper J. M., Norring S. A., Schaffer L., North S. J., Jang-Lee J., Gilmartin T., Head S. R., Haslam S. M., Dell A., Marth J. D., Bennett E. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16517–16522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stocker P. J., Bennett E. S. (2006) J. Gen. Physiol. 127, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deutscher S. L., Nuwayhid N., Stanley P., Briles E. I., Hirschberg C. B. (1984) Cell 39, 295–299 [DOI] [PubMed] [Google Scholar]
- 35. Martinez-Duncker I., Dupré T., Piller V., Piller F., Candelier J. J., Trichet C., Tchernia G., Oriol R., Mollicone R. (2005) Blood 105, 2671–2676 [DOI] [PubMed] [Google Scholar]
- 36. Bennett E., Urcan M. S., Tinkle S. S., Koszowski A. G., Levinson S. R. (1997) J. Gen. Physiol. 109, 327–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ufret-Vincenty C. A., Baro D. J., Lederer W. J., Rockman H. A., Quinones L. E., Santana L. F. (2001) J. Biol. Chem. 276, 28197–28203 [DOI] [PubMed] [Google Scholar]
- 38. Shi G., Kleinklaus A. K., Marrion N. V., Trimmer J. S. (1994) J. Biol. Chem. 269, 23204–23211 [PubMed] [Google Scholar]
- 39. Klemic K. G., Kirsch G. E., Jones S. W. (2001) Biophys. J. 81, 814–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klemic K. G., Shieh C. C., Kirsch G. E., Jones S. W. (1998) Biophys. J. 74, 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lvov A., Greitzer D., Berlin S., Chikvashvili D., Tsuk S., Lotan I., Michaelevski I. (2009) J. Biol. Chem. 284, 28276–28291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kurata H. T., Soon G. S., Eldstrom J. R., Lu G. W., Steele D. F., Fedida D. (2002) J. Biol. Chem. 277, 29045–29053 [DOI] [PubMed] [Google Scholar]
- 43. Martens J. R., Navarro-Polanco R., Coppock E. A., Nishiyama A., Parshley L., Grobaski T. D., Tamkun M. M. (2000) J. Biol. Chem. 275, 7443–7446 [DOI] [PubMed] [Google Scholar]
- 44. Mohapatra D. P., Misonou H., Pan S. J., Held J. E., Surmeier D. J., Trimmer J. S. (2009) Channels 3, 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dai X. Q., Kolic J., Marchi P., Sipione S., Macdonald P. E. (2009) J. Cell Sci. 122, 775–779 [DOI] [PubMed] [Google Scholar]
- 46. Yoshida M., Nakata M., Yamato S., Dezaki K., Sugawara H., Ishikawa S. E., Kawakami M., Yada T., Kakei M. (2010) Biochem. Biophys. Res. Commun. 396, 304–309 [DOI] [PubMed] [Google Scholar]
- 47. Yoshida M., Dezaki K., Yamato S., Aoki A., Sugawara H., Toyoshima H., Ishikawa S. E., Kawakami M., Nakata M., Yada T., Kakei M. (2009) FEBS Lett. 583, 2225–2230 [DOI] [PubMed] [Google Scholar]
- 48. Frankenhaeuser B., Hodgkin A. L. (1957) J. Physiol. 137, 218–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hille B., Ritchie J. M., Strichartz G. R. (1975) J. Physiol. 250, 34P–35P [PubMed] [Google Scholar]
- 50. Hang H. C., Yu C., Kato D. L., Bertozzi C. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14846–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prescher J. A., Dube D. H., Bertozzi C. R. (2004) Nature 430, 873–877 [DOI] [PubMed] [Google Scholar]
- 52. Laughlin S. T., Baskin J. M., Amacher S. L., Bertozzi C. R. (2008) Science 320, 664–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jaeken J., Matthijs G. (2007) Annu. Rev. Genomics Hum. Genet. 8, 261–278 [DOI] [PubMed] [Google Scholar]
- 54. Marquardt T., Denecke J. (2003) Eur. J. Pediatr. 162, 359–379 [DOI] [PubMed] [Google Scholar]
- 55. Leroy J. G. (2006) Pediatr. Res. 60, 643–656 [DOI] [PubMed] [Google Scholar]
- 56. Kranz C., Basinger A. A., Güçsavaş-Calikoğlu M., Sun L., Powell C. M., Henderson F. W., Aylsworth A. S., Freeze H. H. (2007) Am. J. Med. Genet. A 143A, 1371–1378 [DOI] [PubMed] [Google Scholar]
- 57. Freeze H. H. (2006) Nat. Rev. Genet. 7, 537–551 [DOI] [PubMed] [Google Scholar]
- 58. Higuchi, Mde L., Benvenuti L. A., Martins, Reis M., Metzger M. (2003) Cardiovasc. Res. 60, 96–107 [DOI] [PubMed] [Google Scholar]
- 59. Sternick E. B., Martinelli M., Sampaio R., Sampaio R. C., Gerken L. M., Teixeira R. A., Scarpelli R., Scanavacca M., Nishioka S. D., Sosa E. (2006) J. Cardiovasc. Electrophysiol. 17, 113–116 [DOI] [PubMed] [Google Scholar]
- 60. Rassi A., Jr., Rassi A., Little W. C. (2000) Clin. Cardiol. 23, 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maguire J. H., Hoff R., Sherlock I., Guimarães A. C., Sleigh A. C., Ramos N. B., Mott K. E., Weller T. H. (1987) Circulation 75, 1140–1145 [DOI] [PubMed] [Google Scholar]
- 62. Yacoub S., Mocumbi A. O., Yacoub M. H. (2008) Heart 94, 244–248 [DOI] [PubMed] [Google Scholar]
- 63. Libby P., Alroy J., Pereira M. E. (1986) J. Clin. Invest. 77, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bustamante J. M., Rivarola H. W., Fretes R., Paglini-Oliva P. A. (2005) Int. J. Cardiol. 102, 211–217 [DOI] [PubMed] [Google Scholar]
- 65. Colbert C. M., Pan E. (1999) J. Neurosci. 19, 8163–8171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Du J., Haak L. L., Phillips-Tansey E., Russell J. T., McBain C. J. (2000) J. Physiol. 522, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]