Abstract
Thus far, the role of the Escherichia coli signal recognition particle (SRP) has only been studied using targeted approaches. It has been shown for a handful of cytoplasmic membrane proteins that their insertion into the cytoplasmic membrane is at least partially SRP-dependent. Furthermore, it has been proposed that the SRP plays a role in preventing toxic accumulation of mistargeted cytoplasmic membrane proteins in the cytoplasm. To complement the targeted studies on SRP, we have studied the consequences of the depletion of the SRP component Fifty-four homologue (Ffh) in E. coli using a global approach. The steady-state proteomes and the proteome dynamics were evaluated using one- and two-dimensional gel analysis, followed by mass spectrometry-based protein identification and immunoblotting. Our analysis showed that depletion of Ffh led to the following: (i) impaired kinetics of the biogenesis of the cytoplasmic membrane proteome; (ii) lowered steady-state levels of the respiratory complexes NADH dehydrogenase, succinate dehydrogenase, and cytochrome bo3 oxidase and lowered oxygen consumption rates; (iii) increased levels of the chaperones DnaK and GroEL at the cytoplasmic membrane; (iv) a σ32 stress response and protein aggregation in the cytoplasm; and (v) impaired protein synthesis. Our study shows that in E. coli SRP-mediated protein targeting is directly linked to maintaining protein homeostasis and the general fitness of the cell.
Keywords: Membrane/Proteins, Protein/Targeting, Bacteria, Membrane, Proteomics, Escherichia coli, Ffh (Fifty-four Homologue), Membrane Protein, Proteomics, Signal Recognition Particle
Introduction
The signal recognition particle (SRP)5 is a ubiquitous ribonucleoprotein particle, found in all three kingdoms of life (1, 2). The SRP was first identified in mammalian cells, where it targets both secretory and membrane proteins in a co-translational fashion to the membrane of the endoplasmic reticulum. When the targeting signal emerges from the exit tunnel in the ribosome, the SRP binds to it. In conjunction with its membrane-associated receptor, the SRP connects the ribosome nascent chain complex with the membrane embedded Sec-translocon, which is a protein-conducting channel that mediates both the translocation of proteins across and integration of proteins into the membrane (3).
The mammalian SRP consists of six polypeptides and a 300-nucleotide RNA, the so-called 7SL RNA (4). A 54-kDa subunit that binds to signal sequences and part of the 7SL RNA forms the core of the mammalian SRP. Two additional subunits, SRP9/14 along with part of the 7SL RNA, form the so-called Alu domain that is involved in translational pausing. The Escherichia coli SRP is one of the simplest SRPs known. It consists of a subunit named Ffh (Fifty-four homologue), which is homologous to the mammalian SRP54, and a 4.5 S RNA component, which is homologous to part of the mammalian 7SL RNA. Ffh, 4.5 S RNA, and the SRP receptor FtsY are essential for viability of E. coli (5, 6). Although it is clear that the E. coli SRP, which lacks an Alu domain, can mediate the co-translational targeting of proteins (7–10), there is no consensus if it can pause translation (11–13).
Thus far, targeted approaches have been used to study the role of the E. coli SRP using only a limited number of model proteins (7, 14–19). In E. coli, SRP-dependent targeting can take place with ribosomes containing short nascent peptides, with or without a signal-anchor sequence, as long as the nascent peptide resides within the peptide exit tunnel (20). When nascent peptides emerge from the ribosome, the targeting complex with ribosomes exposing a signal-anchor sequence is maintained, whereas ribosomes exposing other sequences are released. This is in keeping with the observation that the E. coli SRP has a preference for the relatively hydrophobic targeting signals present in nascent cytoplasmic membrane proteins as shown by in vitro cross-linking studies (16, 22).
The SRP-targeting pathway delivers ribosome nascent chain complexes that synthesize membrane proteins either to the SecYEG-translocon/YidC insertion site or to independent YidC in the cytoplasmic membrane (23, 24). The SecYEG-translocon is a protein-conducting channel homologous to the eukaryotic Sec-translocon. The cytoplasmic membrane protein YidC has been identified as an indispensable factor that assists in the integration, folding, and assembly of cytoplasmic membrane proteins both in association with the SecYEG-translocon and as a separate entity (23, 24).
It has been shown for only a handful of E. coli cytoplasmic membrane proteins that their insertion into the cytoplasmic membrane is negatively affected in cells with a compromised SRP-targeting pathway (24, 25). Depletion of SRP usually does not lead to a complete block of insertion (19, 26). This is remarkable considering that in the in vivo biogenesis experiments the proteins under study are usually overexpressed, which most likely enhances the effects of a compromised SRP-targeting pathway. Based on these observations it has been suggested that in E. coli, besides the SRP pathway, alternative pathways are operational that can mediate the targeting of cytoplasmic membrane proteins (23).
In the yeast Saccharomyces cerevisiae and the Gram-positive bacterium Streptococcus mutans, the SRP is not essential for viability (27, 28). In S. cerevisiae, the absence of SRP negatively affects protein synthesis and translocation rates in the endoplasmic reticulum. Furthermore, this induces a heat shock response, which is diagnostic for a protein misfolding/aggregation problem in the cytoplasm. However, S. cerevisiae can cope with these effects and even adapt to them over time (27). In S. mutans, the deletion of the genes encoding the constituents of the SRP-targeting pathway leads to down-regulation of the protein synthesis machinery and the induction of the heat shock response (29). S. mutans has, unlike E. coli, an extra copy of YidC, YidC2, that has been shown to offer an alternative co-translational targeting pathway (30).
Just like in yeast and S. mutans, in E. coli depletion of SRP causes a heat shock response, also known as the σ32 stress response (26). In addition, it has been shown that in E. coli, SRP levels can be lowered dramatically without affecting viability (15, 26). Interestingly, cells where the σ32 stress response-induced proteases Lon and ClpQ cannot be expressed are more sensitive to SRP depletion than cells that can express these proteases. Based on this observation, it has been proposed that in E. coli the SRP plays a role in preventing toxic accumulation of aggregated mistargeted cytoplasmic membrane proteins in the cytoplasm.
To complement the targeted studies that have been used so far to study the role of SRP in E. coli, we have characterized cells depleted for the SRP component Ffh in a global manner. Our analysis of the (sub)proteomes of cells depleted of Ffh using different gel electrophoresis techniques combined with mass spectrometry and immunoblotting shows that the role of the E. coli SRP in protein targeting is directly linked to maintaining protein homeostasis and the general fitness of the cell.
EXPERIMENTAL PROCEDURES
Strain and Culture Conditions
In E. coli strain WAM121, the chromosomal copy of the gene encoding Ffh is placed under control of the promoter of the araBAD operon (18). WAM121 was cultured in Luria-Bertani (LB) medium supplemented with arabinose (0.2% w/v) and kanamycin (50 μg/ml) at 37 °C in an Innova 4330 (New Brunswick Scientific) shaker at 180 rpm. Overnight cultures were washed in LB medium without arabinose and then diluted to A600 = 0.04 in LB medium without arabinose to deplete cells of Ffh (“Ffh-depleted cells”) or medium containing 0.2% arabinose to induce expression of Ffh (“control cells:). Growth was monitored by measuring the A600 with a Shimadzu UV-1601 spectrophotometer.
SDS-PAGE and Immunoblot Analysis
Immunoblot analysis was used to monitor the protein levels of DegP, Ffh, FtsQ, FtsY, Lep, SecA, SecB, Skp, and PspA in whole cell lysates, aggregates, and/or cytoplasmic membranes. Whole cells (0.1 A600 unit), aggregates (isolated from 2 A600 units), and purified cytoplasmic membranes (5 μg of protein) were solubilized in Laemmli solubilization buffer and separated by SDS-PAGE. Proteins were transferred from the polyacrylamide gels to a polyvinylidene fluoride (PVDF) membrane (Millipore). Membranes were blocked and decorated with antisera to the components listed above essentially as described before (31). Proteins were detected with HRP-conjugated secondary antibodies (Bio-Rad) using the ECL system (according to the instructions of the manufacturer, GE Healthcare) and a Fuji LAS 1000-Plus CCD camera. Blots were quantified using the Image Gauge 3.4 software (Fuji). Experiments were repeated with three independent samples.
Flow Cytometry
Analysis of Ffh-depleted and control cells using flow cytometry was carried out using a FACSCalibur (BD Biosciences) instrument essentially as described previously (32, 33). To assess viability, cells were incubated in the dark at room temperature with 30 μm propidium iodide (PI) for 15 min (34). For staining of the cytoplasmic membrane, cells were cultured at 37 °C for 30 min with 2 μm of the cytoplasmic membrane-specific fluorophore FM4-64 (35). Cultures were diluted in ice-cold PBS to a final concentration of ∼106 cells per ml. A low flow rate was used throughout data collection with an average of 250 events/s. Forward and side scatter acquisition was used for comparison of cell morphology. Data acquisition was performed using CellQuest software (BD Biosciences), and data were analyzed with FlowJo software (Tree Star).
Protein Translocation Assay
Translocation of OmpA was monitored essentially as described previously (32). Cultures corresponding to 0.4 A600 unit were labeled with [35S]methionine (60 μCi/ml, 1 Ci = 37 GBq) for 30 s followed by precipitation in 10% trichloroacetic acid (TCA). TCA-precipitated samples were washed with acetone, resuspended in 10 mm Tris-HCl, pH 7.5, 2% SDS, and immunoprecipitated with antiserum to OmpA. The OmpA precipitate was subjected to standard SDS-PAGE analysis. Gels were scanned in a Fuji FLA-3000 phosphorimager and quantified as described above.
Two-dimensional Gel Electrophoresis (2DE)
Whole cell lysates (1 A600 unit) were analyzed by 2DE using isoelectric focusing in the first dimension and SDS-PAGE in the second dimension (32). Gels used for comparative analysis of whole cell lysates were stained with high sensitivity silver stain (36).
Isolation and Analysis of Protein Aggregates
Protein aggregates were extracted from whole cells essentially as described before (37). Cells corresponding to 75 A600 units were used for each aggregate extraction. To determine the fraction of the total cellular protein that was aggregated, the protein content of cell lysates and aggregate extracts was determined with the BCA assay according to the instructions of the manufacturer (Pierce). In addition, whole cell lysates (0.1 A600 unit) and aggregate fractions corresponding to 4 A600 units were solubilized in Laemmli solubilization buffer and separated by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250, and the total intensities of the lanes were quantified using the Image Gauge 3.4 software (Fuji). Aggregates isolated from 15 A600 units were analyzed by SDS-PAGE using 24-cm-long 8–16% acrylamide gradient gels and 2DE. Gels were stained with Coomassie Brilliant Blue R-250, and proteins were identified by mass spectrometry (MS) as described below. The aggregate fraction was also subjected to in-solution digest followed by nano-liquid chromatography electrospray tandem MS (nanoLC-ESI-MS/MS) essentially as described before (38).
Isolation of Cytoplasmic Membranes
Cytoplasmic membranes were isolated essentially as described before (39). Membrane fractions used for immunoblot analysis were prepared from nonradiolabeled cultures. Membrane fractions used for analysis by two-dimensional BN/SDS-PAGE were prepared from a mixture of labeled and unlabeled cells (33). Cells corresponding to 300 A600 units were cultured as described above. An aliquot of 6 A600 units of cells was transferred to minimal M9 medium and labeled with [35S]methionine (60 μCi/ml, 1 Ci = 37 GBq) for 30 s, followed by a chase of 5 min with cold methionine (final concentration 10 mg/ml). Labeled cells were subsequently collected by centrifugation, and cell pellets were snap-frozen in liquid nitrogen. The remainder of the cells (294 A600 units) was harvested by centrifugation and washed once with buffer K (50 mm triethanolamine (TEA), 250 mm sucrose, 1 mm EDTA, 1 mm dithiothreitol (DTT), pH 7.5). The cell pellets were snap-frozen in liquid nitrogen and stored at −80 °C. Before breaking the cells, labeled and unlabeled cells from the same culture were pooled in a 1:50 ratio. The resulting mixture was resuspended in 8 ml of buffer K supplemented with 0.1 mg/ml Pefabloc protease inhibitor and 5 μg/ml DNase I and lysed by two cycles of French press (18,000 p.s.i.). The lysate was cleared of unbroken cells by centrifuging twice at 10,000 × g for 20 min, and the total membrane fraction was collected by centrifugation at 100,000 × g for 1 h. The membrane pellet was resuspended in 1 ml of buffer M (50 mm TEA, 1 mm EDTA, 1 mm DTT, pH 7.5) and loaded on top of a six-step sucrose gradient (from bottom to top) as follows: 0.5 ml of 55%, 1.5 ml of 50%, 1.5 ml of 45%, 2.5 ml of 40%, 2.5 ml of 35%, 2.5 ml of 30% (w/w sucrose in buffer M). After centrifugation at 210,000 × g for 15 h, the cytoplasmic membrane fraction was collected from the 35% sucrose layer. The collected fractions were diluted in TEA buffer (50 mm TEA, 1 mm DTT, pH 7.5) to a sucrose concentration below 10%. Cytoplasmic membranes were collected by centrifugation at 170,000 × g for 1 h and subsequently resuspended in buffer L (50 mm TEA, 250 mm sucrose, 1 mm DTT, pH 7.5). The cytoplasmic membrane fraction was snap-frozen in liquid nitrogen. Protein concentrations were determined using the BCA assay (Pierce). Samples were stored at −80 °C.
Analysis of Cytoplasmic Membrane Fractions by Two-dimensional BN/SDS-PAGE
Comparative two-dimensional BN/SDS-PAGE was performed as described previously (33, 39, 40). In short, [35S]methionine-labeled cytoplasmic membranes (125 μg of protein) were solubilized in 0.5% (w/v) n-dodecyl β-maltoside and subjected to BN electrophoresis in the first dimension and denaturing SDS-PAGE in the second dimension. For calibration, ferritin (440 and 880 kDa), aldolase (158 kDa), and albumin (66 kDa) (GE Healthcare) were used as molecular weight markers. Gels were stained with Coomassie Brilliant Blue R-250 (32). To assess the phosphorimage scans with [35S]methionine-labeled membranes, using both the BCA assay and Coomassie staining/densitometry, it was verified that equal amounts of protein had been loaded (see below).
Image Analysis and Statistics
Image analysis of one- and two-dimensional gels and statistics was essentially done as described previously (32, 33). Stained gels were scanned using a GS-800 densitometer from Bio-Rad. Radiolabeled gels were scanned in a Fuji FLA-3000 phosphorimager. Two-dimensional gels spots were detected, matched, and quantified using PDQuest software version 8.0 (Bio-Rad). The analysis of Coomassie-stained and [35S]methionine-labeled cytoplasmic membrane proteins was done on the same set of gels. In all cases, each analysis set consisted of at least three gels in each replicate group (i.e. Ffh-depleted cells and the control). Each gel in a set represented an independent sample (i.e. from a different bacterial colony, culture, and membrane preparation). Independent samples were subjected to 2DE or two-dimensional BN/SDS-PAGE and image analysis in parallel, i.e. en groupe. Quantities of stained spots were normalized using the “total intensity of valid spots” method to compensate for nonexpression-related variations in spot quantities between gels (there were no significant variations in the total spot quantity between the two groups, Ffh depletion and control).
Mass Spectrometry-based Identification of Proteins
Coomassie-stained protein spots or bands were excised, washed, and digested with modified trypsin, and peptides were extracted manually or automatically (ProPic and Progest, Genomic Solutions, Ann Arbor, MI). Peptides were applied to the matrix-assisted laser desorption/ionization mass spectrometry (MALDI) target plate as described previously (41). Mass spectra were obtained automatically by MALDI-TOF MS in reflectron mode (Voyager-DE-STR; PerSeptive Biosystems, Framingham, MA), followed by automatic internal calibration using tryptic peptides from autodigestion. The spectra were analyzed for monoisotopic peptide peaks (m/z range 850–5000) using the software MoverZ from Genomic Solutions with a signal to noise ratio threshold of 3.0. Matrix and/or autoproteolytic trypsin fragments were not removed. Spectral annotations (in particular assignments of monoisotopic masses) were verified by manual inspection for a large number of measurements. The resulting peptide mass lists were used to search the SwissProt 56.0 data base (release 11/09) for E. coli with Mascot (version 2.0) in automated mode, using the following search parameters/criteria: significant protein MOWSE score at p < 0.05; no missed cleavages allowed; variable methionine oxidation; fixed carbamidomethylation of cysteines; minimum mass accuracy 50 ppm. The search results pages were extracted and analyzed by an additional in-house filter6 applying the following three criteria for positive identification: (i) minimum MOWSE score ≥50; (ii) ≥ four matching peptides with an error distribution within ±25 ppm; and (iii) ≥15% sequence coverage. False-positive rates were less than 1%, as determined by searching with the .pkl list against the E. coli data base (SwissProt 56.0) mixed with a randomized version of the E. coli data base, generated using a Perl script from Matrix Science.
Aggregate fractions isolated from whole cells were subjected to in solution digest with modified trypsin. The resulting peptide mixture was analyzed by nano-LC-ESI-MS/MS in automated mode on a quadruple/orthogonal acceleration TOF tandem mass spectrometer (Q-TOF, Micromass) (see Friso et al. (42) for details). The spectra were used to search SwissProt 56.0 data base (downloaded locally) automated using Mascot (version 2.0). When searching Mascot, the maximum precursor and fragment errors were 1.2 and 0.8 Da. Probability-based MOWSE score, number of matching peptides, and highest peptide score were extracted from the Mascot peptide summary report pages using in-house written software.6 Minimal criteria for identification were as follows: (i) one matching peptide with a peptide score higher than the minimally significant (p < 0.05) individual ion score; (ii) two matching peptides higher than 21; or (iii) three matching peptides with peptide score of 20 or higher. All significant MS/MS identifications by Mascot were manually verified for spectral quality and matching y and b ion series.
Oxygen Consumption Measurements
For oxygen consumption measurements in whole cells, cells were cultured as described above. Cells were harvested at 3.000 × g, resuspended in ice-cold PBS, and adjusted to a concentration of 200 A units/ml. The protein concentration of the samples was determined with the BCA assay according to the instructions of the manufacturer (Pierce). Oxygen consumption in whole cells was measured using a Hansatech Oxytherm oxygraph for 2 min. The reaction was started by the addition of 3 A unit cells to 1 ml of PBS at 25 °C.
Kinetics of the Integration of Lep and Lep-inv into the Cytoplasmic Membrane
WAM121 cells harboring plasmid pMS119 (43) containing either the gene encoding Lep or inverted Lep (Lep-inv) were cultured in the presence and absence of arabinose and transferred to minimal M9 medium for labeling with [35S]methionine essentially as described above. Ampicillin (final concentration 100 μg/ml) was added to the culture media to maintain the plasmids. Expression of both Lep and Lep-inv was induced for 15 s with isopropyl 1-thio-β-d-galactopyranoside (final concentration 10 μm). Notably, plasmid pMS119 expresses the LacI repressor. LacI is needed for controlling the expression of proteins from the isopropyl 1-thio-β-d-galactopyranoside-inducible tac promoter in WAM121, which is derived from the lac operon-deficient strain MC4100 (18). Cells were labeled with [35S]methionine (60 μCi/ml, 1 Ci = 37 GBq) for 30 s, followed by chases of 0, 1, 3, and 5 min with cold methionine (final concentration 10 mg/ml). Subsequently, cells were converted to spheroplasts. For spheroplasting, cells were collected at 16,100 × g, resuspended in ice-cold buffer (40% w/v sucrose, 33 mm Tris, pH 8.0), and incubated with lysozyme (final concentration 5 μg/ml) and 1 mm EDTA for 15 min on ice. Aliquots of the spheroplast suspension were incubated on ice for 1 h either in the presence or absence of proteinase K (final concentration 0.3 mg/ml). Subsequently, phenylmethylsulfonyl fluoride (PMSF) was added to the spheroplast suspension (final concentration 0.33 mg/ml) to inhibit the protease. After addition of PMSF, samples were precipitated with trichloroacetic acid (final concentration 10%), washed with acetone, resuspended in 10 mm Tris, 2% SDS, immunoprecipitated with antisera to Lep, OmpA (a periplasmic control), and AraB/bandX (a cytoplasmic control), washed, and analyzed by standard SDS-PAGE (19). Gels were scanned in a Fuji FLA-3000 phosphorimager and quantitated as described above. The experiments were repeated three times.
RESULTS
Characterization of the Ffh Depletion Strain WAM121
In E. coli strain WAM121, the gene encoding the essential SRP core component Ffh is under control of the araBAD promoter (18). Cells were cultured aerobically in LB medium in the presence of arabinose to induce expression of Ffh (these cells will be further referred to as “control cells”) and in the absence of arabinose to deplete cells of Ffh. Growth was monitored by measuring the A600 (Fig. 1A). As expected, growth of WAM121 cells cultured in the absence of arabinose was negatively affected upon longer depletion compared with the control cells.
FIGURE 1.
Effect of Ffh depletion on growth, Ffh levels, integrity of the cytoplasmic membrane, OmpA translocation, and SecA levels/distribution. A, effect of Ffh depletion on cell growth. Growth of WAM121 cells cultured in the presence (control) and absence (Ffh depletion) of 0.2% arabinose was monitored by measuring the A600. After 4 h of depletion, 0.2% arabinose was added to part of the culture (readdition). B, Ffh levels in WAM121 cells grown in the presence (control) and absence (Ffh depl) of arabinose and in MC4100 cells from which WAM121 was derived. Whole cells (0.02 A600 units) were subjected to SDS-PAGE followed by immunoblot analysis with antibodies to Ffh. C, histograms representing the fluorescence of cultures stained with the fluorophore PI, whose uptake is an indicator for the integrity of the cytoplasmic membrane. D, effect of Ffh depletion on OmpA translocation. Ffh-depleted and control cells were labeled with [35S]methionine for 30 s. OmpA was immunoprecipitated and subjected to standard SDS-PAGE analysis, and labeled material was detected by phosphorimaging. To calibrate the system with an OmpA precursor, CM124 cells (SecE depletion strain, in which the secE gene is under control of the araBAD promotor) grown in the absence of arabinose (SecE depl) was used. E, SecA levels and distribution. Whole cells (WC; 0.1 A600 unit per lane) and cytoplasmic membranes (CM; 5 μg of protein) were separated by means of SDS-PAGE and subsequently subjected to immunoblot analysis with antibodies to SecA.
It is important to maximize the depletion of Ffh, while minimizing pleiotropic effects. After 2 h of Ffh depletion, the cells still contained a considerable amount of SRP. Therefore, we explored longer depletion times. After 4 h of growth in the absence of arabinose, growth was hardly impaired, and cells contained less than 1% of the amount of Ffh of the control cells (Fig. 1B). The levels of Ffh in WAM121 cells grown in the absence of arabinose were similar after 4, 6, and 8 h of culturing, indicating that, after 4 h of culturing in the absence of arabinose, Ffh levels were already “minimal” (results not shown). Notably, the Ffh accumulation levels in WAM121 cells grown in the presence of arabinose were similar to Ffh accumulation levels in MC4100 cells from which WAM121 was derived (Fig. 1B). Thus, WAM121 cells grown in the presence of arabinose serve as a proper control.
The morphology, i.e. the size and granularity (membrane and/or aggregate content), of Ffh-depleted and control cells was monitored using flow cytometry. Cultures of both Ffh-depleted and control cells consisted of a homogeneous population of cells, and the size and granularity of the cells did not change upon Ffh depletion (results not shown). To monitor the membrane content per cell directly, cells were labeled with the cytoplasmic membrane-specific fluorophore FM4-64 and subsequently analyzed using flow cytometry. The membrane content per cell did not change upon Ffh depletion (results not shown), which is in keeping with the unaffected granularity of cells. The integrity of the cytoplasmic membrane was assessed by measuring the uptake of the dye PI using flow cytometry. In less than 1% of the cells, the uptake of PI was enhanced compared with control cells (Fig. 1C). This indicates that the effect of Ffh depletion on the integrity of the cytoplasmic membrane is negligible at the 4-h time point.
To further assess if 4 h of depletion is a good time point to study the effects of Ffh depletion, we monitored the SecYEG-translocon function in two different ways because it plays a key role in the insertion of membrane proteins into the cytoplasmic membrane (23). First we monitored the translocation of the SecYEG-translocon-dependent outer membrane protein OmpA across the cytoplasmic membrane in Ffh-depleted and control cells (Fig. 1D). OmpA is one of the most abundant and highly expressed proteins in E. coli, and it is targeted to the SecYEG-translocon by the chaperone SecB rather than by the SRP pathway (32). OmpA expression levels and translocation and SecB levels were not affected upon Ffh depletion (Figs. 1D and 2B). This indicates that SecB-mediated protein targeting and SecYEG-translocon-mediated protein translocation are not affected upon the depletion of Ffh.
FIGURE 2.
Analysis of whole-cell lysates of Ffh-depleted and control cells by 2DE and immunoblotting and FtsY levels and distribution in the cell. A, comparative 2DE analysis of total lysates from control cells and cells depleted of Ffh for 4 h. Proteins from 1 A600 unit of solubilized cells were separated by 2DE. Proteins were visualized by silver staining, and differences between Ffh-depleted and control cells were analyzed using PDQuest. 18 spots were significantly (p < 0.01) affected by Ffh depletion. Proteins were identified by MALDI-TOF MS and peptide mass fingerprinting using spots excised from gels stained with Coomassie Brilliant Blue and/or matching with reference gels/maps (supplemental Table 1). The numbers in the figure correspond to the ones in supplemental Table 1. B, PspA, DegP, Skp, and SecB levels. Whole cells (0.1 A600 unit per lane) were separated by means of SDS-PAGE and subsequently subjected to immunoblot analysis with antibodies to the aforementioned components. C, FtsY levels and distribution in the cell. Whole cells (WC; 0.1 A600 unit per lane) and cytoplasmic membranes (CM; 5 μg of protein) were separated by means of SDS-PAGE and subsequently subjected to immunoblot analysis with antibodies to FtsY.
To verify further the proper function of the SecYEG-translocon upon Ffh depletion, we monitored the levels and distribution of SecA in the cell (Fig. 1E). SecA is an auxiliary subunit of the SecYEG-translocon, which is involved in the translocation of secretory proteins and sizeable periplasmic loops of cytoplasmic membrane proteins (23). SecA is located both in the cytoplasm and at the cytoplasmic membrane, and its levels are up-regulated when protein translocation via the SecYEG-translocon across the membrane is compromised (44). We found that neither SecA levels nor its distribution in the cell were changed upon Ffh depletion. This is another indication that the depletion of Ffh does not affect the function of the SecYEG-translocon. These observations are in keeping with the two-dimensional BN/SDS-PAGE analysis of the cytoplasmic membrane proteome of cells depleted of Ffh showing that the level of the SecYEG-translocon was not affected upon Ffh depletion (see below). Notably, the two-dimensional BN/SDS-PAGE analysis also showed that the level of YidC, which just like the SecYEG-translocon plays a key role in the biogenesis of cytoplasmic membrane proteins, was also not affected upon Ffh depletion (see below).
Taken together, after 4 h of Ffh depletion, Ffh levels were minimal, and the function and levels of the SecYEG-translocon and the levels of YidC were not affected. Furthermore, the morphology and cytoplasmic membrane content of the cells were not changed. Finally, the integrity of the cytoplasmic membrane was not affected upon Ffh depletion.
Ffh Depletion Leads to the Induction of σ32 Stress and PspA Responses
It has been shown by immunoblotting that upon Ffh depletion the levels of both DnaK and GroEL are increased, indicating the induction of the σ32 stress response (26). To study this in more detail and to monitor if Ffh depletion has other effects on the soluble proteome, whole cell lysates of cells depleted of Ffh for 4 h and control cells were compared by 2DE and immunoblot analysis. Proteins were separated by denaturing immobilized pH gradient strips, pH 4–7, in the first dimension and by Tricine-SDS-PAGE in the second dimension. Gels were stained with silver, and spot volumes were compared using PDQuest. This analysis demonstrated that the volumes of 18 spots were significantly (p < 0.01) changed in the lysates of Ffh-depleted cells compared with the control; the intensity increased in 12 spots and decreased in 6 spots. The affected spots were excised and used for protein identification by MALDI-TOF MS and peptide mass finger printing and/or matching with reference maps (Fig. 2A and supplemental Table 1). Spot statistics and MS data are provided in supplemental Table 1.
Many changes identified by 2DE were indeed linked to the σ32 stress response. Upon Ffh depletion, the accumulation levels of the following σ32-inducible cytoplasmic chaperones were increased: DnaK (2.48-fold change), GroEL (2.19-fold change), GroES (1.84-fold change), ClpB (4.47-fold change), and IbpA (“on” response, i.e. the protein is expressed in cells depleted of Ffh but not in control cells) (Fig. 2A and supplemental Table 1).
Analysis of the 2DE gels also showed that Ffh depletion results in increased accumulation levels of phage shock protein A (PspA) (on response) (Fig. 2A and supplemental Table 1). This was confirmed by immunoblotting (Fig. 2B).
Using a promotor probe approach, it has been shown that Ffh depletion does not lead to cell envelope stress, i.e. protein misfolding in the cell envelope (45). Indeed, as shown by immunoblotting, accumulation levels of the cell envelope stress-induced periplasmic proteins DegP and Skp were not affected upon Ffh depletion (Fig. 2B), and no DegP and Skp precursors accumulated. This is another indication that Ffh depletion did not affect the SecYEG-translocon capacity. Finally, the levels of FtsY and its distribution in the cell were monitored using immunoblotting (Fig. 2C). Both did not change upon Ffh depletion. Taken together, Ffh depletion induces σ32 stress and the PspA responses.
Ffh Depletion Leads to the Formation of Protein Aggregates in the Cytoplasm
The σ32 stress response in cells depleted of Ffh indicated that reduced Ffh levels lead to protein misfolding/aggregation in the cytoplasm. Indeed, we could isolate aggregated proteins from cells depleted of Ffh for different periods of time (Fig. 3A). Based on cell fractionation combined with SDS-PAGE, Coomassie staining, gel densitometry, and the BCA assay, it was shown that the aggregated protein constituted only around 1% of the total cellular protein (supplemental Fig. 1). This is in keeping with no detectable changes in the granularity of cells depleted of Ffh. Notably, the composition of the aggregates isolated from cells depleted of Ffh for different periods of time was similar.
FIGURE 3.
Characterization of aggregates isolated from cells depleted of Ffh for different periods of time and control cells, and characterization of aggregates isolated from cells depleted for Ffh for 4 h to which 0.2% arabinose was added and cultured further (Ffh readd). A, aggregates were isolated from 75 A600 units of cells, and material corresponding to 15 A600 units was loaded per lane. The protein content of the loaded material was analyzed by SDS-PAGE on a 24-cm-long 8–16% gradient gel stained by Coomassie Brilliant Blue R-250, and proteins were identified by mass spectrometry as described under “Experimental Procedures” (supplemental Table 2). Proteins were also identified by nanoLC-ESI-MS/MS of solubilized aggregates digested with trypsin (supplemental Table 2). B, presence of FtsQ and Lep in aggregates. Aggregates (material isolated from 2 A600 units of cells loaded per lane) were analyzed using a combination of SDS-PAGE and immunoblotting with antibodies to FtsQ and Lep. Ffh depl, aggregates isolated from cells depleted of Ffh for 4 h.
Characterization of the aggregates of a secB null mutant strain enabled us to identify SecB substrates (32). Therefore, the content of the protein aggregates isolated from cells depleted of Ffh for 4 h was analyzed by nanoLC-ESI-MS/MS of solubilized aggregates digested with trypsin and one- and two-dimensional gel electrophoresis followed by in-gel digestion and analysis by MALDI-TOF MS/peptide mass fingerprinting (Fig. 3A and supplemental Table 2). In total, 111 proteins were identified as follows: 5 secretory proteins, 97 cytoplasmic proteins, and only 9 cytoplasmic membrane proteins (supplemental Table 2). Among the cytoplasmic membrane proteins was the established SRP-dependent subunit of cytochrome bo3, CyoA (25, 46–48).
It is possible that the number of identified cytoplasmic membrane proteins in aggregates isolated from cells depleted of Ffh is under-represented because the typical absence of lysine and arginine in the transmembrane domain regions leads to few and large peptides upon trypsin digestion (49). To complement this approach, we probed the aggregates isolated from cells depleted of Ffh for 4 h with antibodies against the known SRP-dependent cytoplasmic membrane proteins FtsQ and Lep (Fig. 3B) (25). We could detect these two cytoplasmic membrane proteins in both the aggregates and the cytoplasmic membrane fraction but not in the soluble fraction. Notably, only minute amounts of the total of these proteins present in the cell was found in aggregates (FtsQ, 1.7 ± 0.2%, and Lep, 0.3 ± 0.1%).
Interestingly, the amount of aggregated protein did not change over time, suggesting that the protein aggregates formed upon Ffh depletion were dynamic, i.e. aggregated proteins are either degraded or reactivated. To study the dynamic nature of the aggregates, SRP expression was restored by adding arabinose to cells depleted of Ffh. Within 2 h after induction of expression of Ffh, the aggregates completely disappeared.
Taken together, Ffh depletion led to the aggregation of proteins in the cytoplasm. However, the aggregated protein constituted only a minor fraction of the total cellular protein and also a minor fraction of the total cytoplasmic membrane protein. The amount and number of cytoplasmic membrane proteins in the aggregates isolated from Ffh-depleted cells was too low to use this information as a starting point for the identification of SRP substrates.
Effects of Ffh Depletion on the Steady-state Composition of the Cytoplasmic Membrane Proteome
As the next step in our analysis, we monitored the effects of Ffh depletion on the steady-state composition of the cytoplasmic membrane proteome. It is of note that besides integral membrane proteins, the cytoplasmic membrane proteome also consists of a variety of other proteins (40, 50). Cytoplasmic membranes from cells depleted of Ffh for 4 h and control cells were isolated using a combination of French press and sucrose gradient centrifugation. Membranes were solubilized using the mild nonionic detergent n-dodecyl β-maltoside. Protein complexes were negatively charged with Coomassie Brilliant Blue G-250 and separated according to their size by BN/PAGE. The subunit composition of the separated protein complexes was subsequently analyzed in the second dimension by SDS-PAGE. Two-dimensional BN/SDS-polyacrylamide gels were stained with colloidal Coomassie to study the steady-state cytoplasmic membrane proteomes. In this study, we used a protocol where the 1st dimension BN gel is cast on a GelBond PAG film. Immobilization of the BN gel prevents distortion of gel lanes when they are cut out and transferred to the second dimension. This lowers spot variation, thereby greatly improving reproducibility of two-dimensional BN/SDS-polyacrylamide gels, which makes it possible to reliably analyze them (33, 39, 40, 51).
Spot volumes in two-dimensional BN/SDS-polyacrylamide gels stained with colloidal Coomassie were compared using the image analysis software PDQuest (Fig. 4A and supplemental Table 3). Each analysis set contained at least three biological replicates, and the threshold for acceptance was 99% significance determined by the Student's t test. Spots were excised and used for protein identification by MALDI-TOF MS/peptide mass fingerprinting and/or matching with reference gels. Upon Ffh depletion, the accumulation levels of the NADH dehydrogenase (Nuo) complex, succinate dehydrogenase (Dhs) complex, the cytochrome bo3 oxidase (Cyo) complex, and the mechanosensitive channel MscS were down by ∼50% (Nuo complex 0.37-fold change, Dhs complex 0.51-fold change, Cyo complex 0.48-fold change, and the MscS homo-oligomer 0.54-fold change; supplemental Table 3). The periplasmic peripheral membrane protein DacA, also known as Pbp5, was up 3-fold (DacA occurs in following two spots: 3.50- and 2.90-fold change, respectively). In addition, the soluble chaperones DnaK and GroEL were up in the cytoplasmic membrane fraction (4.66- and 4.49-fold change, respectively). This despite the fact that Ffh depletion does not induce cell envelope stress, i.e. protein misfolding in the cell envelope (45). Accumulation levels of the other identified proteins/complexes in the cytoplasmic membrane fractions, including the two key players in membrane protein biogenesis the SecYEG-translocon and YidC, were not significantly affected upon Ffh depletion.
FIGURE 4.
Analysis of the steady-state cytoplasmic membrane proteomes of Ffh-depleted and control cells by two-dimensional BN/SDS-PAGE and oxygen consumption measurements in whole cells. A, analysis of the steady-state cytoplasmic membrane proteomes by two-dimensional BN/SDS-PAGE. The cytoplasmic membrane fractions of WAM121 cells grown in the presence (control) and absence (depleted of Ffh for 4 h) of arabinose were isolated by density centrifugation as described under “Experimental Procedures.” The cytoplasmic membrane fractions were analyzed by two-dimensional BN/SDS-PAGE. Proteins were identified by MALDI-TOF MS and peptide mass fingerprinting (supplemental Table 3) using spots excised from Coomassie Brilliant Blue-stained gels. Differences in the cytoplasmic membrane proteomes of Ffh-depleted and control cells were analyzed using PDQuest. Significantly affected (p < 0.01) proteins are indicated in supplemental Table 3. Representative two-dimensional BN/SDS-polyacrylamide gels with proteins detected by staining with colloidal Coomassie Brilliant Blue (protein steady-state levels) are shown. Nuo, NADH dehydrogenase complex; Dhs, succinate dehydrogenase complex; Cyo, cytochrome bo3 oxidase complex; and MscS, MscS complex. B, oxygen consumption measurements in whole cells grown in the presence (control) and absence (depleted of Ffh for 4 h) of arabinose. Experiments were done in triplicate. Oxygen consumption of control cells was set to 100.
To monitor the impact of the lowered levels of the aforementioned respiratory complexes, we measured oxygen consumption rates in whole cells depleted of Ffh for 4 h and control cells. Upon Ffh depletion oxygen consumption rates were ∼25% lower (Fig. 4B).
Taken together, analysis of the steady-state composition of the cytoplasmic membrane proteome upon Ffh depletion showed that the accumulation levels of complexes involved in respiration were negatively affected, and the accumulation levels of the soluble chaperones DnaK and GroEL were increased.
Effects of Ffh Depletion on the Kinetics of the Biogenesis of the Cytoplasmic Membrane Proteome
The effects of Ffh depletion on the kinetics of the biogenesis of the cytoplasmic membrane proteome were also monitored. To this end, cytoplasmic membranes from cells depleted of Ffh for 4 h and control cells were isolated from [35S]methionine-labeled cultures and analyzed using a combination of two-dimensional BN/SDS-PAGE and phosphorimaging. Unfortunately, the variation of spots of [35S]methionine-labeled proteins is too strong to allow comparative analysis. This is mainly due to shrinking/deformation of the gels upon drying, prior to phosphorimaging, and variations in phosphorimage plates. It is impossible to unambiguously map most spots in the phosphorimage scans to the Coomassie-stained steady-state gels. The reason for this is that, especially upon short labeling/chase times, spots from complex assembly intermediates occur that are not detectable in steady-state Coomassie-stained gels. Notably, as described under “Experimental Procedures,” the steady-state composition and the kinetics of the biogenesis of the cytoplasmic membrane proteomes were studied using the same cytoplasmic membrane isolates.
The cytoplasmic membrane fractions of cells pulse-labeled for 30 s with [35S]methionine showed that the total intensities of the spots in the gels from Ffh-depleted cells were only 8.2 ± 1.2% as compared with the control cells. However, despite the difference in intensities, the spot patterns in scans of two-dimensional BN/SDS-polyacrylamide gels with cytoplasmic membranes isolated from cells depleted of Ffh were similar to the control (Fig. 5A, upper panel). Is the difference in total intensities due to differences in the uptake and incorporation of [35S]methionine in Ffh-depleted versus control cells? To address this question, incorporation of [35S]methionine in whole cells was monitored. Incorporation of [35S]methionine in cells depleted of Ffh was 77 ± 3.8% compared with the control cells (Fig. 5B). There are some differences in the banding patterns between Ffh-depleted and control cells, which are most likely due to Ffh depletion-induced stress. At any rate, the big difference in intensity between the two-dimensional BN/SDS-polyacrylamide gels containing the cytoplasmic membrane fractions from [35S]methionine-labeled Ffh-depleted and control cells cannot be explained by differences in the uptake and incorporation of [35S]methionine.
FIGURE 5.
Analysis of biogenesis kinetics of the cytoplasmic membrane proteomes of Ffh-depleted and control cells by two-dimensional BN/SDS-PAGE and the protein synthesis in whole cells. A, WAM121 cells grown in the presence (control) and absence (depleted of Ffh for 4 h) of arabinose were labeled with [35S]methionine for 30 s (top) and labeled for 30 s followed by a chase for 5 min with cold methionine (bottom). The cytoplasmic membrane fractions were isolated by density centrifugation from a mixture of labeled and nonlabeled cells as described under “Experimental Procedures.” The cytoplasmic membrane fractions were analyzed by two-dimensional BN/SDS-PAGE and phosphorimaging. Representative two-dimensional BN/SDS-polyacrylamide gels with proteins detected by phosphorimaging (protein incorporation) are shown. Notably, the 30-s pulse and the 5-min chase experiments represent two different experiments. B, WAM121 cells grown in the presence (control) and absence (depleted of Ffh for 4 h) of arabinose were labeled with [35S]methionine for 30 s. Total cell lysates (0.02 A600 unit) were analyzed by one-dimensional SDS-PAGE and phosphorimaging. The intensities of with [35S]methionine-labeled proteins were quantified using Image Gauge 3.4 software (Fuji).
After a pulse of 30 s, the subsequent addition of an excess of cold methionine to stop the labeling of proteins with [35S]methionine and a chase of 5 min, incorporation of labeled protein in the cytoplasmic membrane proteomes from cells depleted of Ffh and control cells was 106 ± 6.4%. Notably, also after the 5-min chase, the spot patterns were similar (Fig. 5A, lower panel). This indicates that most of the protein incorporated into the cytoplasmic membrane proteome after the 5-min chase was already synthesized after the 30-s pulse but did not co-fractionate with the cytoplasmic membrane fraction after the 30-s pulse. Taken together, Ffh depletion results in a minor reduction of protein synthesis and slows down the kinetics of the biogenesis of the cytoplasmic membrane proteome as a whole.
Effects of Ffh Depletion on Kinetics of Integration of Lep and Lep-inv into the Cytoplasmic Membrane
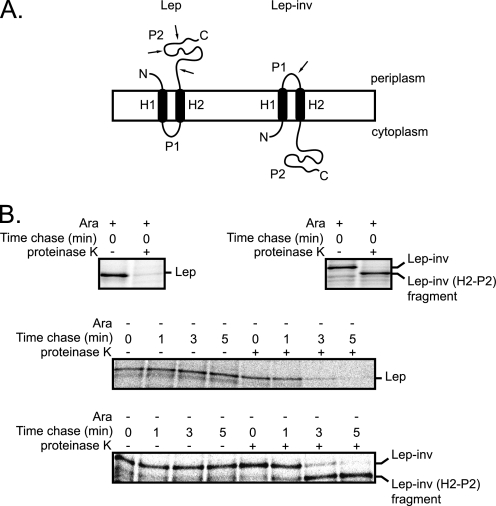
To study the effect of Ffh depletion on the kinetics of the biogenesis of the cytoplasmic membrane proteome in more detail, the kinetics of the integration of the model cytoplasmic membrane proteins Lep and Lep-inv into the cytoplasmic membrane were monitored using a pulse-chase approach (Fig. 6A). The kinetics of integration of Lep and Lep-inv into the cytoplasmic membrane were studied in cells depleted of Ffh for 4 h and control cells. Notably, the set-up of the pulse-chase experiments was designed in such a way that the expression levels of Lep and Lep-inv were kept very low to minimize any secondary effects caused by their expression (see “Experimental Procedures”). The pulse-chase experiments showed that integration of both Lep and Lep-inv into the cytoplasmic membrane was severely slowed down upon Ffh depletion (Fig. 6B). These observations along with the effect of Ffh depletion on the kinetics of the biogenesis of the whole cytoplasmic membrane proteome show that Ffh depletion has a major impact on the kinetics of the biogenesis of the cytoplasmic membrane proteome.
FIGURE 6.
Ffh depletion impairs the kinetics of integration of the cytoplasmic model proteins Lep and Lep-inv into the cytoplasmic membrane. A, orientation of Lep and Lep-inv in the cytoplasmic membrane. The Lep-inv mutant was derived from Lep by adding 3 lysine codons between codons 4 and 5, inserting 10 codons encoding the sequence Gly-Gln-Ser-Leu-Asn-Ala-Pro-Thr-Ser-Gly between codons 22 and 24, deleting residues 30–52, and changing Lys56 to Asn and Glu61 to Val (21, 74). In spheroplasts, proteinase K degrades the P2 domain of Lep and the P1 loop of Lep-inv (arrows). For Lep-inv, this treatment gives rise to a protease-resistant H2-P2 fragment that can be immunoprecipitated with a Lep antiserum, whereas no immunoprecipitable material remains when the P2 domain in Lep has been digested. B, WAM121 cells harboring pMS119Lep or pMS119Lep-inv were cultured in the presence of arabinose (control, Ara+) and absence of arabinose (depleted of Ffh for 4 h, Ara−). Expression of Lep and Lep-inv was induced, and cells were pulse-labeled with [35S]methionine and chased with cold methionine as described under “Experimental Procedures.” Cells were converted to spheroplasts immediately and 1, 3, and 5 min after the nonradioactive methionine was added and further processed as described under “Experimental Procedures.” Antibodies to the P2 domain of Lep were used for the Lep/Lep-inv immunoprecipitations. Immunoprecipitations with antibodies to OmpA and bandX were carried out to check the spheroplasting (supplemental Fig. 2).
DISCUSSION
Thus far, targeted approaches have been used to study the role of the E. coli SRP, which is essential for viability. To complement these targeted studies, we characterized cells depleted of the SRP component Ffh in a global manner; i.e. the subproteomes of cells depleted of Ffh and control cells were analyzed using a proteomics approach.
After 4 h of depletion, cells contained less than 1% of Ffh compared with the control cells, and Ffh levels were not lowered any further upon prolonged depletion. Using flow cytometry, we showed that 4 h of Ffh depletion had no effect on the morphology of the cells and the integrity of the cytoplasmic membrane. Furthermore, Ffh depletion did not affect the abundance and function of the SecYEG-translocon, the abundance of YidC, and the abundance and distribution of the SRP receptor, FtsY.
As expected, Ffh depletion induces the σ32 stress response (26). This response is an indication for protein misfolding/aggregation in the cytoplasm (52). We were able to isolate protein aggregates from the cytoplasm of Ffh-depleted cells. However, they constituted only around 1% of the total cellular protein. The aggregates contained a large variety of proteins, including some cytoplasmic membrane proteins. Among them were the known SRP-dependent proteins CyoA, FtsQ, and Lep (25, 46–48). For FtsQ and Lep, we showed that the aggregates contained only minute amounts of the total of these proteins and that steady-state levels in the cytoplasmic membrane were not detectably affected. In contrast, the accumulation level of CyoA in the cytoplasmic membrane was significantly decreased upon Ffh depletion. These observations suggest that if a cytoplasmic membrane protein is mistargeted upon Ffh depletion, only a small fraction of the total of this protein will be mistargeted and/or that the mistargeted protein will efficiently be degraded. At any rate, the characterization of the aggregates indicates that upon Ffh depletion mistargeted cytoplasmic membrane proteins can aggregate. The aggregated membrane proteins will titrate chaperones thereby reducing their availability. This will cause misfolding/aggregation of chaperone substrates in the cytoplasm (39). Protein aggregation levels remained constant over time, and upon readdition of arabinose to cells depleted for Ffh, the aggregates disappeared. Apparently, aggregate formation was halted, and the cells were cleared of the aggregated protein. It has been shown that SRP depletion is more toxic in a strain background lacking the genes encoding the σ32 stress response-induced Lon and ClpQ proteases (26). These proteases could very well be involved in the degradation of mistargeted cytoplasmic membrane proteins and/or the turnover of protein aggregates thereby controlling the aggregate content in the cell. It is possible that at least some aggregated proteins are reactivated after extraction from the aggregates (53). Even though essential proteins were identified in the aggregates, the aggregated fraction of the total cellular protein is probably too insignificant to account for the essential nature of the E. coli SRP. Indeed, growth of E. coli cells with impaired Trigger Factor/DnaKJ chaperone activities was not hampered even though 10% of the total cellular protein was aggregated (54). Thus, in contrast to what has been suggested previously, aggregation of proteins is most likely not a major reason for the toxicity of the depletion of SRP (26).
Analysis of the steady-state cytoplasmic membrane proteomes of cells depleted of Ffh and control cells using two-dimensional BN/SDS-PAGE showed that Ffh depletion has a negative effect on the levels of the NADH dehydrogenase complex, succinate dehydrogenase complex, the cytochrome bo3 oxidase complex, and the mechanosensitive channel MscS. The only common feature of the integral membrane proteins that are part of these complexes was that all but one are part of the complexes involved in respiration. The oxygen consumption rates in whole cells depleted of Ffh were lower than in control cells, which is in keeping with the decreased levels of respiratory complexes. Impaired respiration hampers the generation of the protonmotive force. Interestingly, expression of PspA is induced upon Ffh depletion. Expression of PspA has been shown to be induced when cells experience problems in maintaining cytoplasmic membrane integrity and/or the protonmotive force (55). Because the membrane integrity was unaffected, the induction of PspA expression upon Ffh depletion is most likely a response to the hampered generation of the protonmotive force.
The other constituents of the cytoplasmic membrane proteome and their organization in complexes were not affected, with the exception of the periplasmic peripheral membrane protein DacA, which is involved in cell wall biogenesis, and the soluble cytoplasmic chaperones DnaK and GroEL. The accumulation levels of these three proteins were increased upon Ffh depletion. Expression of DacA is positively affected by the transcriptional regulator BolA, whose expression is induced upon various stresses, including protein misfolding in the cytoplasm (56). As discussed below, DnaK and GroEL may partially substitute for SRP.
To study the kinetics of the biogenesis of the cytoplasmic membrane proteome, a pulse-chase approach was used. After a 30-s pulse followed by a 5-min chase, there was much less of a difference between the amount of [35S]methionine incorporated into the cytoplasmic membrane proteomes isolated from Ffh-depleted and control cells than after a 30-s pulse. This indicates that the kinetics of the biogenesis of the cytoplasmic membrane proteome are impaired upon Ffh depletion. Pulse-chase experiments with model cytoplasmic membrane proteins confirmed that the kinetics of the biogenesis of the cytoplasmic membrane proteome are indeed impaired upon Ffh depletion. However, complex formation still occurred upon Ffh depletion, which is in keeping with the observation that the SecYEG-translocon capacity and YidC levels were not affected and that no cell envelope stress responses are induced upon Ffh depletion.
It is possible that upon Ffh depletion, cytoplasmic membrane proteins integrate improperly into the membrane. However, although the formation of complexes is delayed upon Ffh depletion, they do form. There are no detectable differences between the organization of the cytoplasmic membrane proteomes of Ffh-depleted and control cells. It is very unlikely that this would be the case if improper integration of inner membrane proteins into the membrane would be a generic problem.
Recently, it was shown that exhaustive depletion of the SRP receptor FtsY leads to the inactivation of ribosomes by the ribosome-inactivating protein ribosome-modulation factor (RMF)(57). Analysis of protein synthesis in whole cells using a pulse-labeling approach indicated that upon Ffh depletion, protein synthesis was only to some extent negatively affected.
Why are the steady-state levels of some complexes negatively affected upon Ffh depletion? Possibly, in the absence of SRP, suboptimal targeting can disturb the coordinated assembly of proteins into stable complexes. For the homo-oligomeric potassium channel Kv1.3, it has been shown that the nascent chains already assemble into an oligomer before their translation is completed, which indicates that targeting, membrane insertion, and complex formation indeed occur in a concerted mechanism (58). However, as explained above, disturbed coordination of assembly of proteins into stable complexes is not a generic problem.
How do the results of our global analysis of E. coli cells depleted of Ffh compare with other more targeted studies in which the SRP-targeting pathway was compromised? Interestingly, SRP levels can be lowered dramatically without notably affecting the viability and fitness of the cell (15). In this respect it should be noted that the number of SRP molecules per cell is only between 40 and 200 (59, 60). It has been shown for various membrane proteins that even upon extensive depletion of Ffh a still significant fraction, up to 50%, of the protein is properly inserted in the cytoplasmic membrane and that the dependence of membrane proteins on SRP can vary a lot (19, 26). Furthermore, for an AcrB-PhoA fusion, it has been shown that upon SRP depletion the insertion into the cytoplasmic membrane of a significant fraction of this particular protein is delayed rather than blocked (26). Based on this observation, it has been suggested that the E. coli SRP plays a role in the kinetics of membrane protein biogenesis. Here, we have shown that the E. coli SRP is key to controlling the kinetics of the biogenesis of the cytoplasmic membrane proteome.
In contrast to E. coli, S. cerevisiae, and S. mutans, mutant strains with a nonfunctional SRP-targeting pathway are viable (27). Protein targeting, synthesis, and homeostasis are, just like in E. coli cells depleted of SRP, all negatively affected in these mutant strains. However, S. cerevisiae and S. mutans can compensate for or tolerate the lack of a functional SRP-targeting pathway. Possibly, in E. coli the SRP is indispensable for the biogenesis of one or a few essential cytoplasmic proteins that we did not detect in our study.
In E. coli, besides the SRP-targeting pathway, there may be alternative mechanisms that can mediate protein targeting to the cytoplasmic membrane. In cells depleted of Ffh, the cytoplasmic membrane fraction is strongly enriched in the chaperones DnaK and GroEL. This is despite the fact that Ffh depletion does not result in any cell envelope stress, i.e. DnaK and GroEL are not attracted to the cytoplasmic membrane by misfolded proteins (45). It has been shown that both the DnaK and GroEL chaperones can connect with the auxiliary SecYEG-translocon component SecA (61–63), which has affinity for signal anchor sequences (64). SecA could provide a point of entry for the insertion of membrane proteins that are targeted by DnaK and GroEL to the cytoplasmic membrane. DnaK can facilitate the insertion of membrane proteins into the cytoplasmic membrane (65), and GroEL can promote post-translational insertion of membrane proteins into the cytoplasmic membrane (66). The similar intensities of the two-dimensional BN/SDS-polyacrylamide gels with [35S]methionine-labeled membranes after the 5-min chase support that post-translational insertion of membrane proteins can indeed occur. It has also been proposed that FtsY rather than SRP targets ribosomes to the cytoplasmic membrane and that the SRP acts after the FtsY-mediated targeting step (67–69). Finally, in various biological systems, including bacteria, it has been shown that mRNA targeting occurs (70–72). In E. coli, the mRNAs encoding integral membrane proteins are enriched in uracil, which is an evolutionarily ancient feature that could be involved in the targeting of these mRNA molecules to cytoplasmic membrane-bound ribosomes (73).
In conclusion, our analysis of the subproteomes of E. coli cells depleted of the SRP component Ffh showed that the SRP enhances the kinetics of the biogenesis of the cytoplasmic membrane proteome and that this is directly linked to maintaining protein homeostasis and the general fitness of the cell. Our study underscores the need of using both targeted and global approaches to identify and further our understanding of the different pathways mediating the targeting of proteins to membranes.
Supplementary Material
Acknowledgment
Proteomics infrastructure was supported by a grant from NYSTAR (to K. J. v. W.).
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01GM081827-03. This work was also supported by the Swedish Research Council, Carl Tryggers Stiftelse, Marianne and Marcus Wallenberg Foundation, Swedish Foundation for Strategic Research-supported Center for Biomembrane Research (to J.-W. d. G.), grants from the Swedish Foundation for International Cooperation in Research and Higher Education (to J.-W. d. G. and K. J. v. W.), and the Wenner-Gren Foundations (to K. J. v. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. S1 and S2.
Q. Sun and K. J. van Wijk, unpublished data.
- SRP
- signal recognition particle
- BN
- blue native
- TEA
- triethanolamine
- 2DE
- two-dimensional gel electrophoresis
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- PI
- propidium iodide.
REFERENCES
- 1. Luirink J., Sinning I. (2004) Biochim. Biophys. Acta 1694, 17–35 [DOI] [PubMed] [Google Scholar]
- 2. Cross B. C., Sinning I., Luirink J., High S. (2009) Nat. Rev. Mol. Cell Biol. 10, 255–264 [DOI] [PubMed] [Google Scholar]
- 3. Osborne A. R., Rapoport T. A., van den Berg B. (2005) Annu. Rev. Cell Dev. Biol. 21, 529–550 [DOI] [PubMed] [Google Scholar]
- 4. Keenan R. J., Freymann D. M., Stroud R. M., Walter P. (2001) Annu. Rev. Biochem. 70, 755–775 [DOI] [PubMed] [Google Scholar]
- 5. Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D. (1990) Cell 63, 591–600 [DOI] [PubMed] [Google Scholar]
- 6. Phillips G. J., Silhavy T. J. (1992) Nature 359, 744–746 [DOI] [PubMed] [Google Scholar]
- 7. Valent Q. A., Scotti P. A., High S., de Gier J. W., von Heijne G., Lentzen G., Wintermeyer W., Oudega B., Luirink J. (1998) EMBO J. 17, 2504–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schierle C. F., Berkmen M., Huber D., Kumamoto C., Boyd D., Beckwith J. (2003) J. Bacteriol. 185, 5706–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kadokura H., Beckwith J. (2009) Cell 138, 1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bowers C. W., Lau F., Silhavy T. J. (2003) J. Bacteriol. 185, 5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Powers T., Walter P. (1997) EMBO J. 16, 4880–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raine A., Ullers R., Pavlov M., Luirink J., Wikberg J. E., Ehrenberg M. (2003) Biochimie 85, 659–668 [DOI] [PubMed] [Google Scholar]
- 13. Avdeeva O. N., Myasnikov A. G., Sergiev P. V., Bogdanov A. A., Brimacombe R., Dontsova O. A. (2002) FEBS Lett. 514, 70–73 [DOI] [PubMed] [Google Scholar]
- 14. Macfarlane J., Müller M. (1995) Eur. J. Biochem. 233, 766–771 [DOI] [PubMed] [Google Scholar]
- 15. Ulbrandt N. D., Newitt J. A., Bernstein H. D. (1997) Cell 88, 187–196 [DOI] [PubMed] [Google Scholar]
- 16. Valent Q. A., de Gier J. W., von Heijne G., Kendall D. A., ten Hagen-Jongman C. M., Oudega B., Luirink J. (1997) Mol. Microbiol. 25, 53–64 [DOI] [PubMed] [Google Scholar]
- 17. Luirink J., High S., Wood H., Giner A., Tollervey D., Dobberstein B. (1992) Nature 359, 741–743 [DOI] [PubMed] [Google Scholar]
- 18. de Gier J. W., Mansournia P., Valent Q. A., Phillips G. J., Luirink J., von Heijne G. (1996) FEBS Lett. 399, 307–309 [DOI] [PubMed] [Google Scholar]
- 19. Fröderberg L., Houben E., Samuelson J. C., Chen M., Park S. K., Phillips G. J., Dalbey R., Luirink J., De Gier J. W. (2003) Mol. Microbiol. 47, 1015–1027 [DOI] [PubMed] [Google Scholar]
- 20. Bornemann T., Jöckel J., Rodnina M. V., Wintermeyer W. (2008) Nat. Struct. Mol. Biol. 15, 494–499 [DOI] [PubMed] [Google Scholar]
- 21. Nilsson I., von Heijne G. (1990) Cell 62, 1135–1141 [DOI] [PubMed] [Google Scholar]
- 22. Beck K., Wu L. F., Brunner J., Müller M. (2000) EMBO J. 19, 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Annu. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 24. Xie K., Dalbey R. E. (2008) Nat. Rev. Microbiol. 6, 234–244 [DOI] [PubMed] [Google Scholar]
- 25. Dalbey R. E., Chen M. (2004) Biochim. Biophys. Acta 1694, 37–53 [DOI] [PubMed] [Google Scholar]
- 26. Bernstein H. D., Hyndman J. B. (2001) J. Bacteriol. 183, 2187–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mutka S. C., Walter P. (2001) Mol. Biol. Cell 12, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hasona A., Crowley P. J., Levesque C. M., Mair R. W., Cvitkovitch D. G., Bleiweis A. S., Brady L. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17466–17471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasona A., Zuobi-Hasona K., Crowley P. J., Abranches J., Ruelf M. A., Bleiweis A. S., Brady L. J. (2007) J. Bacteriol. 189, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Funes S., Hasona A., Bauerschmitt H., Grubbauer C., Kauff F., Collins R., Crowley P. J., Palmer S. R., Brady L. J., Herrmann J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fröderberg L., Röhl T., van Wijk K. J., de Gier J. W. (2001) FEBS Lett. 498, 52–56 [DOI] [PubMed] [Google Scholar]
- 32. Baars L., Ytterberg A. J., Drew D., Wagner S., Thilo C., van Wijk K. J., de Gier J. W. (2006) J. Biol. Chem. 281, 10024–10034 [DOI] [PubMed] [Google Scholar]
- 33. Baars L., Wagner S., Wickström D., Klepsch M., Ytterberg A. J., van Wijk K. J., de Gier J. W. (2008) J. Bacteriol. 190, 3505–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hewitt C. J., Nebe-Von-Caron G. (2004) Adv. Biochem. Eng. Biotechnol. 89, 197–223 [DOI] [PubMed] [Google Scholar]
- 35. Fishov I., Woldringh C. L. (1999) Mol. Microbiol. 32, 1166–1172 [DOI] [PubMed] [Google Scholar]
- 36. Oakley B. R., Kirsch D. R., Morris N. R. (1980) Anal. Biochem. 105, 361–363 [DOI] [PubMed] [Google Scholar]
- 37. Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B. (2001) Mol. Microbiol. 40, 397–413 [DOI] [PubMed] [Google Scholar]
- 38. Peltier J. B., Ytterberg A. J., Sun Q., van Wijk K. J. (2004) J. Biol. Chem. 279, 49367–49383 [DOI] [PubMed] [Google Scholar]
- 39. Wagner S., Baars L., Ytterberg A. J., Klussmeier A., Wagner C. S., Nord O., Nygren P. A., van Wijk K. J., de Gier J. W. (2007) Mol. Cell. Proteomics 6, 1527–1550 [DOI] [PubMed] [Google Scholar]
- 40. Klepsch M., Schlegel S., Wickström D., Friso G., van Wijk K. J., Persson J. O., de Gier J. W., Wagner S. (2008) Methods 46, 48–53 [DOI] [PubMed] [Google Scholar]
- 41. Peltier J. B., Emanuelsson O., Kalume D. E., Ytterberg J., Friso G., Rudella A., Liberles D. A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K. J. (2002) Plant Cell 14, 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Friso G., Giacomelli L., Ytterberg A. J., Peltier J. B., Rudella A., Sun Q., Wijk K. J. (2004) Plant Cell 16, 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Balzer D., Ziegelin G., Pansegrau W., Kruft V., Lanka E. (1992) Nucleic Acids Res. 20, 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakatogawa H., Murakami A., Ito K. (2004) Curr. Opin. Microbiol. 7, 145–150 [DOI] [PubMed] [Google Scholar]
- 45. Shimohata N., Nagamori S., Akiyama Y., Kaback H. R., Ito K. (2007) J. Cell Biol. 176, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Bloois E., Haan G. J., de Gier J. W., Oudega B., Luirink J. (2006) J. Biol. Chem. 281, 10002–10009 [DOI] [PubMed] [Google Scholar]
- 47. Celebi N., Yi L., Facey S. J., Kuhn A., Dalbey R. E. (2006) J. Mol. Biol. 357, 1428–1436 [DOI] [PubMed] [Google Scholar]
- 48. du Plessis D. J., Nouwen N., Driessen A. J. (2006) J. Biol. Chem. 281, 12248–12252 [DOI] [PubMed] [Google Scholar]
- 49. Wu C. C., Yates J. R., 3rd. (2003) Nat. Biotechnol. 21, 262–267 [DOI] [PubMed] [Google Scholar]
- 50. Stenberg F., Chovanec P., Maslen S. L., Robinson C. V., Ilag L. L., von Heijne G., Daley D. O. (2005) J. Biol. Chem. 280, 34409–34419 [DOI] [PubMed] [Google Scholar]
- 51. Wagner S., Klepsch M. M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K. J., Slotboom D. J., Persson J. O., de Gier J. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14371–14376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arsène F., Tomoyasu T., Bukau B. (2000) Int. J. Food Microbiol. 55, 3–9 [DOI] [PubMed] [Google Scholar]
- 53. Mogk A., Deuerling E., Vorderwülbecke S., Vierling E., Bukau B. (2003) Mol. Microbiol. 50, 585–595 [DOI] [PubMed] [Google Scholar]
- 54. Deuerling E., Patzelt H., Vorderwülbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B. (2003) Mol. Microbiol. 47, 1317–1328 [DOI] [PubMed] [Google Scholar]
- 55. Darwin A. J. (2005) Mol. Microbiol. 57, 621–628 [DOI] [PubMed] [Google Scholar]
- 56. Santos J. M., Lobo M., Matos A. P., De Pedro M. A., Arraiano C. M. (2002) Mol. Microbiol. 45, 1729–1740 [DOI] [PubMed] [Google Scholar]
- 57. Bürk J., Weiche B., Wenk M., Boy D., Nestel S., Heimrich B., Koch H. G. (2009) J. Bacteriol. 191, 7017–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lu J., Robinson J. M., Edwards D., Deutsch C. (2001) Biochemistry 40, 10934–10946 [DOI] [PubMed] [Google Scholar]
- 59. Jensen C. G., Pedersen S. (1994) J. Bacteriol. 176, 7148–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wikström P. M., Björk G. R. (1988) J. Bacteriol. 170, 3025–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bochkareva E. S., Solovieva M. E., Girshovich A. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Randall L. L., Hardy S. J. (2002) Cell. Mol. Life Sci. 59, 1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Phillips G. J., Silhavy T. J. (1990) Nature 344, 882–884 [DOI] [PubMed] [Google Scholar]
- 64. Papanikou E., Karamanou S., Economou A. (2007) Nat. Rev. Microbiol. 5, 839–851 [DOI] [PubMed] [Google Scholar]
- 65. Chen Y., Song J., Sui S. F., Wang D. N. (2003) Protein Expr. Purif. 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 66. Bochkareva E., Seluanov A., Bibi E., Girshovich A. (1996) J. Biol. Chem. 271, 22256–22261 [DOI] [PubMed] [Google Scholar]
- 67. Herskovits A. A., Bibi E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Herskovits A. A., Bochkareva E. S., Bibi E. (2000) Mol. Microbiol. 38, 927–939 [DOI] [PubMed] [Google Scholar]
- 69. Herskovits A. A., Shimoni E., Minsky A., Bibi E. (2002) J. Cell Biol. 159, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nicchitta C. V. (2002) Curr. Opin. Cell Biol. 14, 412–416 [DOI] [PubMed] [Google Scholar]
- 71. Pyhtila B., Zheng T., Lager P. J., Keene J. D., Reedy M. C., Nicchitta C. V. (2008) RNA 14, 445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ramamurthi K. S., Schneewind O. (2003) Mol. Microbiol. 50, 1189–1198 [DOI] [PubMed] [Google Scholar]
- 73. Prilusky J., Bibi E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6662–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. von Heijne G. (1989) Nature 341, 456–458 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.